Abstract
The general transcription factor TATA-binding protein (TBP) is a key initiation factor involved in transcription by all three eukaryotic RNA polymerases. In addition, the related metazoan-specific TBP-like factor (TLF/TRF2) is essential for transcription of a distinct subset of genes. Here we characterize the vertebrate-specific TBP-like factor TBP2, using in vitro assays, in vivo antisense knockdown, and mRNA rescue experiments, as well as chromatin immunoprecipitation. We show that TBP2 is recruited to promoters in Xenopus oocytes in the absence of detectable TBP recruitment. Furthermore, TBP2 is essential for gastrulation and for the transcription of a subset of genes during Xenopus embryogenesis. In embryos, TBP2 protein is much less abundant than TBP, and moderate overexpression of TBP2 partially rescues an antisense knockdown of TBP levels and restores transcription of many TBP-dependent genes. TBP2 may be a TBP replacement factor in oocytes, whereas in embryos both TBP and TBP2 are required even though they exhibit partial redundancy and gene selectivity.
TATA-binding protein (TBP) is a key component of the general transcription machinery that is involved in transcription by all three eukaryotic RNA polymerases. TBP is essential in yeast, binds to a variety of TATA boxes, and is recruited to TATA-less promoters by means of protein–protein interactions (1, 2). In recent years, TBP-related factors (TRFs) have been identified in metazoans. These factors facilitate nonuniversal gene-selective roles of the transcription machinery (reviewed in refs. 3–5).
TRF1 was identified in Drosophila, where it plays a role in transcription of the tudor gene and tRNA genes (6–8). Vertebrate and lower metazoan genomes do not encode a TRF1 ortholog; however, the mosquito genome does, suggesting that TRF1 is an insect-specific factor. In contrast, another TBP-like factor (TLF, also referred to as TRF2) is found in all metazoan genomes examined (9). The domain organization of TLF varies between species. In vertebrates, TLF consists of a single domain that is distantly related to the core domain of TBP, which interacts with the TATA box. Because of critical differences in the core domain, TLF does not bind to canonical TATA boxes, but interacts with TFIIA and TFIIB (10–12). TLF is essential for the transcription of a subset of genes during early embryogenesis in worms, fish, and frogs, whereas in mice TLF is required for spermatogenesis but not embryogenesis (13–18). TBP itself is also required for early development, although there is significant TBP-independent embryonic transcription in Xenopus, zebrafish, and mice (15, 16, 19), some of which cannot be attributed to TLF (19).
Here we characterize TBP2, a recently identified, vertebrate-specific TBP paralog from Xenopus that constitutes the third member of the vertebrate TBP family. By chromatin association and transcription assays and by a loss-of-function approach in vivo, we show that TBP and TBP2 play both specialized and redundant roles in gene regulation in the early embryo.
Materials and Methods
Chromatin Immunoprecipitation (ChIP). ChIP analysis was performed essentially as described (20, 21). For oocytes, 0.2 ng of DNA of each plasmid (pHSVtk-CAT, pH2B-Luc, pBluescript-Gsc as control) was injected into the germinal vesicle (GV), and the oocytes were incubated for 24 h at 18°C in Modified Barth's solution (MBS) (22). Fifty oocytes per ChIP extract were fixed in MBS containing 1% formaldehyde for 20 min. Embryos (n = 300) were fixed in 1% formaldehyde for 15–60 min. The oocytes and embryos were washed in 125 mM Glycine (30 min) and twice in MBS or 25% Marc's modified Ringer's solution, respectively (22), homogenized in 2 ml of low-salt whole-cell extract buffer (23), sonicated six times for 10 sec on ice, and centrifuged, and supernatants were frozen. ChIP extract (100 μl) was diluted with 100 μl of IP buffer (50 mM Tris·HCl, pH 8/100 mM NaCl/2mM EDTA/1 mM DTT/1% Nonidet P-40, and protease inhibitors) and incubated with 1 μg of antibody for 2 h on ice; 8 μl of protein G Sepharose beads (Amersham Pharmacia) was added and incubated for 18 h on a rotating wheel at 4°C. The beads were subsequently washed in ChIP1 buffer (IP buffer plus 0.1% sodium deoxycholate), ChIP2 buffer (ChIP1 buffer with 500 mM NaCl final concentration), ChIP3 buffer (ChIP1 buffer with 250 mM LiCl), ChIP1 buffer, and TE buffer (10 mM Tris, pH 8/1 mM EDTA). The material was eluted in 1% SDS in sodium carbonate, digested with proteinase K at 65°C for 6 h, phenol extracted, and precipitated, and the recovery of specific DNA sequences was determined by quantitative PCR using SYBR Green PCR core reagents (Applied Biosystems) and an Applied Biosystems GeneAmp 5700 PCR machine. Anti-TBP antibodies were as described (24). Mouse monoclonal antibodies against the Xenopus TBP2 N-terminal domain were generated by A & G Pharmaceutical (Columbia, MD).
Constructs. The Xenopus TBP2 cDNA (GenBank accession no. AW638001) was PCR amplified (primers 5′-GCTGTACCCGGGAATTCCCTCAAAATGGATGGAGAG-3′ and 5′-GTTGCACCCGGGTCTGTCTTAATGGAGGGACTTATG-3′), digested with SmaI, and cloned into the EcoRV site of pT7TS (25). Capped TBP2 mRNA was transcribed by an in vitro RNA synthesis kit (Ambion, Austin, TX). Four point mutations (caatgacAccTtaTgaCga, mutations in capitals) were introduced in the TBP2 construct by using a site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. The Xenopus TBP construct (pSP64A-xTBP) and the promoter plasmids (pH2B-Luc; pG5-HSV-tk-CAT) have been described (23, 26).
In Vitro Assays. In vitro transcription using Xenopus egg extracts has been described (23). Electrophoretic mobility shift assays were as described in refs. 27 and 28.
Modified Oligonucleotides. The TBP2 antisense N,N-diethylethylenediamine (DEED)-modified oligonucleotide (TBP2-AS167) 5′ -T+C+A+T+C+GTATGGA+G+T+C+A+T-3′ (DEED-modified linkages indicated with “+”) was synthesized as described (29). Between 1 and 3 ng of DEED-modified oligonucleotide was injected per embryo. The sequence of the TBP antisense morpholino-modified oligonucleotide (TBP-AS, Gene Tools, Philomath, OR) is as follows: 5′-CAAAGAAGACTCTCCATCCATTTTG-3′. Between 15 and 25 ng of TBP-AS was injected per embryo.
Northern Blotting. RNA was isolated by using Trizol (Invitrogen) extraction and LiCl precipitation. Northern blot analysis was performed with three embryo-equivalents (≈20 μg) of total RNA per sample, Hybond N-plus (Amersham Pharmacia) membranes, and Hybrisol I (Chemicon International) as blocking and hybridization solution. Probes were made by random primed labeling.
Western Blotting. Oocyte and embryo extracts were prepared as described (23). The antibodies used were anti-TBP 58C9 and anti-TFIIB C18 (both from Santa Cruz Biotechnology). Western blotting signals were detected by using peroxidase-conjugated secondary antibodies (DAKO) and enhanced chemiluminescence (Amersham Pharmacia).
Results
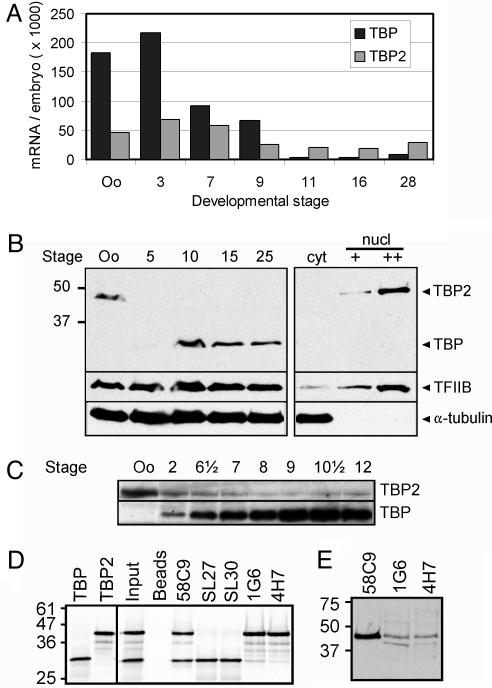
TBP2 Is a Basal Transcription Factor Highly Abundant in Oocyte Nuclei. By comparative EST and genomic database analysis, we identified a cDNA encoding a previously unknown TBP paralog from Xenopus laevis, here referred to as TBP2. This factor exists in human, rat, mouse, zebrafish, pufferfish, and Xenopus, but was not found in Ciona, Drosophila, Anopheles, Caenorhabditis elegans, or unicellular eukaryotes (unpublished data; see refs. 30 and 31). This vertebrate-specific TBP paralog is 95% identical to TBP in the core domain, whereas the N-terminal domains of TBP and TBP2 are much less conserved, showing 26% identity. We quantified the expression levels of TBP2 mRNA during Xenopus development and compared these to the levels of TBP mRNA by quantitative RT-PCR (qRT-PCR). TBP2 mRNA was found at similar levels in oocytes and at all stages of embryogenesis examined (Fig. 1A), whereas TBP mRNA levels varied significantly as observed before (23).
Fig. 1.
Characterization of TBP2. (A) Quantitation of TBP and TBP2 mRNA levels in oocytes and embryos by qRT-PCR, expressed as molecules per oocyte or embryo. (B) Western blot analysis of TBP and TBP2 expression (antibody 58C9). TBP2 protein is abundant in oocytes where it localizes to the nucleus, as does the TFIIB transcription factor. TBP is more abundant during embryogenesis, but low levels of TBP2 protein persist during embryogenesis (see C). Oo, stage VI oocyte, numbers refer to Nieuwkoop–Faber stages of development; Cyt, oocyte cytoplasm; nucl, oocyte nucleus (GV). (C) Long exposure of a Western blot reveals low levels of TBP2 persisting during embryogenesis (antibody 58C9). (D) Immunoprecipitation experiments using reticulocyte lysates programmed with TBP or TBP2 mRNA in the presence of [35S]methionine (first two lanes). Input, 20% of the amount of mixed TBP and TBP2 lysate used in immunoprecipitations. Beads, no-antibody control (nonspecific interaction with beads). 58C9 precipitates both TBP and TBP2, and SL27 and SL30 selectively precipitate TBP, whereas 1G6 and 4H7 preferentially precipitate TBP2. (E) Western blot analysis using oocyte GV extracts. 58C9, 1G6, and 4H7 all bind to TBP2.
Because we knew that TBP mRNA is subject to translational regulation, we examined the protein expression profiles of TBP and TBP2. Using 58C9, a monoclonal antibody that recognizes the core domain of TBP, we detected a protein with an apparent molecular mass of ≈45 kDa in oocyte whole cell and nuclear extracts (Fig. 1B), whereas TBP (33 kDa) was detected in embryonic extracts (Fig. 1B, stages 10–25). TBP accumulates during early embryogenesis as a result of regulated translation of the maternal mRNA (23). Higher exposure of a 58C9 Western blot showed relatively low levels of the 45-kDa protein in embryos (Fig. 1C). Using overexpressed TBP2 and reticulocyte lysates programmed with TBP2 mRNA, we determined that 58C9 binds to TBP and TBP2 equally well, and that TBP2 exhibits the same electrophoretic mobility on SDS/PAGE as the endogenous 45-kDa protein (Fig. 1D and data not shown). We raised antibodies against the TBP2 N-terminal domain, and two of these (1G6 and 4H7) recognized the oocyte 45-kDa protein on Western blots (Fig. 1E). These antibodies also bound with high affinity to TBP2 in immunoprecipitation experiments and were not reactive with TBP (Fig. 1D). Collectively, these experiments indicate that the endogenous 45-kDa protein is TBP2, that it is relatively abundant in oocytes where it localizes to the nucleus, and that it is expressed in embryos at lower levels (Fig. 1 B and C).
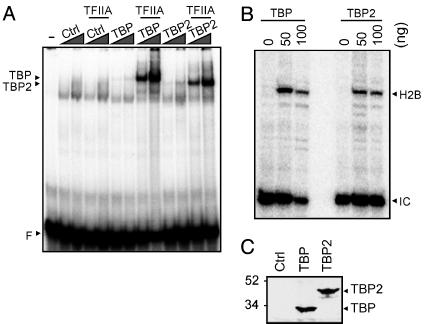
Oocytes are transcriptionally active, but meiotic maturation (release from a prophase of meiosis I arrest) is accompanied by transcriptional repression. As a result, unfertilized eggs (arrested in metaphase of meiosis II) and cleavage stage embryos are transcriptionally quiescent; the onset of embryonic transcription occurs at the mid-blastula transition (MBT) (reviewed in ref. 32). Interestingly, TBP and TBP2 are not abundant in transcriptionally quiescent eggs and cleavage stage embryos (Fig. 1C and data not shown), in contrast to transcriptionally active oocytes and post-MBT embryos, where TBP2 or TBP, respectively, are abundant (Fig. 1B). This finding raised the question whether TBP2 is an oocyte replacement factor for TBP. This would require similar biochemical properties of these proteins. Because of the extensive sequence homology between TBP and TBP2 in the core domain, which in the case of TBP is known to bind to the TATA box, we predicted that TBP2 would be capable of binding to the TATA box. This was found to be the case (Fig. 2A). TBP does not bind to the TATA box efficiently by itself; however, binding of TBP to the TATA box is stabilized by the basal transcription factor TFIIA under gel shift conditions (33). Similarly, TFIIA stabilized TBP2 binding to the TATA box (Fig. 2 A).
Fig. 2.
TBP2 is a TATA-binding basal transcription factor. (A) Gel-shift assay with control (Ctrl), TBP, and TBP2 in vitro translation lysates (2 and 6 μl, compare with C), with or without the addition of recombinant TFIIA (25 ng). F, free probe, Adenovirus Major Late promoter TATA box. (B) In vitro transcription in Xenopus egg extract with recombinant TBP or TBP2. Eggs (matured oocytes) are naturally devoid of TBP, so that TBP stimulates transcription >50-fold in egg extracts (23). H2B, correctly initiated transcript from the H2B promoter as detected by primer extension. IC, internal control for primer extension. (C) In vitro translation of TBP and TBP2 using reticulocyte lysates and [35S]methionine, showing that TBP and TBP2 were used in approximately equal amounts in the gel-shift experiment (A).
To assess its functional properties, we tested the ability of TBP2 to initiate transcription from the Xenopus histone H2B.1 promoter in an egg extract. We have shown previously that transcription in this system depends on exogenous TBP, which was explained by the low levels of TBP in oocytes, fertilized eggs, and cleavage stage embryos (23). We asked whether TBP2 could substitute for TBP in this developmentally relevant in vitro system. We found that transcription initiated correctly from the H2B.1 promoter in a TBP- or TBP2-dependent manner (Fig. 2B). Therefore, TBP2 is a functional basal transcription factor that can bind to the TATA box in a similar fashion as TBP.
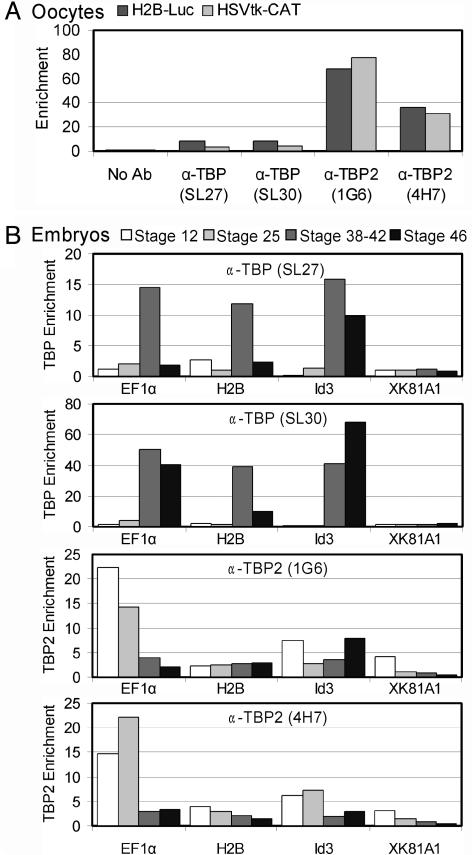
TBP2 Is Recruited to Promoters in Oocytes and Early Embryos. We examined whether TBP2 is recruited to promoters in vivo by ChIP using two TBP2-specific (1G6 and 4H7) and two TBP-specific (SL27 and SL30) monoclonal antibodies (Fig. 1D). We first examined recruitment of TBP2 to promoter constructs injected into the oocyte GV (oocyte nucleus). TBP2 rather than TBP associated with promoter DNA in oocytes (Fig. 3A), in line with the high abundance of TBP2 and low abundance of TBP in this cell (Fig. 1C). The TBP2 recruitment in oocytes was promoter-specific, because tests involving a coinjected promoter-less plasmid or a region 1,400 bp away from the promoter on the same plasmid gave signals close to background (data not shown). The TBP antibodies were effective in precipitating TBP-bound chromatin as judged from experiments using tadpole chromatin (Fig. 3B). We then repeated the promoter injection experiment in gastrula embryos, asking whether recruitment preferences might correlate with the relative abundance of TBP and TBP2 (Fig. 1). Strikingly, both TBP and TBP2 were recruited to the injected H2B.1 promoter in the gastrula, despite the low levels of TBP2 (data not shown).
Fig. 3.
Recruitment of TBP and TBP2 as assayed by ChIP. (A) TBP2 is recruited to promoters injected into oocyte nuclei. ChIP assay with two TBP antibodies (SL27, SL30) and two TBP2 antibodies (1G6, 4H7) on episomal chromatin isolated from GV-injected oocytes. The no-antibody control defines the nonspecific binding of chromatin to the protein G beads. Enrichment is the signal relative to background (no-antibody control). (B) Recruitment to endogenous promoters during embryogenesis. A switch in recruitment of TBP and TBP2 is observed at the EF1α promoter during development (stages 12, 24–28, 38–42, and 46).
We next tested recruitment of TBP and TBP2 to several endogenous loci in chromatin of different embryonic stages. A distinct difference was observed between gastrula (stages 12–13) and tailbud (stages 24–28) embryos as compared to tadpoles (stages 38–42 and 46), especially with respect to TBP recruitment. At gastrula and tailbud, 2- to 6-fold enrichment above background of both TBP and TBP2 was observed on the endogenous H2B.1, MyoD, and Id3 promoters, with higher enrichment of TBP2 at the elongation factor 1α (EF1α) promoter (Fig. 3B and data not shown). In tadpoles, TBP recruitment to the endogenous histone H2B.1, MyoD, and EF1α promoters was much higher (10- to 40-fold above background), whereas TBP2 remained similar or lower than in the early embryos. At all stages, the XK81A1 promoter showed low recruitment of either factor. These in vivo binding results can only partially be explained by factor abundance: although TBP is much more abundant than TBP2 in tadpoles, explaining the results at this stage, the same is true in gastrula and tailbud embryos that nevertheless showed either similar recruitment of both factors or higher recruitment of TBP2. Recruitment at the EF1α promoter undergoes the greatest change: TBP2 binds at high levels in gastrula and tailbud stages and then decreases, whereas TBP recruitment was undetectable in gastrula and tailbud stages but increased dramatically in tadpoles (Fig. 3B). These data suggest a switch in basal transcription factor recruitment during development at several promoters, most prominently at the EF1α promoter. Notably, this switch does not occur at the transition from oogenesis to embryogenesis when the relative levels of TBP and TBP2 change drastically (Fig. 1 B and C), but rather in a gradual fashion during subsequent development.
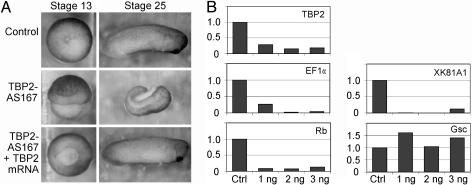
TBP2 Is Required for Embryonic Development and Transcription of a Subset of Genes. The gel shift and in vitro transcription data underlined the similarity between TBP2 and TBP, suggesting that these proteins are functionally redundant, at least in vitro (Fig. 2). Therefore, these in vitro data could not explain the functional significance of a transition in recruitment of TBP and TBP2 (Fig. 3). Because low levels of TBP2 persist during embryogenesis, it appeared possible that TBP2 might play a specialized role in development. To address this question, TBP2 was knocked down in vivo by using an antisense oligonucleotide-based approach, similar to what we have done for TBP and TLF (15). A chemically modified antisense oligonucleotide was used which contains a backbone of cationic DEED groups at each of the six most 5′ and 3′ linkages. These DEED-modified oligonucleotides are protected against exonucleases in vivo and can mediate RNaseH cleavage of the targeted mRNA (34). A TBP2 antisense oligonucleotide (TBP2-AS167) was injected into fertilized eggs, and the development of the embryos was monitored over a 48-h period. Injection of TBP2-AS167 significantly reduced the level of TBP2 mRNA (Fig. 4B), and caused abnormal gastrulation and failure of blastopore closure in 59% of the embryos (n = 174); the phenotype is not fully penetrant and varied between experiments (Fig. 4A and Table 1). When TBP2-AS167-injected embryos were able to complete gastrulation, they typically died at larval stages. To examine the specificity of the TBP2-AS167 antisense oligo, four silent mutations were introduced at third codon positions in the antisense target region of the TBP2 cDNA to produce mRNA that is resistant to TBP2-AS167-mediated cleavage. Coinjection of antisense-resistant TBP2 mRNA restored normal gastrulation to most embryos (76%, n = 174), supporting the view that the gastrulation phenotype resulted from a specific degradation of endogenous TBP2 mRNA (Fig. 4A and Table 1).
Fig. 4.
TBP2 is required for gastrulation and embryonic transcription. (A) Effects of TBP2 knockdown on development. (Top) Control embryos at gastrula and larval stages (stages 13 and 25, respectively). (Middle) Typical examples of embryos injected with 1 ng of TBP2-AS167 oligonucleotide at the same stages. (Bottom) Embryos injected with 1 ng of TBP2-AS167 and 0.05–0.2 ng of synthetic antisense-resistant TBP2 mRNA. (B) Expression analysis of TBP2 knockdown embryos by qRT-PCR. mRNA levels of TBP2, EF1α, Rb, XK81A1, and Goosecoid were determined in control embryos (Ctrl) and embryos injected with TBP2-AS167 oligonucleotide (1, 2, or 3 ng). Control embryos had reached stage 12.5 when samples were taken for RNA isolation.
Table 1. TBP2 is required for gastrulation.
| Experiment | Control (%) | TBP2-AS167 (%) | TBP2-AS167 + TBP2 mRNA (%) |
|---|---|---|---|
| 1 | 40 of 41 (98) | 0 of 33 (0) | 20 of 22 (91) |
| 2 | 50 of 50 (100) | 6 of 17 (35) | 21 of 40 (53) |
| 3 | 50 of 50 (100) | 3 of 9 (33) | 11 of 13 (85) |
| 4 | 100 of 100 (100) | 50 of 77 (65) | 60 of 70 (86) |
| 5 | 50 of 50 (100) | 12 of 38 (32) | 20 of 29 (69) |
| Total | 290 of 291 (100) | 71 of 174 (41) | 132 of 174 (76) |
Table indicates number and percentage of normally gastrulating embryos.
Gene expression in TBP2 knockdown embryos was analyzed by using qRT-PCR. Genes were selected for expression analysis on the basis of previous experiments (15), ChIP experiments (Fig. 3), and preliminary data obtained with a low density microarray (unpublished data). In TBP2-AS167-injected embryos the levels of mRNA encoding the Xenopus translation elongation factor EF1α, the retinoblastoma (Rb) tumor suppressor protein, and the XK81A1 keratin protein were significantly lower compared to control embryos (Fig. 4B). In contrast, Goosecoid (Gsc) expression was not affected by TBP2 knockdown. Because TBP2 is recruited to the EF1α promoter in vivo (Fig. 3B), and EF1α expression depends on normal TBP2 levels (Fig. 4B), transcription from the EF1α promoter is most likely mediated by an initiation complex containing TBP2. EF1α expression is also affected in TBP knockdown embryos (Fig. 5), presumably because of indirect effects. These data show that TBP2 is required for gastrulation and embryonic transcription of a subset of genes.
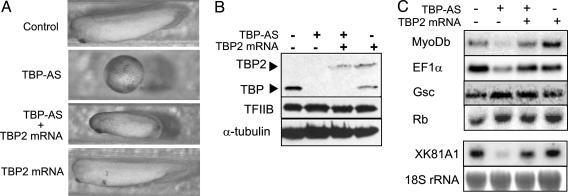
Fig. 5.
TBP2 can partly substitute for TBP function. (A) TBP2 overexpression partially rescues the TBP knockdown phenotype. Control embryos, TBP-AS injected embryos, embryos injected with both TBP-AS and TBP2 mRNA, and TBP2 overexpression embryos are shown at the time when control embryos reached stage 30. (B) Coinjection of TBP-AS and TBP2 mRNA replaces TBP with similar levels of TBP2. Western blotting of injected stage 30 embryos by using 58C9, a monoclonal antibody that recognizes the core domain of TBP and TBP2. Similar results were obtained when extracts from gastrula embryos (stage 12) were used. Antibodies against TFIIB and α-tubulin were used as loading control. (C) Northern blotting analysis using probes to determine expression of MyoDb, EF1α, Gsc, and XK81A1. The RNA used for this analysis was isolated from stage 12 embryos, except for the analysis of XK81A1 expression, in which case stage 18 embryos were used. Methylene blue-stained 18S rRNA was used as loading control.
TBP2 Can Partly Substitute for TBP Function. We have shown before that TBP is required for gastrulation and transcription of a subset of genes in the embryo (15). TBP knockdown embryos die before completing gastrulation, illustrating the important role of TBP in embryonic development. We asked whether the TBP-dependent genes identified in these experiments selectively require TBP, or whether transcription of these genes could be mediated by TBP2 as well; if the latter were correct, we would assume that the endogenous TBP2 levels are too low in TBP knockdown embryos to allow expression of these genes (Fig. 1 B and C). To address this question we depleted TBP from embryos and replaced it by similar levels of TBP2, by coinjecting TBP antisense oligonucleotide and TBP2 mRNA. In these experiments, we used a TBP Morpholino antisense oligonucleotide (TBP-AS) that has similar effects as the previously used DEED antisense oligonucleotide, except that the embryos arrested slightly later (Fig. 5A). Immunoblotting shows the depletion of TBP and the expression of TBP2 in the injected embryos (Fig. 5B). Injection of 20 ng of TBP-AS led to developmental arrest of 87% of the embryos (n = 128, Fig. 5A and Table 2). Strikingly, coinjection of 0.3 ng of TBP2 mRNA could overcome the developmental arrest so that the rescued embryos developed further, albeit at a slower rate (Fig. 5A); 85% of this group developed up to stage 23, then died at the time when control embryos had reached stage 30 (n = 103). Injection of TBP2 mRNA itself had no effect on development (Fig. 5A and Table 2).
Table 2. TBP2 can partly substitute for TBP function.
| Experiment | Control (%) | TBP-AS (%) | TBP-AS + TBP2 mRNA (%) | TBP2 mRNA (%) |
|---|---|---|---|---|
| 1 | 13 of 13 (100) | 5 of 55 (9) | 40 of 49 (81) | 57 of 57 (100) |
| 2 | 9 of 9 (100) | 4 of 30 (13) | 20 of 23 (87) | 28 of 28 (100) |
| 3 | 18 of 18 (100) | 7 of 43 (16) | 27 of 31 (87) | 32 of 32 (100) |
| Total | 40 of 40 (100) | 16 of 128 (13) | 87 of 103 (85) | 117 of 117 (100) |
Table indicates number and percentage of embryos developing further than stage 16. Water injection served as control.
To determine whether the phenotypes matched the transcriptional competence of these embryos, we analyzed their RNA by Northern blotting. MyoDb, EF1α, and XK81A1 are dependent on TBP for their expression in vivo (15) and EF1α and XK81A1 are also affected by TBP2 knockdown (Fig. 4B). The expression of these genes in TBP-depleted embryos could be restored by TBP2 mRNA injection (Fig. 4C). The behavior of these genes is contrasted by Gsc, which was not influenced by TBP2-AS167 or TBP-AS (Figs. 4B and 5C), and by Rb, which is negatively affected by TBP2-AS167 but not TBP-AS (Figs. 3B, 4C, and 5B). These results demonstrate that TBP2 can replace TBP during early development of Xenopus for the expression of some but not all genes. The embryos do not survive under these conditions, suggesting that compensation of reduced levels of TBP by increased levels of TBP2 is not sufficient for normal development and that TBP and TBP2 are not fully interchangeable during embryogenesis.
Discussion
Here, we have reported on the function of TBP2, a unique, vertebrate-specific TBP paralog. Recently, this factor, also known as TRF3, was shown to be expressed in many mammalian cell lines and tissues, and was found to play a role during zebrafish embryonic development (30, 31). Here we show that this TATA-binding basal transcription factor is highly abundant in Xenopus oocytes but is also present in embryos at relatively low levels (Fig. 1). It is specifically recruited to promoters in oocytes and embryos (Fig. 3) and plays an essential, specialized role in embryonic gene regulation as shown by antisense knockdown experiments (Fig. 4). The successful execution of gastrulation depends on TBP2, and so does transcription of genes like EF1α, Rb, and XK81A1, even though relatively high levels of TBP are present (Fig. 1). Furthermore, EF1α appears to be a target gene of TBP2, because this factor was found at relatively high levels at the EF1α promoter during early development (Fig. 3). Another major conclusion is that there is substantial functional redundancy between TBP and TBP2 in the embryo. TBP2 can substitute for TBP in the transcription of several genes, and TBP2 overexpression partially rescues the developmental arrest caused by a TBP knockdown (Fig. 5). The rescue is incomplete, suggesting that some TBP-dependent embryonic genes cannot be transcribed by TBP2, or that the functional redundancy between TBP and TBP2 is restricted to early development. We have observed gene-selective roles of TBP and TBP2 in gene expression (Figs. 4 and 5; see ref. 15), as well as developmental stage specificity in expression and promoter recruitment of TBP and TBP2 (Figs. 1 and 3), highlighting their specialized roles in development. These specialized roles may reduce the constraints on gene regulation in the organism if transcription is regulated by a combination of gene-specific transcription factors and different basal transcription factors.
There have been a number of reports on TBP-independent transcription. In some cases, the TBP-independent transcription could be attributed to the function of TLF (15, 16), the more distantly related TBP family member found in all metazoans. However, in mouse TBP knockout embryos, RNA polymerase II transcription was detected in the absence of detectable TLF expression (19). It has been speculated that this TBP-independent transcription involves TFTC, a complex containing several TBP-associated factors, but no TBP. This complex is capable of directing transcription from a core promoter in vitro in the absence of TBP (35). TFTC is similar to the yeast Spt3-Ada-Gcn5-acetyltransferase (SAGA) complex and contains histone acetyl transferase (HAT) activity implicated in transcriptional activation (36). TFTC may be involved in facilitating TBP-independent STAT2-mediated transcription during viral infection (37). Here, we identify an additional potential mechanism for TBP-independent transcription, in which transcription is facilitated by TBP2 rather than TBP, TLF, or TFTC.
The major differences between TBP and TBP2 are in the N-terminal domain. The function of this domain in TBP has remained enigmatic. It shows a fair amount of sequence homology between vertebrate TBP genes, suggesting an important function in gene regulation. This domain affects both the binding of the core domain to the TATA box (38) and the recruitment of TBP to a U6 snRNA promoter in vitro (39). Deletion of the TBP N terminus from the mouse genome uncovered a role in vivo, although there is no dramatic transcriptional phenotype associated with this deletion (40, 41). Our data on the redundant and specialized roles of TBP and TBP2 in embryonic transcription further highlight the pivotal role of the core domain in transcription initiation, whereas the N-terminal domain may play a more subtle role that is important for only a subset of genes. Future work will need to examine differential protein–protein interactions and the cellular and developmental pathways affected by these basal transcription factors. Our studies contribute to the view that the basal transcription machinery consists of general factors and nonuniversal, variable basal transcription factors. Different combinations of general and variable basal transcription factors are required for correct initiation of transcription at different promoters during development.
Acknowledgments
We thank Henk Stunnenberg and Huiqing Zhou (Department of Molecular Biology, Radboud University Nijmegen, Nijmegen, The Netherlands) for recombinant TFIIA and antibodies. This work was supported financially by the KNAW, the Royal Netherlands Academy of Arts and Sciences, and NWO–ALW, the Netherlands Organization for Scientific Research–Research Council for Earth and Life Sciences.
Abbreviations: TBP, TATA-binding protein; TRF, TBP-related factor; TLF, TBP-like factor; ChIP, chromatin immunoprecipitation; GV, germinal vesicle; DEED, N,N-diethylethylenediamine; qRT-PCR, quantitative RT-PCR; EF1α, elongation factor 1α.
References
- 1.Hernandez, N. (1993) Genes Dev. 7, 1291–1308. [DOI] [PubMed] [Google Scholar]
- 2.Roeder, R. G. (1996) Trends Biochem. Sci. 21, 327–335. [PubMed] [Google Scholar]
- 3.Veenstra, G. J. C. & Wolffe, A. P. (2001) Trends Biochem. Sci. 26, 665–671. [DOI] [PubMed] [Google Scholar]
- 4.Hochheimer, A. & Tjian, R. (2003) Genes Dev. 17, 1309–1320. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, I. (2003) Trends Biochem. Sci. 28, 391–398. [DOI] [PubMed] [Google Scholar]
- 6.Hansen, S. K., Takada, S., Jacobson, R. H., Lis, J. T. & Tjian, R. (1997) Cell 91, 71–83. [DOI] [PubMed] [Google Scholar]
- 7.Takada, S., Lis, J. T., Zhou, S. & Tjian, R. (2000) Cell 101, 459–469. [DOI] [PubMed] [Google Scholar]
- 8.Holmes, M. C. & Tjian, R. (2000) Science 288, 867–870. [DOI] [PubMed] [Google Scholar]
- 9.Dantonel, J. C., Wurtz, J. M., Poch, O., Moras, D. & Tora, L. (1999) Trends Biochem. Sci. 24, 335–339. [DOI] [PubMed] [Google Scholar]
- 10.Rabenstein, M. D., Zhou, S., Lis, J. T. & Tjian, R. (1999) Proc. Natl. Acad. Sci. USA 96, 4791–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teichmann, M., Wang, Z., Martinez, E., Tjernberg, A., Zhang, D., Vollmer, F., Chait, B. T. & Roeder, R. G. (1999) Proc. Natl. Acad. Sci. USA 96, 13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore, P. A., Ozer, J., Salunek, M., Jan, G., Zerby, D., Campbell, S. & Lieberman, P. M. (1999) Mol. Cell. Biol. 19, 7610–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaltenbach, L., Horner, M. A., Rothman, J. H. & Mango, S. E. (2000) Mol. Cell 6, 705–713. [DOI] [PubMed] [Google Scholar]
- 14.Dantonel, J. C., Quintin, S., Lakatos, L., Labouesse, M. & Tora, L. (2000) Mol. Cell 6, 715–722. [DOI] [PubMed] [Google Scholar]
- 15.Veenstra, G. J. C., Weeks, D. L. & Wolffe, A. P. (2000) Science 290, 2312–2235. [DOI] [PubMed] [Google Scholar]
- 16.Muller, F., Lakatos, L., Dantonel, J., Strahle, U. & Tora, L. (2001) Curr. Biol. 11, 282–287. [DOI] [PubMed] [Google Scholar]
- 17.Martianov, I., Fimia, G.-M., Dierich, A., Parvinen, M., Sassone-Corsi, P. & Davidson, I. (2001) Mol. Cell 7, 509–515. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, D., Penttila, T.-L., Morris, P. L., Teichmann, M. & Roeder, R. G. (2001) Science 292, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 19.Martianov, I., Viville, S. & Davidson, I. (2002) Science 298, 1036–1039. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., Lin, Q., Yoon, H.-G., Huang, Z.-Q., Strahl, B. D., Allis, C. D. & Wong, J. (2002) Mol. Cell. Biol. 22, 5688–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rietveld, L. E., Caldenhoven, E. & Stunnenberg, H. G. (2002) EMBO J. 21, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, H. B. (1991) Methods Cell Biol. 36, 657–662. [PubMed] [Google Scholar]
- 23.Veenstra, G. J. C., Destrée, O. H. J. & Wolffe, A. P. (1999) Mol. Cell. Biol. 19, 7972–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruppert, S. M., McCulloch, V., Meyer, M., Bautista, C., Falkowski, M., Stunnenberg, H. G. & Hernandez, N. (1996) Hybridoma 15, 55–68. [DOI] [PubMed] [Google Scholar]
- 25.Zorn, A. M. & Krieg, P. A. (1997) Genes Dev. 11, 2176–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kass, S. U., Landsberger, N. & Wolffe, A. P. (1997) Curr. Biol. 7, 157–165. [DOI] [PubMed] [Google Scholar]
- 27.Ma, D., Watanabe, H., Mermelstein, F., Admon, A., Oguri, K., Sun, X., Wada, T., Imai, T., Shiroya, T., Reinberg, D., et al. (1993) Genes Dev. 7, 2246–2257. [DOI] [PubMed] [Google Scholar]
- 28.Mitsiou, D. J. & Stunnenberg, H. G. (2000) Mol. Cell 6, 527–537. [DOI] [PubMed] [Google Scholar]
- 29.Dagle, J. M., Littig, J. L., Sutherland, L. B. & Weeks, D. L. (2000) Nucleic Acids Res. 28, 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persengiev, S. P., Zhu, X., Dixit, B. L., Maston, G. A., Kittler, E. L. W. & Green, M. R. (2003) Proc. Natl. Acad. Sci. USA 100, 14887–14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartfai, R., Balduf, C., Hilton, T., Rathmann, Y., Hadzhiev, Y., Tora, L., Orban, L. & Muller, F. (2004) Curr. Biol. 14, 593–598. [DOI] [PubMed] [Google Scholar]
- 32.Veenstra, G. J. C. (2002) in Advances in Developmental Biology and Biochemistry: Gene Expression at the Beginning of Animal Development, ed. DePamphilis, M. L. (Elsevier, Amsterdam), Vol. 12.
- 33.Maldonado, E., Ha, I., Cortes, P., Weis, L. & Reinberg, D. (1990) Mol. Cell. Biol. 10, 6335–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dagle, J. M. & Weeks, D. L. (2001) Differentiation (Berlin) 69, 75–82. [DOI] [PubMed] [Google Scholar]
- 35.Wieczorek, E., Brand, M., Jacq, X. & Tora, L. (1998) Nature 393, 187–191. [DOI] [PubMed] [Google Scholar]
- 36.Brand, M., Moggs, J. G., Oulad-Abdelghani, M., Lejeune, F., Dilworth, F. J., Stevenin, J., Almouzni, G. & Tora, L. (2001) EMBO J. 20, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulson, M., Press, C., Smith, E., Tanese, N. & Levy, D. E. (2002) Nat. Cell Biol. 4, 140–147. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, X. & Herr, W. (2002) Cell. 108, 615–627. [DOI] [PubMed] [Google Scholar]
- 39.Mittal, V. & Hernandez, N. (1997) Science 275, 1136–1140. [DOI] [PubMed] [Google Scholar]
- 40.Hobbs, N. K., Bondareva, A. A., Barnett, S., Capecchi, M. R. & Schmidt, E. E. (2002) Cell 110, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, E. E., Bondareva, A. A., Radke, J. R. & Capecchi, M. R. (2003) J. Biol. Chem. 278, 6168–6174. [DOI] [PubMed] [Google Scholar]