Abstract
Fabry disease is caused by mutations in the GLA gene and is characterized by a large genotypic and phenotypic spectrum. Missense mutations pose a special problem for graduating diagnosis and choosing a cost-effective therapy. Some mutants retain enzymatic activity, but are less stable than the wild type protein. These mutants can be stabilized by small molecules which are defined as pharmacological chaperones. The first chaperone to reach clinical trial is 1-deoxygalactonojirimycin, but others have been tested in vitro. Residual activity of GLA mutants has been measured in the presence or absence of pharmacological chaperones by several authors. Data obtained from transfected cells correlate with those obtained in cells derived from patients, regardless of whether 1-deoxygalactonojirimycin was present or not. The extent to which missense mutations respond to 1-deoxygalactonojirimycin is variable and a reference table of the results obtained by independent groups that is provided with this paper can facilitate the choice of eligible patients. A review of other pharmacological chaperones is provided as well. Frequent mutations can have residual activity as low as one-fourth of normal enzyme in vitro. The reference table with residual activity of the mutants facilitates the identification of non-pathological variants.
Keywords: Fabry disease/drug therapy, α-galactosidase, pharmacological chaperones, 1-deoxynojirimycin
1. Introduction
Fabry disease (FD, OMIM #301500) is a rare pathology, but accounts for 8.8% of the patients affected by inherited disorders of metabolism [1] and is the second most common lysosomal storage disorder [2]. FD is caused by those mutations in the GLA gene that result in a deficiency of the protein product, lysosomal α-galactosidase (AGAL Uniprot: AGAL_HUMAN P06280; EC: 3.2.1.22), and the accumulation of its substrates. The real incidence of FD is difficult to establish. It was estimated at 1 in 100,000 [3].
Screening of various at-risk populations, patients with renal failure [4,5], stroke [6], and cardiomyopathy [7,8], have shown a significant prevalence of FD in symptomatic population. GLA gene variations have been found in newborn screening with a frequency as high as 1 in 1200 or 1 in 3100 [9,10]. Some of the found variations remain unclear with respect to clinical significance.
Although GLA is located on X chromosome (Xq22.1), heterozygous females can be symptomatic. This is due to random inactivation and lack of cross-correction that occurs in other lysosomal storage disorders such as mucopolysaccharidosis type II [11]. Random X-chromosome inactivation in heterozygous females leads to a mosaic of cells, half of which express wild-type AGAL. Under these circumstances, female patients have mild or no signs of the disease. In some cases, however, a skewed inactivation, which occurs for unknown reasons, leads to the preferential expression either of the chromosome carrying the wild type or the mutant GLA. Under these circumstances, female patients can be as severely affected as much as the male patients carrying the same mutation [12].
AGAL is a homodimeric glycoprotein with 429 amino acids per chain and shares structural similarities with the other lysosomal glycosidases. It catalyzes the removal of α-galactosyl residues from glycosphingolipids, in particular globotriaosylceramide, Gb3 or GL-3 (also known as ceramide trihexoside). Its products are lactosylceramide and galactose. Gb3 mainly occurs in the endothelial, kidney, heart, and nervous cells and there is evidence suggesting its involvement in the renal pathology [13,14], but the underlying mechanism remains largely unknown [15]. Gb3 and its isoforms based on ceramide modification are detectable in blood and urinary samples [16,17] from the patients for the use as diagnostic and prognostic biomarkers following and supporting genetic testing. Meanwhile, a deacylated metabolite of Gb3, globotriaosylsphingosine (lyso-Gb3) has emerged as a superior biomarker demonstrating higher sensitivity than Gb3 and a good correlation to the FD phenotype [18].
FD is characterized by a large phenotypic spectrum, with mildly and severely affected patients, and shares many symptoms with common diseases. In severe cases, often referred to as classic FD, the first specific signs appearing in childhood or adolescence are angiokeratoma, cornea verticillata, neuropathic pain, acroparesthesias and hypohidrosis. These are followed by progressive proteinuric renal insufficiency, rhythm and conductance disorders with progressive hypertrophic cardiomyopathy and cerebrovascular stroke [19,20,21]. In mild cases, often referred to as atypical FD, only some symptoms are present, usually the cardiac ones. The Mainz Severity Score Index (MSSI) [22,23] was developed to measure the severity of FD and to monitor the clinical course of the disease in response to therapy. The MSSI includes four components or sub-scores that assess the general, neurological, cardiovascular and renal signs and symptoms. Although the MSSI score is able to differentiate FD from other severe debilitating diseases, a minor, still significant overlap, in particular for cardiac sub-scores, between healthy and FD affected persons was observed. MSSI was originally developed for classic FD. Other tools, such as Fabry Disease Severity Scoring System (DS3) [24] and Fabry STabilization indEX (FASTEX) [25], were subsequently developed to cover a broader range of cases.
The broad heterogeneous symptom spectrum might be due in part to genetic modifiers and other extra-genetic (epigenetic, environmental) factors that are currently discussed [26,27].
Enzyme replacement therapy (ERT) has been approved for the last 15 years. There exist two formulations of the recombinant AGAL, agalsidase α or β, that are commercialized by Shire, Lexington, MA, USA and Genzyme, a Sanofi company, Cambridge, MA, USA respectively [28]. ERT may decrease cardiac mass [29,30,31,32] and reduce the accumulation of the substrate Gb3 in the kidney [33,34,35], but the effects on nervous system and renal function have not been definitively assessed [36,37,38,39]. Early start of ERT has been suggested because irreversible organ damage, cardiac fibrosis or severe renal dysfunction, would render the therapy ineffective [34,36,40]. Recommendations for initiation and cessation of enzyme replacement therapy in patients with Fabry disease have recently been provided by the European Fabry Working Group consensus document [28]. The effect of ERT on patients with mild mutations, which retain some residual AGAL activity, has not been considered separately [41]. This is unfortunate because ERT is not the only possible therapy for FD. A new approach with pharmacological chaperones (PC) has been proposed and a small molecular weight molecule is on the verge of being approved with the commercial name of Galafold™. This drug is an iminosugar, which closely resembles the natural product of AGAL galactose, and has been known by different names, 1-deoxygalactonojirimycin (DGJ), migalastat, AMIGAL, AT1001. DGJ inhibits reversibly AGAL at nanomolar concentrations, but stabilizes the wild type enzyme in vitro against thermal [42] and chemical induced denaturation [43] too. DGJ can be used in synergy with ERT either co-administrating both drugs intravenously or one orally (DGJ) and the other intravenously (recombinant enzyme). DGJ prolongs the half-life of AGAL in vivo, both in mouse models and in humans and leads to an improved clearance of Gb3 [44,45,46].
DGJ can be used for a stand-alone oral therapy of FD for specific missense genotypes. The efficacy of DGJ was tested in vitro, ex vivo, in cells derived from patients, and in vivo. Oral administration of DGJ reduces Gb3 in kidney, heart and skin of Fabry transgenic mice carrying the responsive human mutation R301Q [47]. When administered with an oral dose of 150 mg, it was well tolerated, increased AGAL activity [48] and decreased plasma lyso-Gb3 [47] in the majority of the patients with responsive GLA mutations. Interestingly, the best results are obtained when an intermittent regimen is used. The results of a clinical trial phase 3 study carried out on males and females affected by FD has been recently published. Patients received 150 mg of Galafold™ or placebo every other day. The study began with six months of double-blind administration and proceeded with 6 + 12 months of open-label administration. Although the authors conclude their abstract stating quite cautiously that “the percentage of patients who had a response at 6 months did not differ significantly between the migalastat (DGJ) group and the placebo group”, promising results are shown. A reduction of the number of Gb3 inclusions per kidney interstitial capillary as well as a reduction of plasma lyso-Gb3 were observed [49].
More than 700 variants have been reported in HGMD for the GLA gene so far and, differently from other lysosomal disorders such as Gaucher, there are not prevalent mutations, on the contrary most are usually found only in a single family. The number of missense mutations, 467 described so far, is a surprisingly high value for a medium size protein, such as AGAL. In order to appreciate this finding it should be considered that more than 70,000 missense mutations affecting proteins associated to human diseases have been reported, with seven variants per protein on average. The large number of missense mutations poses several problems for making a diagnosis and initiating the most appropriate therapy. Recently, it was proposed to use residual activity measured in vitro to classify mutations. We wish to contribute to the evaluation of such a proposal with the first meta-analysis of the residual activity of GLA missense mutations measured by several independent research groups employing different protocols, either ex vivo, in cells derived from patients, or in vitro, in transiently transfected cells. Results covering 317 of missense mutants, mostly cases reported in HGMD and associated to FD, were collected. Data were obtained in the absence or in the presence of DGJ. For this reason, our analysis provides an independent perspective on the amenability to pharmacological chaperones. In addition to this we reviewed other small molecules that were reported to have a stabilizing effect on some GLA missense mutations in vitro and might be developed to act in synergy or as an alternative to DGJ.
2. Results
Meta-Analysis of Data Reporting Residual Activity and Responsiveness to DGJ of GLA Missense Mutations
Several independent groups have tested the effect of DGJ on AGAL mutants, administering the drug to cells derived from patients, or most frequently, to HEK293 or COS cell transiently transfected with expression plasmids. The enhancement of enzyme levels and that of the total enzyme activity is monitored in the cells extracts and is regarded as a proof of the stabilization of the mutant in the cell by DGJ. Residual activity is normalized by the total amount of protein in the cell and should not be confused with specific activity, which is normalized by the amount of AGAL. Residual activity is influenced by the stability of the mutant in the cell and by its specific activity. In general, a fixed concentration of DGJ was used, usually 20 μM, in some cases, however, IC50 was determined and the optimal concentration was used. The results gathered from literature are reported in Supplementary File S1 and the methods employed in each study are summarized in Table 1.
Table 1.
Experimental conditions under which DGJ responsiveness has been assessed.
| Reference | Cell Type | Concentration and Incubation Time |
|---|---|---|
| Ishii_2000 [50] | Transfection COS1 | 20 μM DGJ 1 day |
| Spada_2006 [10] | Transfection COS7 | 20 μM DGJ 72 h |
| Shin_2007 [51] | T-cells and fibroblasts | 20 μM DGJ 3 or 4 days |
| Ishii_2007 [52] | Lymphoblasts and fibroblasts | 20 μM DGJ 5 days |
| Shin_2008 [53] | T-cells | 20 μM DGJ 3 days |
| Park_2009 [54] | Transfection COS7 | 20 μM DGJ 2 days |
| Benjamin_2009 [55] | Lymphoblasts and fibroblasts | Depending on EC50 5 days |
| Filoni_2010 [56] | Transfection COS1 and lymphocytes | 20 μM DGJ 72 h |
| Wu_2011 [57] | Transfection HEK293 | Depending on EC50 4 to 5 days |
| Andreotti_2011 [58] | Transfection COS7 | 20 μM DGJ 48 h |
| Lukas_2013 [2] | Transfection HEK293H | 20 μM DGJ 60 h |
| Giugliani_2013 [59] | Transfection HEK293 | 10 μM DGJ |
| Lukas_2016 [60] | Transfection HEK293 | 20 μM DGJ 60 h |
The last criteria for responsiveness were adopted for the clinical trial phase 3 published in 2016 [49] and require a relative increase in AGAL activity ≥1.2-fold above baseline and an absolute increase in AGAL ≥ 3% of wild type after incubation with 10 μM DGJ. The concentration of 10 μM is the Cmax concentration in plasma when patients are treated with 150 mg of DGJ, as was the case in clinical trials [47,49]. Ten micromolar, however, is not the highest concentration that can be safely reached in plasma [59,61] and the data obtained before 2016 with 20 μM DGJ, can still be useful to choose eligible patients.
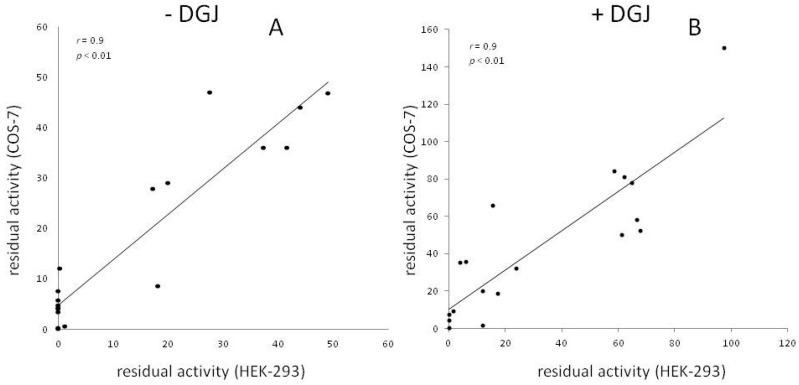
In vitro results are robust and do not depend on the type of recipient cells used for transfection (Figure 1).
Figure 1.
The residual activity of mutants transiently transfected and expressed in COS7 and in HEK293 is shown. In the case that multiple reports are available for a given mutant and a given recipient cell type, the average value was plotted. Results in the absence of DGJ (A) or in the presence of DGJ (B) are reported.
On the other hand, residual activity measured ex vivo varies among individuals and type of cells. A few examples of the levels measured in white blood cells are provided with the average, standard deviation and number of individuals: E66Q 42.3 ± 12.5 (n = 9) [62]; A143T 35.9 ± 7.2 (n = 4), R112H 7.2 ± 7.0 (n = 5), R301Q 7.3 ± 2.7 (n = 6), R356W 1.2 ± 1.9 (n = 4) (Supplementary File S1).
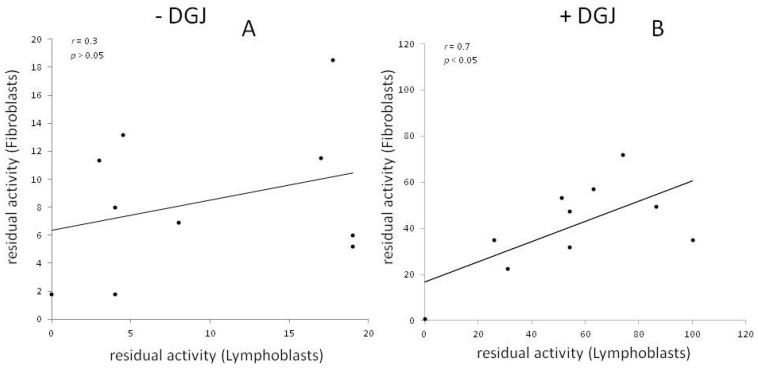
Figure 2 shows the average residual activity measured in lymphoblasts or in fibroblasts harboring the same mutation. A moderate yet statistically significant correlation of the data is observed only in the presence of DGJ.
Figure 2.
The residual activity of mutants measured in fibroblasts and lymphoblasts derived from patients is shown. In case that multiple reports are available for a given mutant and a given cell type, the average value was plotted. Results in the absence of DGJ (A) or in the presence of DGJ (B) are reported.
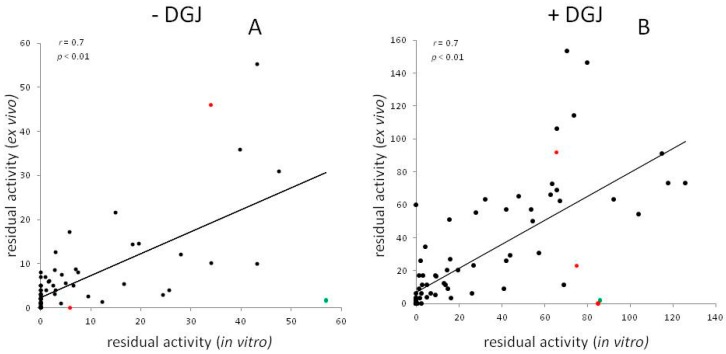
Figure 3 compares the residual activity measured ex vivo in cells derived from patients, (mostly lymphocytes or, in a few cases, fibroblasts), with that measured, in vitro, in transfected cells (HEK293, COS7 or COS1). For each mutant, the averages among results obtained by different authors without (Figure 3A) or with DGJ (Figure 3B) was determined. It can be observed that the residual activity measured in cells derived from patients tends to be lower than that measured in transfected cells, in particular in the absence of DGJ. A few examples are provided indicating in vitro result in HEK293H cells, ex vivo results in leucocytes with average, standard deviation and number of individuals: L180F: 32.4%, 6.0 ± 2.0 (n = 2); N215S: 39.5%, 27.1 ± 16.3 (n = 10); and I253T: 73.0%, 22.6 ± 6.9 (n = 3).
Figure 3.
The residual activity of mutants transiently transfected and expressed in vitro and derived from patient cells is shown. In case that multiple reports are available for a given mutant, the average value was plotted. Results in the absence of DGJ (A) or in the presence of DGJ (B) are reported. Mutations affecting a site of splicing and corresponding to G183, are represented by red symbols, G128E is represented by a green symbol.
However, the residual activities in vitro and ex vivo correlate (Figure 3A r = 0.7, p < 0.01; Figure 3B r = 0.7, p < 0.01). Thus, it can be concluded that tests in vitro can generally recapitulate the residual activity ex vivo and responsiveness to DGJ.
When this manuscript had been completed, we became aware of a recent publication that reports residual activity of AGAL mutants expressed in HEK293 cells and tested with DGJ 10 μM [63]. These data correlate with those reported in Supplementary File S1 (−DGJ r = 0.8, p < 0.000001; +DGJ r = 0.7, p < 0.000001).
Test in vitro have a limitation because they cannot account for the effect of exonic mutation on splicing. In fact, mutants are encoded by plasmids that do not contain introns. It is interesting to analyze the case of the mutations affecting a site of splicing and corresponding to G183 represented by red symbols in Figure 3. Substitution of GLY by SER results in a mutant that does not retain activity in cells derived from patients and does not recover activity with DGJ. The same mutant recovers activity with DGJ in vitro. We can hypothesize that the drug has a stabilizing effect on the protein, but cannot correct the effect on splicing. On the other hand, G183A and G183D are responsive to the drug both in vitro and ex vivo suggesting that these mutations mainly affect the protein, but not the splicing. K213M might be another example where splicing could play a role to explain the in vitro and ex vivo differences. Mutations not occurring at splicing sites can have effect on the maturation of RNA too. We suspect that this might be the case for G128E, green symbol in Figure 3, because very low residual activity was measured by several authors in cells derived from patients, either in the presence or in the absence of DGJ, whereas the mutation retains residual activity and is responsive to DGJ in vitro. Although a putative consensus for an exonic splicing enhancer including the triplet 128 was found, further experiments are needed to confirm the influence of the mutation on RNA processing.
We report the score obtained by a position specific substitution matrix (PSSM) that measures whether the mutation was tolerated during the evolution of homologous proteins in Supplementary File S2. Mutations affecting the active site, as expected, have no residual activity and do not respond to DGJ, mutations occurring at non-conserved sites tend to be responsive.
Predictions were obtained with Web based Polyphen2 using HumDiv or HumVar as the training set and are reported in Supplementary File S2. Both sets use disease mutations in UniprotKB as positive controls, but differ for the negative control set. HumDiv uses differences between human proteins and their closely related mammalian homologs, whereas HumVar uses common human SNPs (MAF > 1%) without annotated involvement in disease. HumVar-trained model is suitable to distinguish mild mutations, the HumDiv-trained model, considers also mild mutations as deleterious one. Although, on average, the residual activity of mutations that are predicted as probably damaging with both training sets is very different from the residual activity of mutations predicted as benign, exceptions can be observed in particular with HumVar-trained model (Table 2).
Table 2.
Putative non-pathological AGAL mutants: features and residual activity in vitro.
| Mutation | No. Hemiz | −DGJ | +DGJ | PSSM | Humdiv | Humvar | Reference |
|---|---|---|---|---|---|---|---|
| L3P | 4 | 117.7 | 129.4 | −3 | Probably damaging | Probably damaging | [60] |
| E66Q | 3 | 47.6 | 53.66 | −2 | Probably damaging | Probably damaging | [68] |
| R118C | 8 | 24.5 | 27.8 | −2 | Probably damaging | Possibly damaging | [10] |
| N139S | 7 | 147.8 | 176.4 | −1 | Benign | Benign | [69] |
| S126G | 18 | 51.3 | 67.4 | −2 | Benign | Benign | [70] |
| A143T | 19 | 39.7 | 63.7 | −1 | Probably damaging | Possibly damaging | [71] |
| I289V | 3 | 79.9 | 95 | 0 | Probably damaging | Possibly damaging | |
| D313Y | 129 | 75.5 | 100.3 | −1 | Probably damaging | Possibly damaging | [71] |
| R363H | 3 | 28 | 65.7 | −1 | Benign | Benign | [72] |
| A368T | 3 | 103.7 | 93.3 | 0 | Benign | Benign | [2] |
| T385A | 36 | 45 | 48.9 | −2 | Possibly damaging | Benign | [2] |
| W399S | 5 | 53 | 51.5 | −4 | Possibly damaging | Benign | [60] |
Other in silico approaches based on the structural features of AGAL, some from our group [64], have been attempted [65,66], to predict the severity of FD genotypes. We believe that data obtained in vitro should always be preferred whenever available.
In Supplementary File S2, all missense variants of GLA described in ExAC [67] are reported. ExAC summarizes exome sequencing data from a wide variety of large-scale sequencing projects. Variants reported in this database, in particular those observed with higher frequency, are likely to be non-pathogenic. The mean residual activity measured in vitro for a subset of ExAC variants, i.e., those observed in more than one male is reported in Table 2. The number of hemizygous individuals reported in ExAC, the PSSM score, Polyphen2 prediction and the reference found in HGMD are reported in the same table.
R118C is the variant with the lowest residual activity, only 24.5% of wild type when tested in transiently transfected cells. It is relatively frequent in the European population, but it is predicted as deleterious both by Polyphen_Humvar and Polyphen_Humdiv. Oliveira and colleagues reviewed the clinical, biochemical and histopathology data obtained from 22 individual carriers and reached the conclusion that it “does not segregate with FD manifestations at least in a highly-penetrant Mendelian fashion”, but might be a risk factor for stroke [73]. In accordance with this, low levels of lyso-Gb3, a biomarker of FD, were measured in the carriers [2]. R118C is considered amenable to DGJ according to galafold amenability table [63]. R118C was tested with DGJ and with Rosiglitazone by Lukas et al. [74]. Although in terms of activity fold increase, the effect of mono-therapy with either drug was small, the combinatorial effect was significantly higher.
A143T has an average residual activity of approximately 39.7% of wild type. Brand and coworkers [75] analyzed 15 females and 10 males carrying this mutation. They observed that female and male A143T carriers showed less organ involvement in comparison to FD patients with other missense mutations and those suffering from stroke/TIA showed no further FD-typical organ manifestations. They came to the conclusion that “A143T seems not to be causal for FD, but rather a genetic variant of unknown significance or a genetic modifier”. A143T is considered amenable for the therapy with DGJ according to the galafold amenability table.
E66Q has specific activity, Vmax, and affinity for the artificial substrate 4-methylumbelliferyl-α-galactopyranoside, Km, similar to those of wild type, but residual activity in transfected cells is approximately one half of the wild type, possibly because the stability at neutral pH is reduced [52]. The mutation is relatively frequent in East Asian population. Sakuraba and coworkers measured the activity in 20 Japanese or Korean male carriers with renal and cardiovascular disorders and found 13% to 26% of the normal mean values for plasma and 24% to 65% of the normal mean values for white blood cells, but the lyso-Gb3 levels were as low as those of healthy controls and no inclusion bodies were found [62]. Hu and co-workers found that the mutation segregated with renal disease in a very large Chinese family, but they did not measure the accumulation of the substrate or of lyso-Gb3 in the same patients [76]. The involvement in cardiovascular disease has also been suspected, but no accumulation of Gb3 was found in the heart of a patient carrying E66Q [77]. The association between E66Q and the risk cerebral small-vessel occlusion is debated [78,79]. In conclusion, pathogenicity of E66Q is still vexata quaestio. E66Q is considered non amenable for DGJ according to galafold amenability table, but an increase in activity upon drug administration was measured by other authors [52,55,57,80].
D313Y was first associated to classic phenotype [71]. Subsequent data clinically and biochemically indicated that D313Y should be considered a variant [81]. Cardiac, nephrological, neurological, laboratory and quality of life data were collected from carriers of D313Y with a 4-year follow up and the results indicated that the mutation is non pathological. Very low levels of lyso-Gb3 were found [2]. The opinion that D313Y is a non-pathological variant is supported by the fact that its frequency of 0.4% in the non-Finnish European population is much higher than the prevalence of FD in the same population. D313Y is considered amenable for DGJ.
For other mutations such as S126G and N139S, the clinical picture did not include specific signs of FD [69,70]. Both mutations are considered amenable for DGJ.
This survey would suggest that a residual activity higher than 25% can indicate a non-pathological variant. Nonetheless, when considering administration of a therapy, if any, clinicians should be aware of the fact that the severity of the disease depends not only on the damage caused to the protein itself by the mutation, but also by other factors, which regrettably have not yet been clarified. It should be considered that the phenotype can differ even among the members of the same family [82] and that the residual activity in plasma or in white blood cells can vary largely in people carrying the same mutation [62].
N215S is a mutation affecting glycosylation of the AGAL enzyme [83]. It presents with a proportionally high residual activity >25% of normal. It is considered a distinct sub-type of FD due to its elevated prevalence compared to non-N215S FD cases and late-onset occurrence [60,84]. Interestingly, this variant has never been scrutinized to cause a pathogenic phenotype. By contrast, it is believed to cause a specific cardiac phenotype. Other cardiac-prone mutations might exist, e.g., the so-called IVS4 + 919G > A splice mutation highly prevalent in the Taiwanese population. A clinical trial investigating the long term clinical course of N215S patients is currently ongoing (clinicaltrials.gov, NCT01429597). In this case, the diagnostic and prognostic value of biomarker lyso-Gb3 can be appreciated. While apparently all (genetically) found patients were identified it was demonstrated that Gb3 was normal in a great fraction of patients. Meehan et al. [85] showed that N215S was present in a patient with renal manifestation and is, thus, suggestive to cause mainly cardiac and renal symptoms. N215S is amenable for DGJ.
3. Future Perspectives for Therapy
DGJ is a promising drug, but it might not be the ideal drug yet. DGJ inhibits AGAL at nanomolar concentration and stabilizes it at micromolar concentrations. Therefore a continuous exposure to the drug can promote AGAL levels, but not AGAL intracellular activity. Reduction of Gb3 concentration was not observed in fibroblasts derived from patients carrying the mutations R301Q or L300P and incubated with DGJ for 10 days, but was observed if the incubation of seven days with the drug was followed by a three day wash-out [55].
The discovery of DGJ was the result of an educated guess, and not of a methodical screening [86]. In fact, DGJ is a glycomimetic with a six-atom ring and very closely resembles the galactose, which is the natural product and inhibitor, and also the first chaperone described for AGAL [87]. A more systematic search was started with the aim of finding other drugs that might have a better ratio between the stabilizing and the inhibitory effect. Most of the molecules considered so far are glycomimetics as DGJ itself. DGJ is an amine and is positively charged at neutral pH. In order to facilitate its diffusion through membranes, alkylation was proposed [88]. Contrary to what was observed for analogous iminosugars active on other lysosomal glycosidases, alkyl-DGJ derivatives had a lower affinity for AGAL and apparently a lower chaperoning potential probably because one important hydrogen-bond, the one established between the heterocyclic NH proton and D170 of AGAL, is lost.
On the contrary, aryl DGJ-derivatives (1-deoxygalactonojirimycin-arylthioureas) that form a hydrogen bond between the aryl-N’H thiourea proton and D231 of AGAL, act as reversible inhibitors and chaperones. When tested at 30 μM concentration on Q279E or R301Q mutants, the best candidate, namely N’-p-methoxyphenyl-DGJ-Aryl thiourea, had a seven fold higher chaperoning activity than DGJ at its optimal concentration [89].
Iminosugars characterized by a smaller, five-atom ring system, have been described [90,91]. 2,5-dideoxy-2,5-imino-d-altritol (DIA) inhibited AGAL and stabilized it against thermal denaturation and acted as a chaperone when tested on Fabry R301Q lymphoblasts although at a concentration 20 times higher than the optimal one for DGJ. The effect on Gb3 accumulation was not tested. One derivative of DIA possessing an aminomethyl group showed a chaperoning effect higher than DGJ when administered to N215S patient lymphocyte cell line at high concentration (100 μM) [92].
DGJ binds and inhibits AGAL both at neutral pH, which is required, and at acidic pH, which is not required [42]. It would be useful to find molecules that bind and stabilize AGAL mutants when they are in the neutral environment of the endoplasmic reticulum, but dissociate when the protein reaches the lysosome. This point was specifically addressed incorporating an orthoester segment into DGJ [93].
Glycomimetics require a precise dosing, whereas non carbohydrate mimetics might offer a larger therapeutic window and an improved therapeutic index. In order to look for chemically diverse drugs, a library of 230,000 diverse compounds was screened but no inhibitors or activators of a-Gal A with an IC50 below 50 μM were identified. Unfortunately the screening procedure relied only on an enzymatic assay carried out at pH 5.9, but not at neutral pH or on assays based on AGAL stabilization [94].
So far reversible inhibitors of AGAL that act as PC have been described. The association between the two effects, inhibition and stabilization, is avoidable because active sites are not the only targets for chaperones. Allosteric ligands might act as pharmacological chaperones, and might be more effective than reversible inhibitors, since they would perform their stabilizing action without competing with the natural substrate. Looking for allosteric PC is difficult because they do not resemble chemically known substrates. Large libraries of structurally diverse compounds should be tested and preliminary screening in silico might be functional. Allosteric ligands do not bind the active site, but one of the many pockets occurring on the surface of a protein. Therefore it is difficult to restrict the area where binding is allowed as required by structure based virtual screening. A recent screening, carried out on 10,000 molecules, showed that it is possible to find molecules that, at least in silico, preferentially bind an allosteric site than the active site. The two sites are located at the opposite sides of the catalytic domain of AGAL [95].
The PC that have been described previously are specific ligands of AGAL. They are effective on missense mutations that cause destabilization of the enzyme and, ultimately, its early degradation. Other small molecules that do not physically interact with AGAL, but have effect on proteostasis, can be considered for the treatment of these cases as well. Proteostasis regulators can be used in synergy with specific PC potentiating their action or allowing lower dosages.
The before mentioned Rosiglitazone, a Peroxisome proliferator-activated receptor gamma (PPARγ) agonist rearranges global cellular ubiquitination by inhibiting the ubiquitin-proteasome system (UPS). It displayed the highest beneficial effect on mutations with a significant residual activity (e.g., R118C and T385A) and was even more effective in combination with a PC [60]. Mechanistic studies are required to explain why other ubiquitination inhibitors such as Pyr-41 failed to increase AGAL activity. This finding might be ascribed to intense adverse effects of cellular ubiquitination inhibition caused by Pyr-41 and associated toxicological aspects.
Ambroxol, a mucolytic agent used in the treatment of respiratory diseases, was identified as an enhancer of AGAL activity. The compound has formerly been demonstrated to act as a PC on mutant Glucocerebrosidase in Gaucher disease. Even though its mechanism of action is not known it was demonstrated to increase cellular AGAL level and activity of most (DGJ−) amenable mutations (E59K, A73V, A143T, A156V, I232T, R301G, R301Q, R356W and R363H) indicating an impact on AGAL proteostasis. Ambroxol was, however, not effective as a monotherapy, but only in the combination with a PC, galactose or DGJ [60].
The synergistic effect of N′-p-methoxyphenyl-DGJ-Aryl thiourea with two proteostasis regulators, 4-phenylbutyric acid and celastrol has been assessed. The latter compound was not effective, but 4-phenylbutyric acid at 0.1 mM concentration was able to enhance the chaperoning activity of the aryl-thiourea (20 μM) on the fibroblasts harbouring Q279E [89].
The effects of lactacystin 2 μM (a proteasome inhibitor) and kifunensine 0.2 mM (an inhibitor of ER α-mannosidase I) on the processing on some mutants assessing the amount of protein was tested by Fan and coworkers [52] They found that some mutants responded to both drugs (F113L, N215S, and M296I), one responded only to lactacystin (E66Q), others only to kifunesine (M72V, I91T, A97V, R112H, L166V, and Q279E), and others had low or no response to either (A20P, A156V, M296V, R356W, G373D, G373S, E59K, and P146S). All mutants that are responsive to kifunensine or lactacystin are also responsive to DGJ, while A156V, M269V, R356W, and E59K are responsive only to DGJ. These results suggest that a different cocktail of drugs might be ideal for specific AGAL mutations.
4. Methods
Residual activities in Supplementary File S1 were obtained from the literature. In those cases where the authors did not report the normalized percentage values, the activity of the mutant in the presence of DGJ was divided by the activity of wild type AGAL multiplied by 100 (+DGJ/wild × 100). The reference wild type activity, measured in the absence of DGJ was obtained for each mutation from the appropriate paper. IC50 values are reported when available.
Pearson correlation coefficients and two tailed p-values were calculated as described by Lowry [96].
PSSM values were calculated as described [97,98]. Active site residues were identified with DrosteP [99]. Predictions were obtained with Web based Polyphen2 using HumDiv or HumVar as the training set under default conditions [100].
Acknowledgments
Funding was provided by Telethon-Italy (Grant no. GGP12108). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Gaetano Viscido and Mario D’Andrea for technical assistance. This work is dedicated to our friend and colleague Maria Malanga.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/12/2010/s1.
Author Contributions
Valentina Citro, Maria Vittoria Cubellis and Giuseppina Andreotti conceived and designed the study and wrote the manuscript. Marco Cammisa analyzed the data and prepared the figures. Chiara Cimmaruta and Ludovica Liguori gathered the data. Jan Lukas contributed to the discussion. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sirrs S., Hollak C., Merkel M., Sechi A., Glamuzina E., Janssen M.C., Lachmann R., Langendonk J., Scarpelli M., Ben Omran T., et al. The frequencies of different inborn errors of metabolism in adult metabolic centres: Report from the ssiem adult metabolic physicians group. JIMD Rep. 2016;27:85–91. doi: 10.1007/8904_2015_435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukas J., Giese A.K., Markoff A., Grittner U., Kolodny E., Mascher H., Lackner K.J., Meyer W., Wree P., Saviouk V., et al. Functional characterisation of α-galactosidase a mutations as a basis for a new classification system in Fabry disease. PLoS Genet. 2013;9:e1003632. doi: 10.1371/journal.pgen.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Germain D.P. Fabry disease. Orphanet. J. Rare Dis. 2010;5:30. doi: 10.1186/1750-1172-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallin E.F., Clatworthy M.R., Pritchard N.R. Fabry disease: Results of the first UK hemodialysis screening study. Clin. Nephrol. 2011;75:506–510. doi: 10.5414/CNP75506. [DOI] [PubMed] [Google Scholar]
- 5.Doi K., Noiri E., Ishizu T., Negishi K., Suzuki Y., Hamasaki Y., Honda K., Fujita T., Tsukimura T., Togawa T., et al. High-throughput screening identified disease-causing mutants and functional variants of α-galactosidase a gene in japanese male hemodialysis patients. J. Hum. Genet. 2012;57:575–579. doi: 10.1038/jhg.2012.68. [DOI] [PubMed] [Google Scholar]
- 6.Rolfs A., Fazekas F., Grittner U., Dichgans M., Martus P., Holzhausen M., Bottcher T., Heuschmann P.U., Tatlisumak T., Tanislav C., et al. Acute cerebrovascular disease in the young: The stroke in young Fabry patients study. Stroke. 2013;44:340–349. doi: 10.1161/STROKEAHA.112.663708. [DOI] [PubMed] [Google Scholar]
- 7.Sachdev B., Takenaka T., Teraguchi H., Tei C., Lee P., McKenna W.J., Elliott P.M. Prevalence of anderson-Fabry disease in male patients with late onset hypertrophic cardiomyopathy. Circulation. 2002;105:1407–1411. doi: 10.1161/01.CIR.0000012626.81324.38. [DOI] [PubMed] [Google Scholar]
- 8.Monserrat L., Gimeno-Blanes J.R., Marin F., Hermida-Prieto M., Garcia-Honrubia A., Perez I., Fernandez X., de Nicolas R., de la Morena G., Paya E., et al. Prevalence of Fabry disease in a cohort of 508 unrelated patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2007;50:2399–2403. doi: 10.1016/j.jacc.2007.06.062. [DOI] [PubMed] [Google Scholar]
- 9.Hwu W.L., Chien Y.H., Lee N.C., Chiang S.C., Dobrovolny R., Huang A.C., Yeh H.Y., Chao M.C., Lin S.J., Kitagawa T., et al. Newborn screening for Fabry disease in taiwan reveals a high incidence of the later-onset GLA mutation C.936 + 919G > A (IVS4 + 919G > A) Hum. Mutat. 2009;30:1397–1405. doi: 10.1002/humu.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spada M., Pagliardini S., Yasuda M., Tukel T., Thiagarajan G., Sakuraba H., Ponzone A., Desnick R.J. High incidence of later-onset Fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006;79:31–40. doi: 10.1086/504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller M., Mellett N., Hein L.K., Brooks D.A., Meikle P.J. Absence of α-galactosidase cross-correction in Fabry heterozygote cultured skin fibroblasts. Mol. Genet. Metab. 2015;114:268–273. doi: 10.1016/j.ymgme.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Echevarria L., Benistan K., Toussaint A., Dubourg O., Hagege A.A., Eladari D., Jabbour F., Beldjord C., de Mazancourt P., Germain D.P. X-chromosome inactivation in female patients with Fabry disease. Clin. Genet. 2016;89:44–54. doi: 10.1111/cge.12613. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann B. Fabry disease: Recent advances in pathology, diagnosis, treatment and monitoring. Orphanet. J. Rare Dis. 2009;4:21. doi: 10.1186/1750-1172-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi A., Maruyama H., Nameta M., Yamamoto T., Matsuda J., Kulkarni A.B., Yoshioka H., Ishii S. A symptomatic Fabry disease mouse model generated by inducing globotriaosylceramide synthesis. Biochem. J. 2013;456:373–383. doi: 10.1042/BJ20130825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin Y.J., Jeon Y.J., Jung N., Park J.W., Park H.Y., Jung S.C. Substrate-specific gene expression profiles in different kidney cell types are associated with Fabry disease. Mol. Med. Rep. 2015;12:5049–5057. doi: 10.3892/mmr.2015.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills K., Johnson A., Winchester B. Synthesis of novel internal standards for the quantitative determination of plasma ceramide trihexoside in Fabry disease by tandem mass spectrometry. FEBS Lett. 2002;515:171–176. doi: 10.1016/S0014-5793(02)02491-2. [DOI] [PubMed] [Google Scholar]
- 17.Auray-Blais C., Cyr D., Ntwari A., West M.L., Cox-Brinkman J., Bichet D.G., Germain D.P., Laframboise R., Melancon S.B., Stockley T., et al. Urinary globotriaosylceramide excretion correlates with the genotype in children and adults with Fabry disease. Mol. Genet. Metab. 2008;93:331–340. doi: 10.1016/j.ymgme.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Smid B.E., van der Tol L., Biegstraaten M., Linthorst G.E., Hollak C.E., Poorthuis B.J. Plasma globotriaosylsphingosine in relation to phenotypes of Fabry disease. J. Med. Genet. 2015;52:262–268. doi: 10.1136/jmedgenet-2014-102872. [DOI] [PubMed] [Google Scholar]
- 19.Linhart A., Elliott P.M. The heart in anderson-Fabry disease and other lysosomal storage disorders. Heart. 2007;93:528–535. doi: 10.1136/hrt.2005.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiffmann R., Moore D.F. Neurological manifestations of Fabry disease. In: Mehta A., Beck M., Sunder-Plassmann G., editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford PharmaGenesis; Oxford, UK: 2006. [PubMed] [Google Scholar]
- 21.Sunder-Plassmann G. Renal manifestations of Fabry disease. In: Mehta A., Beck M., Sunder-Plassmann G., editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford PharmaGenesis; Oxford, UK: 2006. [PubMed] [Google Scholar]
- 22.Whybra C., Bahner F., Baron K. Measurement of disease severity and progression in Fabry disease. In: Mehta A., Beck M., Sunder-Plassmann G., editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford PharmaGenesis; Oxford, UK: 2006. [PubMed] [Google Scholar]
- 23.Whybra C., Kampmann C., Krummenauer F., Ries M., Mengel E., Miebach E., Baehner F., Kim K., Bajbouj M., Schwarting A., et al. The mainz severity score index: A new instrument for quantifying the anderson-Fabry disease phenotype, and the response of patients to enzyme replacement therapy. Clin. Genet. 2004;65:299–307. doi: 10.1111/j.1399-0004.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- 24.Giannini E.H., Mehta A.B., Hilz M.J., Beck M., Bichet D.G., Brady R.O., West M., Germain D.P., Wanner C., Waldek S., et al. A validated disease severity scoring system for Fabry disease. Mol. Genet. Metab. 2010;99:283–290. doi: 10.1016/j.ymgme.2009.10.178. [DOI] [PubMed] [Google Scholar]
- 25.Mignani R., Pieruzzi F., Berri F., Burlina A., Chinea B., Gallieni M., Pieroni M., Salviati A., Spada M. Fabry stabilization index (fastex): An innovative tool for the assessment of clinical stabilization in Fabry disease. Clin. Kidney J. 2016;9:739–747. doi: 10.1093/ckj/sfw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teitcher M., Weinerman S., Whybra C., Beck M., Sharon N., Elstein D., Altarescu G. Genetic polymorphisms of vitamin D receptor (VDR) in Fabry disease. Genetica. 2008;134:377–383. doi: 10.1007/s10709-008-9245-y. [DOI] [PubMed] [Google Scholar]
- 27.Altarescu G., Moore D.F., Schiffmann R. Effect of genetic modifiers on cerebral lesions in Fabry disease. Neurology. 2005;64:2148–2150. doi: 10.1212/01.WNL.0000166000.24321.4F. [DOI] [PubMed] [Google Scholar]
- 28.Biegstraaten M., Arngrimsson R., Barbey F., Boks L., Cecchi F., Deegan P.B., Feldt-Rasmussen U., Geberhiwot T., Germain D.P., Hendriksz C., et al. Recommendations for initiation and cessation of enzyme replacement therapy in patients with Fabry disease: The european Fabry working group consensus document. Orphanet. J. Rare Dis. 2015;10:36. doi: 10.1186/s13023-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banikazemi M., Bultas J., Waldek S., Wilcox W.R., Whitley C.B., McDonald M., Finkel R., Packman S., Bichet D.G., Warnock D.G., et al. Agalsidase-β therapy for advanced Fabry disease: A randomized trial. Ann. Intern. Med. 2007;146:77–86. doi: 10.7326/0003-4819-146-2-200701160-00148. [DOI] [PubMed] [Google Scholar]
- 30.Bierer G., Balfe D., Wilcox W.R., Mosenifar Z. Improvement in serial cardiopulmonary exercise testing following enzyme replacement therapy in Fabry disease. J. Inherit. Metab. Dis. 2006;29:572–579. doi: 10.1007/s10545-006-0361-5. [DOI] [PubMed] [Google Scholar]
- 31.Hughes D.A., Elliott P.M., Shah J., Zuckerman J., Coghlan G., Brookes J., Mehta A.B. Effects of enzyme replacement therapy on the cardiomyopathy of anderson-Fabry disease: A randomised, double-blind, placebo-controlled clinical trial of agalsidase alfa. Heart. 2008;94:153–158. doi: 10.1136/hrt.2006.104026. [DOI] [PubMed] [Google Scholar]
- 32.Rombach S.M., Smid B.E., Linthorst G.E., Dijkgraaf M.G., Hollak C.E. Natural course of Fabry disease and the effectiveness of enzyme replacement therapy: A systematic review and meta-analysis: Effectiveness of ert in different disease stages. J. Inherit. Metab. Dis. 2014;37:341–352. doi: 10.1007/s10545-014-9677-8. [DOI] [PubMed] [Google Scholar]
- 33.Thurberg B.L., Rennke H., Colvin R.B., Dikman S., Gordon R.E., Collins A.B., Desnick R.J., O’Callaghan M. Globotriaosylceramide accumulation in the Fabry kidney is cleared from multiple cell types after enzyme replacement therapy. Kidney Int. 2002;62:1933–1946. doi: 10.1046/j.1523-1755.2002.00675.x. [DOI] [PubMed] [Google Scholar]
- 34.Germain D.P., Waldek S., Banikazemi M., Bushinsky D.A., Charrow J., Desnick R.J., Lee P., Loew T., Vedder A.C., Abichandani R., et al. Sustained, long-term renal stabilization after 54 months of agalsidase β therapy in patients with Fabry disease. J. Am. Soc. Nephrol. 2007;18:1547–1557. doi: 10.1681/ASN.2006080816. [DOI] [PubMed] [Google Scholar]
- 35.Tondel C., Bostad L., Larsen K.K., Hirth A., Vikse B.E., Houge G., Svarstad E. Agalsidase benefits renal histology in young patients with Fabry disease. J. Am. Soc. Nephrol. 2013;24:137–148. doi: 10.1681/ASN.2012030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rombach S.M., Smid B.E., Bouwman M.G., Linthorst G.E., Dijkgraaf M.G., Hollak C.E. Long term enzyme replacement therapy for Fabry disease: Effectiveness on kidney, heart and brain. Orphanet. J. Rare Dis. 2013;8:47. doi: 10.1186/1750-1172-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buechner S., Moretti M., Burlina A.P., Cei G., Manara R., Ricci R., Mignani R., Parini R., di Vito R., Giordano G.P., et al. Central nervous system involvement in anderson-Fabry disease: A clinical and MRI retrospective study. J. Neurol. Neurosurg. Psychiatry. 2008;79:1249–1254. doi: 10.1136/jnnp.2008.143693. [DOI] [PubMed] [Google Scholar]
- 38.Jardim L., Vedolin L., Schwartz I.V., Burin M.G., Cecchin C., Kalakun L., Matte U., Aesse F., Pitta-Pinheiro C., Marconato J., et al. Cns involvement in Fabry disease: Clinical and imaging studies before and after 12 months of enzyme replacement therapy. J. Inherit. Metab. Dis. 2004;27:229–240. doi: 10.1023/B:BOLI.0000028794.04349.91. [DOI] [PubMed] [Google Scholar]
- 39.Jardim L.B., Aesse F., Vedolin L.M., Pitta-Pinheiro C., Marconato J., Burin M.G., Cecchin C., Netto C.B., Matte U.S., Pereira F., et al. White matter lesions in Fabry disease before and after enzyme replacement therapy: A 2-year follow-up. Arq. Neuropsiquiatr. 2006;64:711–717. doi: 10.1590/S0004-282X2006000500002. [DOI] [PubMed] [Google Scholar]
- 40.Weidemann F., Niemann M., Stork S., Breunig F., Beer M., Sommer C., Herrmann S., Ertl G., Wanner C. Long-term outcome of enzyme-replacement therapy in advanced Fabry disease: Evidence for disease progression towards serious complications. J. Intern. Med. 2013;274:331–341. doi: 10.1111/joim.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollak C.E., Weinreb N.J. The attenuated/late onset lysosomal storage disorders: Therapeutic goals and indications for enzyme replacement treatment in gaucher and Fabry disease. Best Pract. Res. Clin. Endocrinol. Metab. 2015;29:205–218. doi: 10.1016/j.beem.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Andreotti G., Monticelli M., Cubellis M.V. Looking for protein stabilizing drugs with thermal shift assay. Drug Test. Anal. 2015;7:831–834. doi: 10.1002/dta.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreotti G., Citro V., Correra A., Cubellis M.V. A thermodynamic assay to test pharmacological chaperones for Fabry disease. Biochim. Biophys. Acta. 2014;1840:1214–1224. doi: 10.1016/j.bbagen.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamin E.R., Khanna R., Schilling A., Flanagan J.J., Pellegrino L.J., Brignol N., Lun Y., Guillen D., Ranes B.E., Frascella M., et al. Co-administration with the pharmacological chaperone AT1001 increases recombinant human α-galactosidase a tissue uptake and improves substrate reduction in Fabry mice. Mol. Ther. 2012;20:717–726. doi: 10.1038/mt.2011.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warnock D.G., Bichet D.G., Holida M., Goker-Alpan O., Nicholls K., Thomas M., Eyskens F., Shankar S., Adera M., Sitaraman S., et al. Oral migalastat HCL leads to greater systemic exposure and tissue levels of active α-galactosidase a in Fabry patients when co-administered with infused agalsidase. PLoS ONE. 2015;10:e0134341. doi: 10.1371/journal.pone.0134341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu S., Lun Y., Brignol N., Hamler R., Schilling A., Frascella M., Sullivan S., Boyd R.E., Chang K., Soska R., et al. Coformulation of a novel human α-galactosidase a with the pharmacological chaperone AT1001 leads to improved substrate reduction in Fabry mice. Mol. Ther. 2015;23:1169–1181. doi: 10.1038/mt.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young-Gqamana B., Brignol N., Chang H.H., Khanna R., Soska R., Fuller M., Sitaraman S.A., Germain D.P., Giugliani R., Hughes D.A., et al. Migalastat hcl reduces globotriaosylsphingosine (lyso-Gb3) in Fabry transgenic mice and in the plasma of Fabry patients. PLoS ONE. 2013;8:e57631. doi: 10.1371/journal.pone.0057631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Germain D.P., Fan J.Q. Pharmacological chaperone therapy by active-site-specific chaperones in Fabry disease: In vitro and preclinical studies. Int. J. Clin. Pharmacol. Ther. 2009;47:S111–S117. [PubMed] [Google Scholar]
- 49.Germain D.P., Hughes D.A., Nicholls K., Bichet D.G., Giugliani R., Wilcox W.R., Feliciani C., Shankar S.P., Ezgu F., Amartino H., et al. Treatment of fabry’s disease with the pharmacologic chaperone migalastat. N. Engl. J. Med. 2016;375:545–555. doi: 10.1056/NEJMoa1510198. [DOI] [PubMed] [Google Scholar]
- 50.Ishii S., Suzuki Y., Fan J.Q. Role of ser-65 in the activity of α-galactosidase a: Characterization of a point mutation (S65T) detected in a patient with Fabry disease. Arch. Biochem. Biophys. 2000;377:228–233. doi: 10.1006/abbi.2000.1743. [DOI] [PubMed] [Google Scholar]
- 51.Shin S.H., Murray G.J., Kluepfel-Stahl S., Cooney A.M., Quirk J.M., Schiffmann R., Brady R.O., Kaneski C.R. Screening for pharmacological chaperones in Fabry disease. Biochem. Biophys. Res. Commun. 2007;359:168–173. doi: 10.1016/j.bbrc.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishii S., Chang H.H., Kawasaki K., Yasuda K., Wu H.L., Garman S.C., Fan J.Q. Mutant α-galactosidase a enzymes identified in Fabry disease patients with residual enzyme activity: Biochemical characterization and restoration of normal intracellular processing by 1-deoxygalactonojirimycin. Biochem. J. 2007;406:285–295. doi: 10.1042/BJ20070479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin S.H., Kluepfel-Stahl S., Cooney A.M., Kaneski C.R., Quirk J.M., Schiffmann R., Brady R.O., Murray G.J. Prediction of response of mutated α-galactosidase a to a pharmacological chaperone. Pharmacogenet. Genom. 2008;18:773–780. doi: 10.1097/FPC.0b013e32830500f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J.Y., Kim G.H., Kim S.S., Ko J.M., Lee J.J., Yoo H.W. Effects of a chemical chaperone on genetic mutations in α-galactosidase a in Korean patients with Fabry disease. Exp. Mol. Med. 2009;41:1–7. doi: 10.3858/emm.2009.41.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamin E.R., Flanagan J.J., Schilling A., Chang H.H., Agarwal L., Katz E., Wu X., Pine C., Wustman B., Desnick R.J., et al. The pharmacological chaperone 1-deoxygalactonojirimycin increases α-galactosidase a levels in Fabry patient cell lines. J. Inherit. Metab. Dis. 2009;32:424–440. doi: 10.1007/s10545-009-1077-0. [DOI] [PubMed] [Google Scholar]
- 56.Filoni C., Caciotti A., Carraresi L., Cavicchi C., Parini R., Antuzzi D., Zampetti A., Feriozzi S., Poisetti P., Garman S.C., et al. Functional studies of new GLA gene mutations leading to conformational Fabry disease. Biochim. Biophys. Acta. 2010;1802:247–252. doi: 10.1016/j.bbadis.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X., Katz E., Della Valle M.C., Mascioli K., Flanagan J.J., Castelli J.P., Schiffmann R., Boudes P., Lockhart D.J., Valenzano K.J., et al. A pharmacogenetic approach to identify mutant forms of α-galactosidase a that respond to a pharmacological chaperone for Fabry disease. Hum. Mutat. 2011;32:965–977. doi: 10.1002/humu.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreotti G., Citro V., de Crescenzo A., Orlando P., Cammisa M., Correra A., Cubellis M.V. Therapy of Fabry disease with pharmacological chaperones: From in silico predictions to in vitro tests. Orphanet. J. Rare Dis. 2011;6:66. doi: 10.1186/1750-1172-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giugliani R., Waldek S., Germain D.P., Nicholls K., Bichet D.G., Simosky J.K., Bragat A.C., Castelli J.P., Benjamin E.R., Boudes P.F. A phase 2 study of migalastat hydrochloride in females with Fabry disease: Selection of population, safety and pharmacodynamic effects. Mol. Genet. Metab. 2013;109:86–92. doi: 10.1016/j.ymgme.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 60.Lukas J., Scalia S., Eichler S., Pockrandt A.M., Dehn N., Cozma C., Giese A.K., Rolfs A. Functional and clinical consequences of novel α-galactosidase a mutations in Fabry disease. Hum. Mutat. 2016;37:43–51. doi: 10.1002/humu.22910. [DOI] [PubMed] [Google Scholar]
- 61.Johnson F.K., Mudd P.N., Jr., Bragat A., Adera M., Boudes P. Pharmacokinetics and safety of migalastat HCL and effects on agalsidase activity in healthy volunteers. Clin. Pharmacol. Drug Dev. 2013;2:120–132. doi: 10.1002/cpdd.1. [DOI] [PubMed] [Google Scholar]
- 62.Togawa T., Tsukimura T., Kodama T., Tanaka T., Kawashima I., Saito S., Ohno K., Fukushige T., Kanekura T., Satomura A., et al. Fabry disease: Biochemical, pathological and structural studies of the α-galactosidase a with E66Q amino acid substitution. Mol. Genet. Metab. 2012;105:615–620. doi: 10.1016/j.ymgme.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 63.Benjamin E.R., Della Valle M.C., Wu X., Katz E., Pruthi F., Bond S., Bronfin B., Williams H., Yu J., Bichet D.G., et al. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet. Med. 2016 doi: 10.1038/gim.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cubellis M.V., Baaden M., Andreotti G. Taming molecular flexibility to tackle rare diseases. Biochimie. 2015;113:54–58. doi: 10.1016/j.biochi.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito S., Ohno K., Sese J., Sugawara K., Sakuraba H. Prediction of the clinical phenotype of Fabry disease based on protein sequential and structural information. J. Hum. Genet. 2010;55:175–178. doi: 10.1038/jhg.2010.5. [DOI] [PubMed] [Google Scholar]
- 66.Riera C., Lois S., Dominguez C., Fernandez-Cadenas I., Montaner J., Rodriguez-Sureda V., de la Cruz X. Molecular damage in Fabry disease: Characterization and prediction of α-galactosidase a pathological mutations. Proteins. 2015;83:91–104. doi: 10.1002/prot.24708. [DOI] [PubMed] [Google Scholar]
- 67.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishii S., Sakuraba H., Suzuki Y. Point mutations in the upstream region of the α-galactosidase a gene exon 6 in an atypical variant of Fabry disease. Hum. Genet. 1992;89:29–32. doi: 10.1007/BF00207037. [DOI] [PubMed] [Google Scholar]
- 69.Havndrup O., Christiansen M., Stoevring B., Jensen M., Hoffman-Bang J., Andersen P.S., Hasholt L., Norremolle A., Feldt-Rasmussen U., Kober L., et al. Fabry disease mimicking hypertrophic cardiomyopathy: Genetic screening needed for establishing the diagnosis in women. Eur. J. Heart Fail. 2010;12:535–540. doi: 10.1093/eurjhf/hfq073. [DOI] [PubMed] [Google Scholar]
- 70.Branton M.H., Schiffmann R., Sabnis S.G., Murray G.J., Quirk J.M., Altarescu G., Goldfarb L., Brady R.O., Balow J.E., Austin Iii H.A., et al. Natural history of Fabry renal disease: Influence of α-galactosidase a activity and genetic mutations on clinical course. Medicine (Baltimore) 2002;81:122–138. doi: 10.1097/00005792-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 71.Eng C.M., Resnick-Silverman L.A., Niehaus D.J., Astrin K.H., Desnick R.J. Nature and frequency of mutations in the α-galactosidase a gene that cause Fabry disease. Am. J. Hum. Genet. 1993;53:1186–1197. [PMC free article] [PubMed] [Google Scholar]
- 72.Shabbeer J., Yasuda M., Luca E., Desnick R.J. Fabry disease: 45 novel mutations in the α-galactosidase a gene causing the classical phenotype. Mol. Genet. Metab. 2002;76:23–30. doi: 10.1016/S1096-7192(02)00012-4. [DOI] [PubMed] [Google Scholar]
- 73.Ferreira S., Ortiz A., Germain D.P., Viana-Baptista M., Caldeira-Gomes A., Camprecios M., Fenollar-Cortes M., Gallegos-Villalobos A., Garcia D., Garcia-Robles J.A., et al. The α-galactosidase a p.Arg118cys variant does not cause a Fabry disease phenotype: Data from individual patients and family studies. Mol. Genet. Metab. 2015;114:248–258. doi: 10.1016/j.ymgme.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukas J., Pockrandt A.M., Seemann S., Sharif M., Runge F., Pohlers S., Zheng C., Glaser A., Beller M., Rolfs A., et al. Enzyme enhancers for the treatment of Fabry and pompe disease. Mol. Ther. 2015;23:456–464. doi: 10.1038/mt.2014.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lenders M., Weidemann F., Kurschat C., Canaan-Kuhl S., Duning T., Stypmann J., Schmitz B., Reiermann S., Kramer J., Blaschke D., et al. α-Galactosidase A p.A143T, a non-Fabry disease-causing variant. Orphanet. J. Rare Dis. 2016;11:54. doi: 10.1186/s13023-016-0441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng H., Xu X., Zhang L., Zhang X., Zheng Y., Luo S., Guo H., Xia K., Li J., Yao H., et al. GLA variation p.E66Q identified as the genetic etiology of Fabry disease using exome sequencing. Gene. 2016;575:363–367. doi: 10.1016/j.gene.2015.09.088. [DOI] [PubMed] [Google Scholar]
- 77.Oikawa M., Sakamoto N., Kobayashi A., Suzuki S., Yoshihisa A., Yamaki T., Nakazato K., Suzuki H., Saitoh S., Kiko Y., et al. Familial hypertrophic obstructive cardiomyopathy with the GLA E66Q mutation and zebra body. BMC Cardiovasc. Disord. 2016;16:83. doi: 10.1186/s12872-016-0262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakamura K., Sekijima Y., Hattori K., Nagamatsu K., Shimizu Y., Yazaki M., Sakurai A., Endo F., Fukushima Y., Ikeda S.I. P.E66Q mutation in the GLA gene is associated with a high risk of cerebral small-vessel occlusion in elderly Japanese males. Eur. J. Neurol. 2014;21:49–56. doi: 10.1111/ene.12214. [DOI] [PubMed] [Google Scholar]
- 79.Satomura A., Yanai M., Fujita T., Nakayama T. Comment on ‘p.E66q mutation in the GLA gene is associated with a high risk of cerebral small-vessel occlusion in elderly japanese males’. Eur. J. Neurol. 2014;21:e62. doi: 10.1111/ene.12459. [DOI] [PubMed] [Google Scholar]
- 80.Shimotori M., Maruyama H., Nakamura G., Suyama T., Sakamoto F., Itoh M., Miyabayashi S., Ohnishi T., Sakai N., Wataya-Kaneda M., et al. Novel mutations of the GLA gene in Japanese patients with Fabry disease and their functional characterization by active site specific chaperone. Hum. Mutat. 2008;29:331. doi: 10.1002/humu.9520. [DOI] [PubMed] [Google Scholar]
- 81.Niemann M., Rolfs A., Giese A., Mascher H., Breunig F., Ertl G., Wanner C., Weidemann F. Lyso-Gb3 indicates that the α-galactosidase a mutation D313Y is not clinically relevant for Fabry disease. JIMD Rep. 2013;7:99–102. doi: 10.1007/8904_2012_154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rigoldi M., Concolino D., Morrone A., Pieruzzi F., Ravaglia R., Furlan F., Santus F., Strisciuglio P., Torti G., Parini R. Intrafamilial phenotypic variability in four families with anderson-Fabry disease. Clin. Genet. 2014;86:258–263. doi: 10.1111/cge.12261. [DOI] [PubMed] [Google Scholar]
- 83.Ioannou Y.A., Zeidner K.M., Grace M.E., Desnick R.J. Human α-galactosidase A: Glycosylation site 3 is essential for enzyme solubility. Biochem. J. 1998;332:789–797. doi: 10.1042/bj3320789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas A., Baker R., Mehta A., Hughes D. The N215S mutation results in a distinct subtype of Fabry disease. Mol. Genet. Metab. 2015;114:S113. doi: 10.1016/j.ymgme.2014.12.258. [DOI] [Google Scholar]
- 85.Meehan S.M., Junsanto T., Rydel J.J., Desnick R.J. Fabry disease: Renal involvement limited to podocyte pathology and proteinuria in a septuagenarian cardiac variant. Pathologic and therapeutic implications. Am. J. Kidney Dis. 2004;43:164–171. doi: 10.1053/j.ajkd.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 86.Fan J.Q., Ishii S., Asano N., Suzuki Y. Accelerated transport and maturation of lysosomal α-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat. Med. 1999;5:112–115. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 87.Okumiya T., Ishii S., Takenaka T., Kase R., Kamei S., Sakuraba H., Suzuki Y. Galactose stabilizes various missense mutants of α-galactosidase in Fabry disease. Biochem. Biophys. Res. Commun. 1995;214:1219–1224. doi: 10.1006/bbrc.1995.2416. [DOI] [PubMed] [Google Scholar]
- 88.Asano N., Ishii S., Kizu H., Ikeda K., Yasuda K., Kato A., Martin O.R., Fan J.Q. In vitro inhibition and intracellular enhancement of lysosomal α-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur. J. Biochem. 2000;267:4179–4186. doi: 10.1046/j.1432-1327.2000.01457.x. [DOI] [PubMed] [Google Scholar]
- 89.Yu Y., Mena-Barragan T., Higaki K., Johnson J.L., Drury J.E., Lieberman R.L., Nakasone N., Ninomiya H., Tsukimura T., Sakuraba H., et al. Molecular basis of 1-deoxygalactonojirimycin arylthiourea binding to human α-galactosidase A: Pharmacological chaperoning efficacy on Fabry disease mutants. ACS Chem. Biol. 2014;9:1460–1469. doi: 10.1021/cb500143h. [DOI] [PubMed] [Google Scholar]
- 90.Kato A., Yamashita Y., Nakagawa S., Koike Y., Adachi I., Hollinshead J., Nash R.J., Ikeda K., Asano N. 2,5-dideoxy-2,5-imino-d-altritol as a new class of pharmacological chaperone for Fabry disease. Bioorg. Med. Chem. 2010;18:3790–3794. doi: 10.1016/j.bmc.2010.04.048. [DOI] [PubMed] [Google Scholar]
- 91.Ayers B.J., Ngo N., Jenkinson S.F., Martinez R.F., Shimada Y., Adachi I., Weymouth-Wilson A.C., Kato A., Fleet G.W. Glycosidase inhibition by all 10 stereoisomeric 2,5-dideoxy-2,5-iminohexitols prepared from the enantiomers of glucuronolactone. J. Org. Chem. 2012;77:7777–7792. doi: 10.1021/jo301243s. [DOI] [PubMed] [Google Scholar]
- 92.Cheng W.C., Wang J.H., Li H.Y., Lu S.J., Hu J.M., Yun W.Y., Chiu C.H., Yang W.B., Chien Y.H., Hwu W.L. Bioevaluation of sixteen admdp stereoisomers toward α-galactosidase A: Development of a new pharmacological chaperone for the treatment of Fabry disease and potential enhancement of enzyme replacement therapy efficiency. Eur. J. Med. Chem. 2016;123:14–20. doi: 10.1016/j.ejmech.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 93.Mena-Barragan T., Narita A., Matias D., Tiscornia G., Nanba E., Ohno K., Suzuki Y., Higaki K., Garcia Fernandez J.M., Ortiz Mellet C. pH-responsive pharmacological chaperones for rescuing mutant glycosidases. Angew. Chem. Int. Ed. Engl. 2015;54:11696–11700. doi: 10.1002/anie.201505147. [DOI] [PubMed] [Google Scholar]
- 94.Motabar O., Liu K., Southall N., Marugan J.J., Goldin E., Sidransky E., Zheng W. High throughput screening for inhibitors of α-galactosidase. Curr. Chem. Genom. 2010;4:67–73. doi: 10.2174/1875397301004010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Citro V., Peña-García J., den-Haan H., Pérez-Sánchez H., del Prete R., Liguori L., Cimmaruta C., Lukas J., Cubellis M.V., Andreotti G. Identification of an allosteric binding site on human lysosomal α-galactosidase opens the way to new pharmacological chaperones for Fabry disease. PLoS ONE. 2016;11:e0165463. doi: 10.1371/journal.pone.0165463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Linear Correlation and Regression. [(accessed on 30 November 2016)]. Available online: http://vassarstats.net/corr_big.html.
- 97.Andreotti G., Guarracino M.R., Cammisa M., Correra A., Cubellis M.V. Prediction of the responsiveness to pharmacological chaperones: Lysosomal human α-galactosidase, a case of study. Orphanet. J. Rare Dis. 2010;5:36. doi: 10.1186/1750-1172-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cammisa M., Correra A., Andreotti G., Cubellis M.V. Fabry_cep: A tool to identify Fabry mutations responsive to pharmacological chaperones. Orphanet. J. Rare Dis. 2013;8:111. doi: 10.1186/1750-1172-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cammisa M., Correra A., Andreotti G., Cubellis M.V. Identification and analysis of conserved pockets on protein surfaces. BMC Bioinform. 2013;14:S9. doi: 10.1186/1471-2105-14-S7-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.