Abstract
NuA4, the only essential histone acetyltransferase complex in Saccharomyces cerevisiae, acetylates the N-terminal tails of histones H4 and H2A. Affinity purification of NuA4 revealed the presence of three previously undescribed subunits, Vid21/Eaf1/Ydr359c, Swc4/Eaf2/Ygr002c, and Eaf7/Ynl136w. Experimental analyses revealed at least two functionally distinct sets of polypeptides in NuA4: (i) Vid21 and Yng2, and (ii) Eaf5 and Eaf7. Vid21 and Yng2 are required for bulk histone H4 acetylation and are functionally linked to the histone H2A variant Htz1 and the Swr1 ATPase complex (SWR-C) that assembles Htz1 into chromatin, whereas Eaf5 and Eaf7 have a different, as yet undefined, role. Mutations in Htz1, the SWR-C, and NuA4 cause defects in chromosome segregation that are consistent with genetic interactions we have observed between the genes encoding these proteins and genes encoding kinetochore components. Because SWR-C-dependent recruitment of Htz1 occurs in both transcribed and centromeric regions, a NuA4/SWR-C/Htz1 pathway may regulate both transcription and centromere function in S. cerevisiae.
The nucleosomes that package eukaryotic DNA into chromatin can prevent the site-specific binding of many proteins to DNA. Two classes of factors have been implicated in regulating access to DNA in this context. The ATP-dependent chromatin-remodeling enzymes use energy derived from ATP hydrolysis to induce nucleosome mobility or disrupt histone–DNA interactions (reviewed in ref. 1). The second class of enzymes catalyzes covalent modification (e.g., lysine acetylation, serine phosphorylation, lysine and arginine methylation, ubiquitylation, and ADP ribosylation) of various histones, usually on their N-terminal tails (reviewed in refs. 2 and 3). Nucleosome variability is also generated by occasional replacement of a major histone with a variant that has a specialized role (reviewed in ref. 4). Among H2A variants is H2A.Z (Htz1 in Saccharomyces cerevisiae), whose insertion into chromatin requires the recently described SWR1 complex (SWR-C) (5–7). H2A.Z is functionally distinct from the major histone H2A. Moreover, the various eukaryotic H2A.Z molecules are more closely related to each other than they are to H2A. This similarity in sequence extends to functional conservation, as H2A.Z from the ciliated protozoan Tetrahymena thermophila can rescue all tested phenotypes associated with S. cerevisiae htz1Δ (8). Htz1 plays a role in transcription (5–7, 9, 10) and may act as a barrier to inhibit the spread of silent telomeric and mating locus hetereochromatin into adjacent transcriptionally active regions (9).
Histone acetylation is carried out by a class of enzymes known as histone acetyltransferases (HATs). These transfer an acetyl group from acetyl-CoA to lysine ε-amino groups on target histone N-terminal tails. Histone acetylation has been linked to transcriptional activation, because actively transcribed regions are characteristically hyperacetylated, whereas silent regions are typically hypoacetylated (reviewed in ref. 11). The acetylation of lysine residues on the N-terminal tails of histones H3 and H4 neutralizes their positive charge, possibly decreasing their affinity for DNA and facilitating chromatin decompaction and disassembly. More important than simple charge neutralization, however, is the specific acetylation pattern at individual lysine residues within the histone N termini (11, 12). At least some acetylated lysines interact with and recruit bromodomain-containing polypeptides (ref. 13 and references therein). The HAT complexes of S. cerevisiae, many of which contain associated bromodomains, include the Gcn5-containing ADA and SAGA complexes, Hat1, Elongator, NuA3, and NuA4. These complexes generally have high specificity for specific lysine residues on specific histone N-terminal tails.
The catalytic subunit of NuA4, Esa1, is a member of the MYST (for MOZ, Ybf2/Sas3, Sas2, and Tip60) family of HATs (reviewed in ref. 14). In S. cerevisiae, NuA4 is required for minor acetylation on histone H2A and the majority of histone H4 acetylation on lysines 5, 8, and 12 (15, 16). Nine other NuA4 subunits have been reported: Esa1 associated factor 3 (Eaf3), a chromodomain-containing factor also found in a Sin3/Rpd3 histone deacetylase complex (17); Eaf5 and Eaf6; Tra1, a member of the phosphatidylinositol kinase family; the actin-related proteins Act1 and Arp4; Yng2, an ING1 tumor suppressor homolog; Epl1, a homolog of the Enhancer of Polycomb-like protein; and Yaf9, a protein related to the AF9/ENL leukemogenic factors.
Here, we report the identification of three NuA4 subunits, namely Vid21/Eaf1/Ydr359c, Swc4/Eaf2/Ygr002c, and Eaf7/Ynl136w. Vid21 and Swc4 both contain SANT (SWI3, ADA2, N-CoR, TFIIIB) domains, also found in several other chromatin remodeling complexes. Swc4, Yaf9, Arp4, and Act1 are also components of the SWR-C (5–7). Microarray and genetic analyses revealed at least two functionally distinct classes of polypeptides in NuA4. The first class contains Vid21 and Yng2, required for most of the acetylation activity of Esa1. The second class, containing Eaf5 and Eaf7, plays a minor role, if any, in acetylation of histone H4 by NuA4.
We showed previously that the SWR-C recruits Htz1 to various regions of the genome (5), and we show here that this is also the case at the centromere. Although NuA4 is not required for assembling Htz1 into chromatin, Htz1 and subunits of NuA4 and the SWR-C are genetically linked to kinetochore components and are important for chromosome stability. Genetic analyses suggest that these factors operate in common, rather than parallel, pathways. These observations imply that NuA4, the SWR-C, and Htz1 function together to regulate chromosome stability and/or transmission as well as transcription in S. cerevisiae.
Experimental Procedures
Tandem affinity purification (TAP)-tagged components of NuA4 were purified on IgG and calmodulin columns from extracts of yeast cells (3 liters) grown in yeast extract/peptone/dextrose (YPD) medium to an OD600 of 1.0–1.5. Complexes were analyzed by SDS/PAGE, matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS, and tandem MS (MS/MS) as described (18). Synthetic genetic array (SGA) analysis was carried out as described (19). Automated analysis of the results was carried out by procedures that are described elsewhere (H. Ding and C. Boone, personal communication). RNA preparation and microarray analysis were as described (20, 21) after isogenic strains were grown in parallel in SC medium at 30°C. Chromatin immunoprecipitation (ChIP) assays using the TAP-tagged strains were performed essentially as described (5, 18). For ChIP analysis in the context of temperature-sensitive alleles of esa1 (esa1-Δ414, esa1-L327S; ref. 22), crosslinking was performed at 25°C or after 3 h at 37°C and compared to an isogenic wild-type strain under the same conditions. IgG agarose for precipitation of TAP-tagged proteins was from Sigma. All primer sequences are listed in Table 2, which is published as supporting information on the PNAS web site. Protein isolation and Western blot procedures were performed as described (23) with modifications described in Supporting Text, which is published as supporting information on the PNAS web site.
Quantitative half-sector analysis was performed as described (24–26). In brief, homozygous diploid strains (YPH982, wild-type; YKB522, eaf1Δ; YKB463, eaf7Δ; YPH493, yaf9Δ; YKB508, swr1Δ; YKB503, htz1Δ) (see Table 3, which is published as supporting information on the PNAS web site) were created containing an ade2ochre allele at the endogous locus and the SUP11 ochre suppressor on a single chromosome fragment [CFIII (CEN3. L) URA3 SUP11]. Strains were plated to single colonies on solid media containing limiting adenine and grown at 25°C for 3 days before the plates were placed at 4°C for optimal red pigment development. Efficient chromosome stability/transmission results in pink colonies. Chromosome loss or 1:0 events were scored as colonies that were half red and half pink, nondisjunction or 2:0 events were scored as colonies that were half red and half white.
Results and Discussion
To further characterize the subunit composition of yeast NuA4, we used single-step transformation to place a TAP tag (17) at the C terminus of several of its known components (Esa1, Epl1, Yng2, Eaf5, and Eaf6). The tagged protein was then purified sequentially on IgG and calmodulin columns and analyzed by SDS/PAGE followed by staining with silver. Protein bands absent from a control preparation and corresponding to the tagged factor and any associated proteins were identified by MALDI-TOF MS and MS/MS.
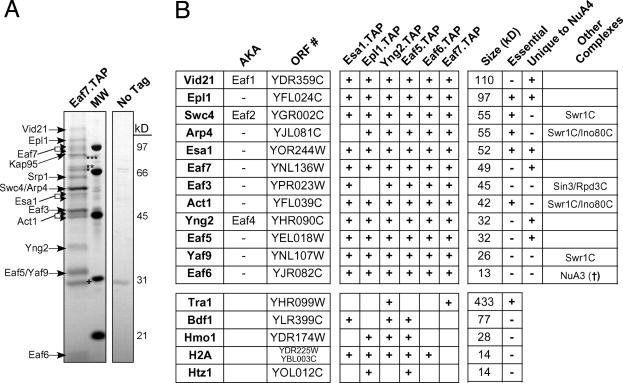
In each case, the tagged protein copurified with all, or almost all, of the known subunits of the NuA4 complex (Fig. 1B and data not shown). Only the 400-kDa subunit Tra1 was not identified by gel band excision and MALDI-TOF MS, even though Tra1 is a well characterized stoichiometric component of the NuA4 complex (16) and silver-stained bands were present at this molecular weight in almost every purified preparation. Tra1 was, however, detected by using liquid chromatography (LC) MS/MS after the purification of Yng2-TAP (Fig. 1B Lower). In each purified preparation, we detected three previously unreported subunits of NuA4, namely Vid21/Eaf1/Ydr359c, Swc4/Eaf2/Ygr002c, and Eaf7/Ynl136w (Fig. 1B). The tagging and purification of Eaf7 confirmed that this subunit is indeed a member of the complex (Fig. 1). In this case, we again detected Tra1 as copurifying with the NuA4 complex. Eaf7-TAP also copurified with Vid21 and Swc4, both of which contain SANT domains that are often present in complexes involved in chromatin metabolism. Vid21 copurified with each tagged subunit of NuA4 that we isolated (Fig. 1B). Interestingly, Swc4 is also a component of the SWR-C chromatin remodeling complex required for replacing histone H2A with the H2A variant, Htz1 (5, 6). Purification of tagged Swc4 led to the isolation of a mixture of NuA4 and the SWR-C (data not shown). Other shared components of the SWR-C and NuA4 include Arp4, Act1, and Yaf9 (Fig. 1B). The human Tip60 HAT complex contains factors homologous to those in both the SWR-C and NuA4 (27–29), consistent with the observation that components are shared between these complexes in S. cerevisiae.
Fig. 1.
Isolation of the NuA4 HAT complex. (A) TAP of the NuA4 complex was carried out on strains containing either TAP-tagged Eaf7 or no tagged protein. The purified protein was analyzed by SDS/PAGE and silver staining. The indicated proteins were identified by trypsin digestion of each stained band followed by MALDI-TOF mass spectrometry or tandem mass spectrometry after subjecting an aliquot of the eluate from the final column directly to trypsin. Contaminating bands are indicated: ***, Sse1; **, Ssa1; *, Ssb1; †, TEV protease. The Srp1/Kap95 importin complex, which transports NLS-containing proteins into the nucleus (56), also copurified with NuA4 and is often found associated with nuclear protein complexes (N.J.K., A.E. and J.F.G., unpublished data). (B) Summary of purified proteins identified by mass spectrometry. Proteins that were present in at least two of the six purifications are represented. Protein size (kDa) was as predicted by amino acid composition (Saccharomyces Genome Database; www.yeastgenome.org) and not determined experimentally. (†, N.J.K., A.E., and J.F.G., unpublished data.)
In three of the six purified preparations, we detected the high-mobility group (HMG) domain protein, Hmo1, by LC/MS/MS (Fig. 1B). There are seven proteins in S. cerevisiae that contain a HMG DNA-binding domain, several of which, including Hmo1, bind to rDNA and are linked to RNA polymerase I transcription (30). An association between NuA4 and Hmo1 may partly explain the observed role of NuA4 in rDNA transcription (31). NuA4 also copurified with small amounts of histone H2A, a known substrate of NuA4, as well as with small amounts of Bdf1 and the histone H2A variant, Htz1, which may represent potential substrates. Bdf1 is also associated with the SWR-C (5) and binds to acetylated histone tails (13).
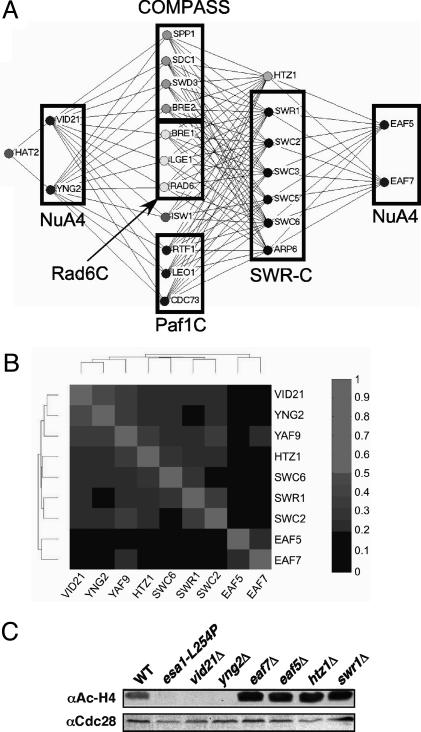
Genes encoding proteins in the same functional pathway should, in principle, have similar effects on gene expression and similar sets of synthetic genetic interactions. To test this concept and provide independent evidence that the identified proteins unique to NuA4 are functionally significant components of the complex, we first used automated SGA analysis (19). For this purpose, a miniarray containing 384 Kanr deletion strains was constructed in which each deletion represents a protein known or suspected to function in some aspect of transcription and/or chromatin modification or remodeling (N.J.K. and J.F.G., unpublished data). Four NatR strains harboring individual gene deletions of nonessential components of NuA4 (VID21, YNG2, EAF5, and EAF7) were then generated and crossed with the 384 deletion strain miniarray to create sets of Kanr Natr haploid double mutants. Genetic interactions identified for vid21Δ and yng2Δ were very similar but dramatically different from those observed with eaf5Δ and eaf7Δ, which, in turn, were very similar to each other (Fig. 2A and data not shown). Strains harboring a deletion of EAF5 or EAF7 were synthetically sick when combined with a deletion of HTZ1 or genes encoding nonessential components of the SWR-C. This finding suggests that these subunits of NuA4 act in a distinct pathway, possibly parallel to that of the SWR-C and Htz1. In contrast, vid21Δ and yng2Δ lacked these interactions, and instead interacted genetically with different genes encoding factors required for histone H3 K4 methylation, as was also seen with htz1Δ and deletion of genes encoding components of the SWR-C (Fig. 2A). This observation included components of the Set1 histone methyltransferase-containing complex, COMPASS (SPP1, SDC1, SWD3, and BRE2) (32); the PAF complex (RTF1, LEO1, and CDC73), which is required for the recruitment of COMPASS to chromatin (33, 34); and the Rad6 complex (RAD6, BRE1, and LGE1) that ubiquitinates K123 of histone H2B, an event required for methylation of H3 K4 by COMPASS (35, 36). Finally, genetic interactions with the Snf2-family member and chromatin remodeler ISW1, the function of which has been linked to histone H3 K4 methylation (37), were also observed. The similar patterns of genetic interactions for the genes encoding the NuA4 subunits Vid21 and Yng2, Htz1, and members of the SWR-C suggested that these proteins function in a common pathway in vivo.
Fig. 2.
Genetic similarity of the SWR-C and a subset of NuA4. (A) Synthetic genetic interactions for the SWR-C and NuA4. SGA technology (19) was used to cross Natr strains harboring individual deletions of genes encoding Htz1 or subunits of the SWR-C or NuA4 with a transcription-targeted array of 384 KanR deletion strains to create sets of Natr Kanr haploid double mutants. Growth rates were assessed by automated image analysis of colony size. Lines connect genes with synthetic genetic interactions. The lengths of lines and proximity of boxes in this diagram and in Fig. 3A are unrelated to the strengths of the indicated synthetic genetic interactions. (B) Microarray analysis of gene expression was performed for the indicated deletion strains. Pearson correlation coefficients were then calculated for each pair of deletions, and the deletions were organized by 2D hierarchical clustering according to the similarities of their effects. (C) Immunoblots of whole-cell protein extracts from wild-type (WT, NJK28), esa1-L245P (LPY3500), vid21Δ (NJK1042), eaf7Δ (NJK1254), yng2Δ (NJK1482), eaf5Δ (NJK1259), htz1Δ (NJK1527), and swr1Δ (NJK1665) cells (see Table 3) grown to log phase in yeast extract/peptone/dextrose at 30°C and transferred to 37°C for 4 h. The membranes were probed with antihyperacetylated histone H4 (Penta) antibodies (α Ac-H4) and then reprobed with antibodies to Cdc28 to verify approximately equal protein loading.
We have also individually crossed NatR versions of each of the 384 deletion strains in our transcription/chromatin modification miniarray against the entire KanR miniarray, thereby creating a 384 × 384 matrix of double mutant strains (N.J.K. and J.F.G., unpublished data). Growth rates were evaluated by automated image analysis, and the results of all of the screens were analyzed by 2D hierarchical clustering. This clustering process groups genes according to the degrees of similarity of their synthetic genetic interactions. One important expectation is that genes with the same or very similar functions would have similar sets of genetic interactions and would fall into the same cluster. The genetic patterns obtained from strains containing deletions of either VID21 or YNG2 were more similar to each other than to those obtained from any of the other 382 genes on the array (i.e., these genes clustered next to each other), implying that they have a very similar role in NuA4 function (data not shown). EAF5 and EAF7 also clustered next to each other, but away from VID21 and YNG2. This finding indicated that Eaf5 and Eaf7 are functionally similar subunits of NuA4, but that their roles are distinct from those of Vid21 and Yng2. Interestingly, HTZ1 and genes encoding components of the SWR-C clustered near VID21 and YNG2, but away from EAF5 and EAF7 (data not shown), again suggesting that only certain components of NuA4 function with the SWR-C in vivo.
To further characterize these seemingly functionally distinct sets of proteins contained within NuA4, we carried out microarray analyses of gene expression in strains containing deletions of VID21, YNG2, EAF5, and EAF7. For comparison purposes, we also conducted microarray experiments on strains deleted for HTZ1 and components of the SWR-C (SWR1, SWC2, and SWC6, as well as YAF9, a subunit of both NuA4 and SWR-C) (raw microarray data are available at www.utoronto.ca/greenblattlab/nua4swrc.xls). Pearson correlation coefficients were calculated for each pair of deletions, and the strains were organized by 2D hierarchical clustering according to the similarities of their effects on gene expression (Fig. 2B).
As was observed with our genetic analyses, the gene expression profiles of strains containing deletions of VID21 and YNG2 were similar to each other and to those of strains containing deletions of HTZ1 or components of the SWR-C. Deletion of YAF9, which encodes a shared component of the SWR-C and NuA4, had a very similar effect on gene expression (Fig. 2B), consistent with a functional role in both complexes. In contrast, EAF5 and EAF7 clustered next to each other but away from the other genes in this gene expression analysis, just as they had in our genetic analysis. Microarray experiments have also been conducted on nearly 200 other deletion strains represented on our 384-strain genetic miniarray (N.J.K., T.R.H., and J.F.G., unpublished data). The clustering of this larger data set (not shown) reflects the data represented in Fig. 2B, in which VID21 and YNG2 clustered next to HTZ1 and genes encoding components of the SWR-C and away from EAF5 and EAF7. Therefore, both the genetic and gene expression data suggest that the Vid21 and Yng2 subunits of NuA4 function in an SWR-C/Htz1 pathway, whereas Eaf5 and Eaf7 are important for a different, perhaps parallel, role of NuA4.
To identify another functional distinction among these NuA4 subunits, we took advantage of the fact that NuA4 is responsible for most histone H4 acetylation in S. cerevisiae. Whole cell extracts were prepared from esa1-L254P, a temperature-sensitive allele with diminished HAT activity (22), or strains containing deletions of various NuA4 subunits and probed with antibody specific for hyperacetylated histone H4. Whereas deletion of EAF5 or EAF7 had no apparent effect on histone H4 acetylation, deletion of VID21 or YNG2 appeared to result in its complete loss (Fig. 2C). It has been reported that Yng2, but not Eaf5, is required for the catalytic activity of NuA4 (38). VID21 and YNG2, but not EAF5 or EAF7, interacted genetically with HAT2, a component of the histone acetyltransferase B complex (39) (Fig. 2A), consistent with the fact that Vid21 and Yng2 are involved in H4 acetylation by NuA4. Acetylation elsewhere (or below the threshold of detection) by NuA4 in the absence of Vid21 or Yng2 must, however, exist, because the catalytic subunit, Esa1, is essential (15), whereas Vid21 and Yng2 are not. Further comparison with the genetic data summarized in Fig. 2A indicated that H4 acetylation by NuA4 (or acetylation elsewhere that also depends on Vid21 and Yng2) and H3 K4 methylation by COMPASS cannot be eliminated simultaneously.
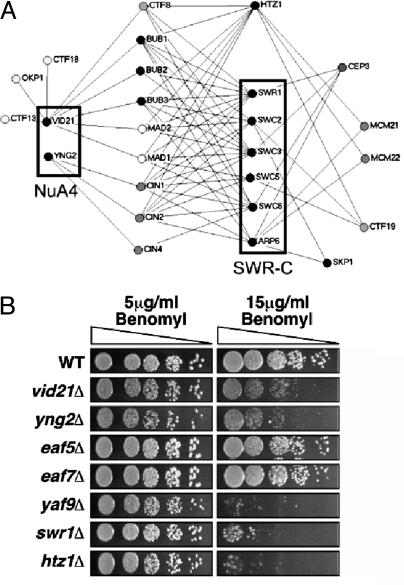
We recently identified VID21 in a ctf13–30/CTF13 genome-wide modifier screen to identify genes whose function impinges on chromosome transmission (26). Deletion strains of VID21 were shown to be sensitive to overexpression of the inner kinetochore genes CTF13 and CBF2 and display a high rate of chromosome loss (26). These observations suggested that NuA4 might be involved in centromere function as well as transcription. Our SGA screen has identified numerous synthetic genetic interactions between Htz1, the SWR-C, NuA4 (VID21 and YNG2), and known kinetochore and spindle checkpoint mutants (Fig. 3A). The kinetochore is a large multiprotein complex that binds centromeric DNA and serves to link chromosomes to spindle microtubules. The kinetochore both mediates and monitors its interaction with the spindle by communicating attachment defects to the checkpoint machinery (reviewed in refs. 40 and 41). In particular, we observed genetic interactions with mutation or deletion of subunits of the C-repeat binding factor 3 inner kinetochore complex [skp1–3 (42), cep3–1 (43), or ctf13–30 (44)] and subunits of the central kinetochore complexes [okp1–5 (45), mcm21Δ, mcm22Δ, or ctf19Δ]. We also detected genetic interactions with mutants of components of the spindle checkpoint, bub1Δ, bub3Δ, mad1Δ, mad2Δ (46), the spindle position checkpoint, bub2Δ (47), and genes required for microtubule stability, cin1Δ, cin2Δ, and cin4Δ (48). Together, these genetic interactions also suggested that Htz1, NuA4, and the SWR-C may be required for chromosome stability in addition to their previously described roles in transcription.
Fig. 3.
Htz1, SWR-C, and the NuA4 complex function to regulate chromosome stability/transmission. (A) SGA analysis (19) using either a transcription-targeted 384-deletion-strain array or genome-wide kinetochore screens (K.B. and V. Measday, unpublished data) identified numerous synthetic genetic interactions between deletions of genes encoding Htz1, SWR-C, or the NuA4 subunits Vid21 and Yng2, and known chromosome stability/transmission factors (see text for details). Genome-wide screens were carried out with four essential kinetochore genes [skp1–3 (42), cep3–1 (43), ctf13–30 (44), and okp1–5 (45)], whereas the nonessential components (mcm21Δ, mcm22Δ, ctf19Δ, bub1Δ, bub3Δ, mad1Δ, and mad2Δ) were present on the targeted miniarray. (B) Effects of the microtubule destabilizing agent benomyl on the growth of wild-type (NJK28), vid21Δ (NJK1042), eaf5Δ (NJK1259), eaf7Δ (NJK1254), yng2Δ (NJK1482), yaf9Δ (NJK1240), htz1Δ (NJK1527), and swr1Δ (NJK1665) strains. Five-fold serial dilutions of strains starting from an OD600 of 0.1 were plated onto yeast extract/peptone/dextrose plates containing 5 or 15 μg/ml benomyl and incubated for 2 days at 30°C.
Numerous kinetochore and spindle checkpoint mutants are hypersensitive to the microtubule destabilizing drug benomyl. This is also the case for certain NuA4 mutants, including esa1–1851, yaf9Δ, yng2Δ, and vid21Δ (refs. 7 and 23 and Fig. 3B). In contrast, eaf5Δ and eaf7Δ strains displayed no sensitivity to benomyl, an observation that is consistent with our microarray and genetic data pointing to different roles for these NuA4 subunits (Fig. 2). Interestingly, benomyl sensitivity is also observed after deletion of HTZ1 or the catalytic subunit of the SWR-C, SWR1 (Fig. 3B and ref. 7). Although the NuA4-SWR-C-Htz1 pathway interacts genetically with the histone H3 K4 methylation pathway (Fig. 2A), deletions of genes encoding subunits of the PAF complex or COMPASS do not cause benomyl sensitivity (ref. 49 and our unpublished data), nor does deletion of RTF1, which encodes a subunit of the PAF complex, cause chromosome loss (50).
To further characterize the roles of the NuA4, the SWR-C, and Htz1 in chromosome stability, we quantified chromosome missegregation in vid21Δ, eaf7Δ, yaf9Δ, swr1Δ, and htz1Δ diploid strains by using colony half-sector analysis (24). In each case, we observed a significant increase in chromosome loss rates compared to wild-type diploids (Table 1). The role of Htz1 in this process has been conserved across species, as deletion of the H2A.Z variant, Pht1, from the fission yeast Schizosaccharomyces pombe is also associated with decreased chromosome stability (51), and mammalian H2A.Z plays a role in chromosome segregation (52). Although the eaf7Δ strain displayed elevated chromosome instability rates, it was benomyl insensitive, indicating that the two phenotypes are not fully correlated (as is also the case for certain kinetochore mutants; our unpublished data). The benomyl sensitivity and chromosome loss defects of a subset of NuA4 mutants, swr1Δ and htz1Δ, suggest that the two complexes have functionally linked roles in chromosome segregation.
Table 1. Rates of chromosome missegregation events.
| Rate
|
|||
|---|---|---|---|
| Genotype | Nondisjunction (2:0 events) | Chromosome loss (1:0 events) | Total colonies |
| Wild type* | 8.7 × 10-5 (1.0) | 8.7 × 10-5 (1.0) | 22,800 |
| vid21Δ | 9.5 × 10-4 (10.9) | 3.6 × 10-3 (41.5) | 16,875 |
| eaf7Δ | 6.7 × 10-4 (7.7) | 1.2 × 10-3 (13.8) | 29,670 |
| yaf9Δ | 1.7 × 10-4 (2.0) | 2.0 × 10-3 (23.0) | 17,400 |
| swr1Δ | 3.0 × 10-4 (3.4) | 6.2 × 10-4 (7.2) | 30,450 |
| htz1Δ | 1.2 × 10-4 (1.4) | 1.3 × 10-3 (14.5) | 32,585 |
Strains were plated to single colonies, and sectoring colonies were scored visually as described in Experimental Procedures. Colonies scored as half-sectored were ≥50% red (24). Strains used were YPH982, YKB522, YKB46, YPH493, YKB508, YKB513, and YKB503. Numbers in parentheses are the factors of increase in rate of missegregation events above wild type. The rates observed are comparable to those described after mutation/deletion of characterized chromosome stability/transmission factors.
Wild-type rates were published in ref. 26.
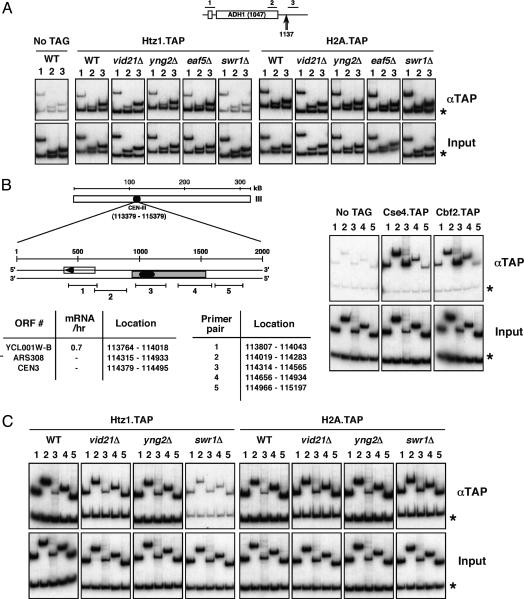
Because of the role of Htz1 in regulating gene expression (5, 9), its effect on chromosomal segregation could, in principle, be indirect. However, our microarray analyses using strains deleted for HTZ1 or components of the SWR-C did not detect any significant changes in the transcription of genes encoding kinetochore proteins (see www.utoronto.ca/greenblattlab/nua4swrc.xls). To ascertain whether Htz1 has a direct role in chromosome segregation, TAP-tagged versions of Htz1 were analyzed by ChIP after proteins were cross-linked in vivo to DNA by using formaldehyde. Yeast cells were lysed and, after isolation and shearing of chromatin, the presence of Htz1 near specific DNA sequences was monitored by immunoprecipitating (IP) with IgG that binds to the protein A component of the TAP tag. After reversal of the cross-links, the coprecipitated DNA was analyzed by PCR amplification with specific primer pairs directed against regions of interest. Each PCR also contained primers directed against a nontranscribed subtelomeric region of chromosome 5, as we have previously determined that Htz1 occupancy is high at this location (5). In each case, we also examined the DNA that coprecipitated with the major histone H2A as a control.
As previously observed (5), Htz1 recruitment at the constitutively transcribed genes ADH1 and PMA1, a Htz1-dependent gene cluster (Htz1 activated domain), and the telomere of chromosome V required Swr1 (Fig. 4A and data not shown). We now show that Htz1 is also recruited to centromeric regions in a Swr1-dependent manner (Fig. 4C), indicating that the SWR-C and Htz1 may directly regulate centromere function as well as transcription. In contrast, recruitment of the major histone H2A is independent of Swr1 at all of these locations (Fig. 4 A and C).
Fig. 4.
Htz1 recruitment is Swr1-dependent and NuA4-independent at all tested regions. (A) ChIP analysis at the ADH1 gene. A schematic of the ADH1 locus is shown with the location of the ChIP primer pairs 1–3 (Upper) and the major polyadenylation/cleavage site at +1137 (Lower) (57). The asterisks indicate a subtelomeric region of chromosome V (9716–9823) that we frequently use as a nontranscribed control when analyzing transcriptionally active regions (5). Chromatin samples from each strain were analyzed after the indicated precipitation. The bottom row is input, used to normalize the PCR amplification efficiency of each primer pair. Equivalent crosslinking of Rpb3 in each case was used as a control for sample integrity (not shown). Whereas swr1Δ leads to a loss of Htz1 occupancy at all tested regions, deletion of members of the NuA4 complex (vid21Δ, yng2Δ, and eaf5Δ) has no detectable effect. (B) A schematic of the chromosome III centromere (CEN-III), with the location of coding sequences, structural elements and ChIP primer pairs depicted. Asterisk indicates a subtelomeric region of chromosome 5 as above. Transcriptional frequency of Ycl001w-b (mRNA/hr) is from ref. 58 (http://web.wi.mit.edu/young/expression/halflife.html). The histone H3 variant Cse4 and the kinetochore component Cbf2 crosslink at the centromere, as reported (59–61). (C) ChIP analysis at CEN-III. Chromatin samples from the indicated strains were analyzed after precipitation via the TAP tag. Htz1 and H2A occupancy at CEN-III (and CEN-V, not shown) is equivalent to that at the subtelomeric (asterisk) region. Htz1 recruitment at this region depends on the SWR-C (indicated by swr1Δ), but independent of NuA4 (indicated by vid21Δ and yng2Δ). H2A recruitment is independent of both complexes. The reduced occupancy at primer pair 3 directly over CEN-III is not unique to Htz1 and is also exhibited by H2A, H2B, H3, and H4 (latter three not shown). This may be a combined function of relative primer efficiency, reduced nucleosome density at this region, and steric occlusion by the kinetochore complex.
Despite the involvement of NuA4 in Htz1 function suggested by our genetic and microarray experiments, Htz1 recruitment at all tested regions was independent of Vid21 and Yng2 (Fig. 4 A and C and data not shown), suggesting that Htz1 recruitment is NuA4-independent. We have also determined that Htz1 or H2A recruitment at all tested locations is unaffected after inactivation of temperature-sensitive alleles of esa1 (esa1-Δ414 and esa1-L327S) (data not shown). It seems likely, therefore, that NuA4 has a postrecruitment role, as yet undefined, in Htz1 function. Because NuA4 is known to acetylate histone H2A, it is tempting to speculate that its role in the SWR-C/Htz1 pathway is to acetylate the histone H2A variant Htz1 after its incorporation into chromatin. H2A.Z N-terminal tail acetylation is essential in Tetrahymena (53, 54), and covalent modification may be required for at least some of the functions performed by this histone variant in S. cerevisiae.
We have shown here that the NuA4 HAT complex has three previously unidentified subunits and that the Vid21 and Yng2 subunits of NuA4 have synthetic genetic interactions and effects on gene expression very similar to those of Htz1 and the SWR-C. We have also found that Htz1 is recruited to centromeric regions and mutations in the genes encoding Htz1, the SWR-C, and subunits of NuA4 lead to a decrease in the fidelity of chromosome segregation. Htz1 may contribute to the overall structure of centromeric chromatin in such a way as to affect centromere function and chromosome transmission (55). Because the HAT activity of NuA4 on histone H4 is unnecessary for recruiting Htz1 into chromatin, we predict that NuA4 directly acetylates Htz1 to influence its function.
Supplementary Material
Acknowledgments
We thank Lorraine Pillus (University of California at San Diego, La Jolla) for strain LPY3500, Jacques Cote for sharing unpublished data, and Affinium Pharmaceuticals for help with MALDI-TOF mass spectrometry. N.J.K. was supported by a Doctoral Fellowship from the Canadian Institutes of Health Research (CIHR), and K.B. was supported by Postdoctoral Fellowships from CIHR and the Michael Smith Foundation for Health Research. This research was supported by grants from the CIHR, the Ontario Genomics Institute, and the National Cancer Institute of Canada with funds from the Canadian Cancer Society (to J.F.G.), and National Institutes of Health Grants GM46498 (to S.B.) and CA16591 (to P.H.). S.B. is a Scholar of the Leukemia and Lymphoma Society.
Abbreviations: SWR-C, SWR1 complex; HAT, histone acetyltransferase; Eaf, Esa1-associated factor; TAP, tandem affinity purification; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; SGA, synthetic genetic array; MS/MS, tandem MS; ChIP, chromatin immunoprecipitation.
References
- 1.Lusser, A. & Kadonaga, J. T. (2003) BioEssays 25, 1192–1200. [DOI] [PubMed] [Google Scholar]
- 2.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 4.Malik, H. S. & Henikoff, S. (2003) Nat. Struct. Biol. 10, 882–891. [DOI] [PubMed] [Google Scholar]
- 5.Krogan, N. J., Keogh, M.-C., Datta, N., Sawa, C., Ryan, O. W., Ding, H., Haw, R. A., Pootoolal, J., Tong, A., Canadien, V., et al. (2003) Mol. Cell 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- 6.Mizuguchi, G., Shwn, X., Landry, J., Hu, W.-H., Sen, S. & Wu, C. (2004) Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- 7.Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., Madhani, H. D. & Rine, J. (2004) Pub. Lib. Sci. Biol. 2, 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson, J. D. & Gorovsky, M. A. (2000) Nucleic Acids Res. 28, 3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meneghini, M. D., Wu, M. & Madhani, H. D. (2003) Cell 112, 725–736. [DOI] [PubMed] [Google Scholar]
- 10.Santisteban, M. S., Kalashnikova, T. & Smith, M. M. (2000) Cell 103, 411–422. [DOI] [PubMed] [Google Scholar]
- 11.Eberharter, A. & Becker, P. B. (2002) EMBO Rep. 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner, B. M. (1993) Cell 75, 5–8. [PubMed] [Google Scholar]
- 13.Matangkasombut, O. & Buratowski, S. (2003) Mol. Cell 11, 353–363. [DOI] [PubMed] [Google Scholar]
- 14.Doyon, Y. & Cote, J. (2004) Curr. Opin. Gen. Dev. 14, 147–154. [DOI] [PubMed] [Google Scholar]
- 15.Smith, E. R., Eisen, A., Gu, W., Sattah, M., Pannuti, A., Zhou, J., Cook, R. G., Lucchesi, J. C. & Allis, C. D. (1998) Proc. Natl. Acad. Sci. USA 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allard, S., Utley, R. T., Savard, J., Clarke, A., Grant, P., Brandl, C. J., Pillus, L., Workman, J. L. & Cote, J. (1999) EMBO J. 18, 5103–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 18.Krogan, N. J., Kim, M., Ahn, S. H., Zhong, G., Kobor, M. S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S. & Greenblatt, J. F. (2002) Mol. Cell Biol. 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong, A. H. Y., Evangalista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W. V., Bussey, H., et al. (2001) Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 21.Peng, W. T., Robinson, M. D., Mnaimneh, S., Krogan, N. J., Cagney, G., Morris, Q., Davierwala, A. P., Grigull, J., Yang, X., Zhang, W., et al. (2003) Cell 113, 919–933. [DOI] [PubMed] [Google Scholar]
- 22.Clarke, A. S., Lowell, J. E., Jacobson, S. J. & Pillus, L. (1999) Mol. Cell. Biol. 19, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Masson, I., Yu, D. Y., Jensen, K., Chevalier, A., Courbeyrette, R., Boulard, Y., Smith, M. M. & Mann, C. (2003) Mol. Cell. Biol. 23, 6086–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshland, D. & Hieter, P. (1987) Methods Enzymol. 155, 351–372. [DOI] [PubMed] [Google Scholar]
- 25.Hyland, K., Kingsbury, J., Koshland, D. & Hieter, P. (1999) J. Cell Biol. 145, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baetz, K. K., Krogan, N. J., Emili, A., Greenblatt, J. F. & Hieter, P. (2004) Mol. Cell. Biol. 24, 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. & Nakatani, Y. (2000) Cell 102, 463–473. [DOI] [PubMed] [Google Scholar]
- 28.Cai, Y., Jin, L., Tomomori-Sato, C., Sato, S., Sorokina, I., Parmely, T. J., Conaway, R. C. & Conaway, J. W. (2003) J. Biol. Chem. 278, 42733–42736. [DOI] [PubMed] [Google Scholar]
- 29.Doyon, Y., Selleck, W., Lane, W. S., Tan, S. & Cote, J. (2004) Mol. Cell. Biol. 24, 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gadal, O., Labarre, S., Boschiero, C. & Thuriaux, P. (2002) EMBO J. 21, 5498–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, J. L., Iyer, V. R., Brown, P. O. & Struhl, K. (2000) Mol. Cell 6, 1297–1307. [DOI] [PubMed] [Google Scholar]
- 32.Hampsey, M. & Reinberg, D. (2003) Cell 113, 429–432. [DOI] [PubMed] [Google Scholar]
- 33.Krogan, N. J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M. A., Dean, K., Ryan, O. W., Golshani, A., Johnston, M., et al. (2003) Mol. Cell 11, 721–729. [DOI] [PubMed] [Google Scholar]
- 34.Ng, H. H., Robert, F., Young, R. A. & Struhl, K. (2003) Mol. Cell 11, 709–719. [DOI] [PubMed] [Google Scholar]
- 35.Dover, J., Schneider, J., Tawiah-Boateng, M. A., Wood, A., Dean, K., Johnston, M. & Shilatifard, A. (2002) J. Biol. Chem. 277, 28368–28371. [DOI] [PubMed] [Google Scholar]
- 36.Sun, Z. W. & Allis, C. D. (2002) Nature 418, 104–108. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa, H., Schneider, R., Bernstein, B. E., Karabetsou, N., Morillon, A., Weise, C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2003) Mol. Cell 12, 1325–1332. [DOI] [PubMed] [Google Scholar]
- 38.Choy, J. S., Tobe, B. T., Huh, J. H. & Kron, S. J. (2001) J. Biol. Chem. 276, 43653–43662. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Garcia, A. B., Sendra, R., Galiana, M., Pamblanco, M., Perez-Ortin, J. E. & Tordera, V. (1998) J. Biol. Chem. 273, 12599–12605. [DOI] [PubMed] [Google Scholar]
- 40.Mcainsh, A. D., Tytell, J. D. & Sorger, P. K. (2003) Annu. Rev. Cell Dev. Biol. 19, 519–539. [DOI] [PubMed] [Google Scholar]
- 41.Measday, V. & Hieter, P. (2004) Nat. Cell Biol. 6, 94–95. [DOI] [PubMed] [Google Scholar]
- 42.Stoler, S., Keith, K. C., Curnick, K. E. & Fitzgerald-Hayes, M. (1995) Genes Dev. 9, 573–586. [DOI] [PubMed] [Google Scholar]
- 43.Strunnikov, A. V., Kingsbury, J. & Koshland, D. (1995) J. Cell Biol. 128, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doheny, K. V., Sorger, P. K., Hyman, A. A., Tugenderich, S., Spencer, F. & Hieter, P. (1993) Cell 73, 761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortiz, J., Stemmann, O., Rank, S. & Lechner, J. (1999) Genes Dev. 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillett, E. S., Espelin, C. W. & Sorger, P. K. (2004) J. Cell Biol. 164, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krishnan, R., Pangilinian, F., Lee, C. & Spencer, F. (2000) Genetics 156, 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming, J. A., Vega, L. R. & Solomon, F. (2000) Genetics 156, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betz, J. L., Chang, M., Washburn, T. M., Porter, S. E., Mueller, C. L. & Jaehning, J. A. (2002) Mol. Genet. Genomics 268, 272–285. [DOI] [PubMed] [Google Scholar]
- 50.Baker, R. E., Harris, K. & Zhang, K. (1998) Genetics 149, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carr, A. M., Dorrington, S. M., Hindley, J., Phear, G. A., Aves, S. J. & Nurse, P. (1994) Mol. Gen. Genet. 245, 628–635. [DOI] [PubMed] [Google Scholar]
- 52.Rangasamy, D., Greaves, I. & Tremethick, D. J. (2004) Nat. Struct. Mol. Biol. 11, 650–655. [DOI] [PubMed] [Google Scholar]
- 53.Ren, Q. & Gorovsky, M. A. (2001) Mol. Cell 7, 1329–1335. [DOI] [PubMed] [Google Scholar]
- 54.Ren, Q. & Gorovsky, M. A. (2003) Mol. Cell. Biol. 23, 2778–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp, J. A. & Kaufman, P. D. (2003) Curr. Top. Microbiol. Immunol. 274, 23–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enenkel, C., Blobel, G. & Rexach, M. (1995) J. Biol. Chem. 270, 16499–16502. [DOI] [PubMed] [Google Scholar]
- 57.Kim, M., Ahn, S. H., Krogan, N. J., Greenblatt, J. F. & Buratowski, S. (2004) EMBO J. 23, 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 59.Meluh, P. B., Yang, P., Glowczewski, L., Koshland, D. & Smith, M. M. (1998) Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- 60.Jiang, W., Lechner, J. & Carbon, J. (1993) J. Cell Biol. 121, 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meluh, P. B. & Koshland, D. (1997) Genes Dev. 11, 3410–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.