Abstract
Common beans (Phaseolus vulgaris) comprise three major geographic genetic pools, one in Mexico, Central America, and Colombia, another in the southern Andes, and a third in Ecuador and northern Peru. Species Rhizobium etli is the predominant rhizobia found symbiotically associated with beans in the Americas. We have found polymorphism in the common nodulation gene nodC among R. etli strains from a wide range of geographical origins, which disclosed three nodC types. The different nodC alleles in American strains show varying predominance in their regional distributions in correlation with the centers of bean genetic diversification (BD centers). By cross-inoculating wild common beans from the three BD centers with soils from Mexico, Ecuador, Bolivia, and Northwestern Argentina, the R. etli populations from nodules originated from Mexican soil again showed allele predominance that was opposite to those originated from Bolivian and Argentinean soil, whereas populations from Ecuadorian soil were intermediate. These results also indicated that the preferential nodulation of beans by geographically related R. etli lineages was independent of the nodulating environment. Coinoculation of wild common beans from each of the three BD centers with an equicellular mixture of R. etli strains representative of the Mesoamerican and southern Andean lineages revealed a host-dependent distinct competitiveness: beans from the Mesoamerican genetic pool were almost exclusively nodulated by strains from their host region, whereas nodules of beans from the southern Andes were largely occupied by the geographically cognate R. etli lineages. These results suggest coevolution in the centers of host genetic diversification.
Keywords: Phaseolus vulgaris–rhizobia coevolution, Rhizobium etli lineages, symbiotic interaction, nodC gene
It is generally accepted that Phaseolus vulgaris L. (the common bean) is native to the Americas. The analysis of natural bean populations, based on diverse criteria such as morphological traits, electrophoresis, and genetic typing of phaseolin, isozymes, restriction fragment length polymorphism (RFLP), and amplified RFLP, has led to the proposal of two major distinguishable areas or centers of bean genetic diversification (BD centers): the Mesoamerican center (Mexico, Central America, and Colombia) and the Andean center in South America (Ecuador, Peru, and Argentina) (1–4). Analysis of the multigenic family coding for phaseolin led Kami et al. (5) to speculate that the ancestral wild P. vulgaris type may be represented by the wild common bean populations from Ecuador and northern Peru and to propose the occurrence of an intermediate gene pool. Compatible with this view, Geffroy et al. (6) more recently investigated anthracnose-resistance specificities in genotypes from the Mesoamerican and Andean gene pools and identified a gene cluster with both Andean and Mesoamerican host-resistance specificities, suggesting that the locus existed before the separation of the two major gene pools of P. vulgaris. However, their results indicated that further coevolution between the pathogen agent and bean resulted in a differentiation for resistance in the three centers of diversification of the host (6).
Soil bacteria of the genus Rhizobium are able to induce nitrogen-fixing symbiotic association with the roots of leguminous plants. P. vulgaris is a promiscuous legume that forms nodules with a diversity of rhizobial genotypes. To date, five Rhizobium species have been recognized as microsymbionts of P. vulgaris: Rhizobium etli bv. phaseoli, Rhizobium leguminosarum bv. phaseoli, Rhizobium tropici, Rhizobium gallicum, and Rhizobium giardini (7). R. etli bv. phaseoli is the predominant species found associated with both wild and cultivated common beans from Mexico, Colombia, and the southern Andes (7), leading to the proposal of coevolution between bean host and rhizobia, although this has not been fully demonstrated and remains a matter of conjecture (8, 9).
Because common beans have become an important crop world-wide, the dissemination of microorganisms harbored by the plants and seeds during the process of export to new regions may have occurred as well. Thus, R. etli is found in regions where beans have been introduced since as early as the 16th century, after the discovery of the Americas. R. etli has been isolated from nodulated, cultivated beans in Spain, France, Austria, Kenya, Tunisia, and Indonesia (7, 10–12).
Species R. etli predominates in nodules formed in wild common beans in northwestern Argentina (NWA); however, population diversity was revealed at the level of sampling, within plants and sites of collection, by using plasmid profile and DNA fingerprinting (13).
More recently, we found the common nodulation gene nodC to be polymorphic among R. etli populations from NWA (14). In this study, we have explored the usefulness of nodC typing as molecular marker to characterize R. etli populations of diverse geographical origin and have assessed its value as an indicator of coevolution between rhizobia and wild beans in the Americas. Toward this aim we first examined the distribution of nodC types among R. etli populations from NWA and also among R. etli strains collected in the three BD centers. Second, rhizobial populations retrieved from soil samples from the same three BD centers by using wild and cultivated common beans as trapping host were nodC-typed to characterize native bean rhizobia. Finally, a set of wild and cultivated accessions representative of the Mesoamerican and Andean groups was analyzed in coinoculation assays for interactions with strains representative of the nodC types found among R. etli isolates.
Methods
Rhizobial Strains. Rhizobia examined in this work were isolated from nodules of wild common beans growing in NWA or were retrieved in the laboratory from field soils. From each plant sampled, three to six nodules were randomly excised and surface-sterilized with ethanol and hydrogen peroxide. Rhizobia were isolated axenically on YEM-Congo red agar medium as described by Vincent (15). Species assignment of bean isolates was done by both the 16S rRNA-encoding DNA–RFLP analysis and the nifH PCR screening procedure as described in ref. 13. Almost all isolates from wild and common bean varieties were found to belong to species R. etli; only very few were characterized as R. leguminosarum bv. phaseoli.
Plant Material. Seeds of natural populations of wild bean plants from NWA were provided by Roberto Neumann [Instituto Nacional de Tecnología Agropecuaria (INTA), Estación Experimental Agropecuaria Salta, Salta, Argentina]. Seeds of 30 wild bean accessions chosen to represent gene pools on the basis of previous results were provided by Daniel Debouck (Unidad de Recursos Genéticos, Centro Internacional de Agricultura Tropical, Cali, Colombia). Seeds of the black bean cultivar Nag12 and the white bean cultivar Alubia Cerrillos were provided by Susana García Medina (INTA, Estación Experimental Agropecuaria Salta), seeds of the Brazilian cultivar Aporé were provided by Mariangela Hungría (Empresa Brasileira de Pesquisa Agropecuaria-Soja, Londrina, Brazil), and seeds of the Mexican cultivar Negro Xamapa were provided by Esperanza Martínez-Romero (Centro de Fijación de Nitrógeno, Cuernavaca, Mexico).
DNA Isolation and RFLP Analysis of PCR-Amplified Symbiotic Genes. DNA from rhizobial cells grown in YEM medium was prepared with a rapid method as described by Alippi and Aguilar (16) and was used as a template in PCR reactions. nodC PCR amplification was performed as described in ref. 17. The primers used for nodA PCR amplification were deduced from the published R. etli nodA sequence (GenBank accession no. M58625), and reaction conditions were identical to those used for nodC PCR. Oligonucleotides nodAf (5′-CGGTGGAAAGTGTGCTGGCAAACGA-3′) and nodAr (5′-ATCGGGGAACCTTGATCGAGCGGAAT-3′) were used as forward and reverse primers, respectively, to amplify ≈560 bp of the 588 bp of the nodA gene. Aliquots of the PCR product were used for restriction pattern analysis with endonucleases AluI, MspI, BclI, HaeIII, DdeI, MboI, CfoI, RsaI, HinfI, and TaqI (nodA) and HinfI, CfoI, MspI, NdeI, RsaI, and HaeIII (nodC), respectively. The RFLP analysis was performed by electrophoresis in 2% agarose gel.
Plant Inoculation with Soil Samples. Soil isolates were recovered from nodules of wild and cultivated beans, which were grown in the laboratory after inoculation with soil suspensions prepared with soil samples. Soil samples were obtained from different field sites from Mexico, Ecuador, Bolivia, and NWA and were provided, respectively, by Jesús Caballero Mellado (Centro de Fijación de Nitrógeno), Gustavo Bernal (Instituto Nacional Autónomo de Investigaciones Agropecuarias, Estación Experimental Santa Catalina, Santa Catalina, Ecuador), Irene Christiansen (Programa Andino Extensivo en Cultivos Hortícola, Santa Cruz de la Sierra, Bolivia), and Mario Chocobar (INTA, Estación Experimental Agropecuaria Salta). One hundred grams of soil was suspended in 1 liter of sterile distilled water, and 8 ml of this suspension per plant was used for inoculation. Plant assays were performed with seeds that were surface-sterilized sequentially with 75% ethanol for 1 min and sodium hypochlorite for 6 min and finally washed with sterile water. Germinated seedlings inoculated with soil suspensions were grown in 300-ml plastic pots filled with sterilized vermiculite and were watered twice with N-free mineral nutrient solution (15) and with sterile distilled water as required.
Coinoculation Experiments. Cultures of R. etli strains showing nodC type α and type δ were grown separately to late exponential phase in YEM broth. Rhizobial mixtures of strains nodC type α and δ (ratio 1:1) were made, and 10 ml of this mixture, containing 108 bacteria, was applied per plant. Mixtures assayed were CFNX5 (isolated in Mexico) plus 55N1 (isolated in NWA), SC15 (isolated in NWA) plus 55N1, and SC15 plus 90N3 (isolated in NWA). At least two plants of each bean accession or cultivar were inoculated with each mixture. In each experiment, a set of plants of both Mesoamerican and south Andean origins was treated in parallel with the same inoculant mixture. Plants were incubated at 26–28°C in a day/night cycle of 16/8 h, watered as described above, and harvested ≈25 days after inoculation. All nodules from each plant were excised, or, alternatively, in cases of numerous nodulation, only nodules located in the upper part of the principal root were removed for further analysis. Nodules were individually surface-sterilized with 96% ethanol and hydrogen peroxide and were crushed, and rhizobia were axenically isolated in YEM-Congo red medium. About 25 nodules per plant and two plants from each bean variety were examined for nodC profile.
Statistics. All data were subjected to variance analysis with Student's t test (18).
Results
RFLP Analysis of PCR-Amplified Symbiotic Genes nodA and nodC in R. etli Isolates from NWA. One hundred and sixty-four R. etli isolates were obtained from nodules of 51 plants of P. vulgaris var. aborigineus collected from 16 different sites in NWA. These were found in virgin lands having no records of previous agricultural management. The common nodulation genes nodA and nodC of each isolate were amplified and analyzed by RFLP analysis. The nodA RFLP patterns were found to be identical in all of the isolates, as well as in the type strain R. etli CFN42. In contrast, RFLP analysis of the nodC PCR product resulted in three distinct patterns that, for convenience, were designated as nodC types α, γ, and δ (Fig. 1). In addition, another distinct nodC pattern was found only in isolates of wild common beans carrying the R. leguminosarum-like allele that codes for the 16S rRNA gene (pattern nodC type β in Fig. 1). These results were in agreement with those from our previous work on indigenous populations isolated from common bean nodules collected in NWA (14). The type strain R. etli CFN 42 showed nodC type α. Because the sole use of only endonuclease HinfI sufficed to yield the same four distinguishable nodC types, hereafter the designation “nodC types” will refer to the patterns resulting from HinfI restriction of the nodC PCR-amplified fragment.
Fig. 1.
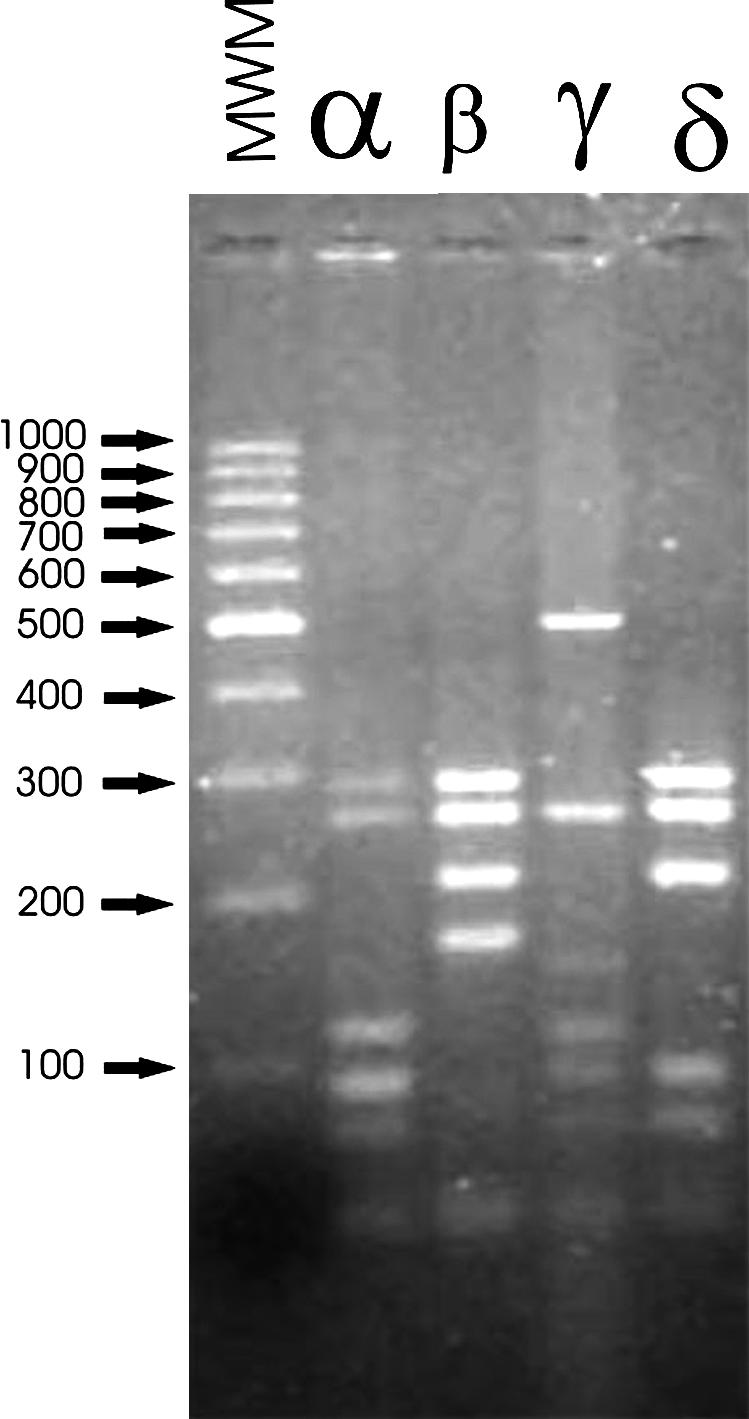
Representative RFLP patterns of nodC types of R. etli isolates from the Americas. Shown are the results of HinfL digestion of PCR-amplified nodC gene. MWM, molecular weight marker (100 bp, Promega).
The three nodC types α, γ, and δ were detected among the R. etli isolates; however, type δ was found to be the most prevalent, occurring in 132 of 164 isolates examined, which represents a probability of 0.80 ± 0.05 at a P = 0.05 level of confidence. The remaining 32 isolates were distributed among types α (n = 24) and γ (n = 8). In our analysis of the nodC gene, we also assayed another collection of isolates of wild common beans from NWA that had been previously characterized by Aguilar et al. (13). Thirteen of 15 isolates having the R. etli 16S rRNA allele exhibited nodC type δ, whereas the other two isolates exhibited nodC type α. The four isolates of collection having the R. leguminosarum 16S rRNA allele exhibited types β (n = 1) and δ (n = 3) (data not shown). These results demonstrated that allele nodC type δ is highly represented among rhizobia isolated from wild common bean nodules of NWA.
Distribution of nodC Alleles in a Worldwide Collection of R. etli Strains. To determine the geographical distribution of nodC types, a collection of 94 R. etli strains from diverse geographical origins other than NWA was assayed. These strains are examples of populations from the United States (n = 5), Mexico (n = 21), Belize (n = 2), Colombia (n = 10), Ecuador (n = 16), Peru (n = 2), Brazil (n = 12), Spain (n = 21), Indonesia (n = 4), and Burundi (n = 1). Some of these strains had been characterized previously for their plasmid profile, DNA fingerprint, and symbiotic ability (8, 10, 12, 19–21). The results shown in Table 1 demonstrate that nodC types α, γ, and δ were represented in the collection, but, unlike the results of isolates from NWA shown in the preceding section, only eight strains showed nodC type δ, namely seven strains from Ecuador and one from Peru. nodC type α and type γ were detected in 27 (31%) and 57 (61%) strains, respectively. Therefore, if we consider these data together with the above results from isolates from NWA there is a clear difference in the distribution of nodC types in the Americas, where the strains that show type δ are found to cluster in the group of strains from Argentina, Ecuador, and Peru. Indeed, these strains had been isolated from the mid- and southern Andean centers of bean domestication. Conversely, all of the strains from the Mesoamerican pool showed only types α and γ. The same was true of strains from regions other than the Americas (Table 1).
Table 1. nodC typing of R. etli strains from different geographical origins.
| Strain | nodC type | Source* |
|---|---|---|
| American origin | ||
| USA | ||
| V23 | γ | P. Graham |
| V33 | γ | P. Graham |
| V36 | γ | P. Graham |
| Kim5S (CIAT7139) | γ | K. Josephson |
| Tal182 | α | E. Martínez-Romero |
| Mexico | ||
| F6 | α | E. Martínez-Romero |
| F8 | γ | E. Martínez-Romero |
| CFN1 | γ | E. Martínez-Romero |
| CFN42 | α | E. Martínez-Romero |
| IE950 | γ | C. Silva (25) |
| IE951 | α | C. Silva (25) |
| IE954 | γ | C. Silva (25) |
| IE994 | α | C. Silva (25) |
| IE2730 | α | C. Silva (25) |
| IE2737 | γ | C. Silva (25) |
| IE4804 | γ | C. Silva (25) |
| IE4810 | γ | C. Silva (25) |
| IE4837 | α | C. Silva (25) |
| CFNX5 | α | S. Brom (9) |
| FNJ-11 | γ | S. Brom (9) |
| UMR1307 | γ | P. Graham |
| UMR1310 | γ | P. Graham |
| UMR1311 | γ | P. Graham |
| UMR1313 | γ | P. Graham |
| LPM×101 | γ | This laboratory |
| LPM×112 | γ | This laboratory |
| Belize | ||
| Olivia 4 | γ | E. Martinez-Romero |
| Viking 1 | γ | K. Josephson |
| Colombia | ||
| CIAT894 | γ | S. Brom (9) |
| CIAT895 | γ | CIAT |
| CIAT903 | α | CIAT |
| CIAT7004 | γ | CIAT |
| CIAT7014 | α | CIAT |
| CIAT7106 | α | CIAT |
| 1002 | γ | P. Graham |
| 1640 | γ | P. Graham |
| Ecuador | ||
| 1373 | δ | P. Graham |
| 1374 | α | P. Graham |
| 1376 | δ | P. Graham (8) |
| 1377 | δ | P. Graham (8) |
| 1378 | γ | P. Graham (8) |
| 1383 | γ | P. Graham |
| 1385 | γ | P. Graham |
| Ec4A | δ | G. Bernal |
| Ec6A | δ | G. Bernal |
| Ec10I | γ | G. Bernal |
| Ec20A | δ | G. Bernal |
| Ec2L | γ | G. Bernal |
| Ec24A | δ | G. Bernal |
| 1468 | α | P. Graham (8) |
| 1478 | α | P. Graham (8) |
| 1481 | γ | G. Bernal |
| Peru | ||
| P-1 | δ | P. Lezama |
| P-4 | γ | P. Lezama |
| Brazil | ||
| Br5 | γ | E. Martínez-Romero |
| CNPAF512 | γ | J. Michiels (18) |
| MH3 | γ | M. Hungría |
| MH9 | α | M. Hungría |
| MH14 | γ | M. Hungría |
| MH16 | α | M. Hungría |
| MH19 | γ | M. Hungría |
| MH31 | γ | M. Hungría |
| MH37 | α | M. Hungría |
| MH39 | γ | M. Hungría |
| MH45 | α | M. Hungría |
| MH48 | γ | M. Hungría |
| Other origins | ||
| Spain | ||
| GR10 | γ | J. Sanjuan (9) |
| GR14 | α | J. Sanjuan (9) |
| GR87 | α | J. Sanjuan (9) |
| 8C3 | α | J. Sanjuan (9) |
| 21NJ2 | α | J. Sanjuan (9) |
| ISP-19 | γ | D. Rodriguez-Navarro (23) |
| ISP-27 | α | D. Rodriguez-Navarro (23) |
| ISP-34 | α | D. Rodriguez-Navarro (23) |
| ISP-37 | γ | D. Rodriguez-Navarro (23) |
| ISP-40 | γ | D. Rodriguez-Navarro (23) |
| ISP-41 | γ | D. Rodriguez-Navarro (23) |
| ISP-42 | α | D. Rodriguez-Navarro (23) |
| ISP-44 | γ | D. Rodriguez-Navarro (23) |
| ISP-45 | γ | D. Rodriguez-Navarro (23) |
| ISP-35 | γ | D. Rodriguez-Navarro (23) |
| ISP-38 | γ | D. Rodriguez-Navarro (23) |
| ISP-52 | α | D. Rodriguez-Navarro (23) |
| ISP-60 | γ | D. Rodriguez-Navarro (23) |
| ISP-64 | γ | D. Rodriguez-Navarro (23) |
| ISP-68 | γ | D. Rodriguez-Navarro (23) |
| ISP-69 | γ | D. Rodriguez-Navarro (23) |
| Indonesia | ||
| GD131 | γ | N. Amarger |
| GD164 | γ | N. Amarger |
| GD261 | γ | N. Amarger |
| CP91 | γ | N. Amarger |
| Burundi | ||
| BC15 | α | N. Amarger |
N. Amarger, Laboratoire de Microbiologie des Sols, Institut Nacional de la Recherche Agronomique, Dijon, France; K. Josephson, Department of Soil, Water and Environmental Science, University of Arizona, Tucson; P. Graham, Rhizobium Research Laboratory, University of Minnesota, St. Paul; E. Martínez-Romero, Centro sobre Fijación de Nitrógeno, UNAM, Cuernavaca, Mexico; G. Bernal, Instituto Nacional Autónomo de Investigaciones Agropecuarias, Estación Experimental Santa Catalina, Quito, Ecuador; M. Hungría, Centro Pesquisa Soja, Embrapa, Londrina, Brazil; P. Lezama, Universidad Privada Antenor Orrego, Trujillo, Perú; CIAT, Centro Internacional de Agricultura Tropical.
Distribution of nodC Alleles in Rhizobial Populations Retrieved from Latin American Soils. To assess whether the prevalence of each nodC type in the collection of R. etli strains isolated from the different BD centers actually reflects the rhizobial populations present in soils, we examined the isolates retrieved by wild and cultivated common beans from soil samples collected in Mexico, Ecuador, Bolivia, and NWA. A soil suspension from each site was used to cross-inoculate wild common beans representative from the Mesoamerican, the intermediate, and the Andean genetic pools, in all combinations. The Mesoamerican cultivar Negro Xamapa (G3645) was also included in this survey. These bean accessions have been characterized previously by other researchers for phaseolin type, RFLP, and amplified RFLP (1–5, 22, 23). Rhizobia that were isolated from nodules formed in plants from either the Mesoamerican or Andean genetic pools and inoculated with a soil suspension from Mexico showed nodC types α and γ (Table 2). In contrast to this, plants inoculated with soil suspensions from Bolivia and from NWA resulted in nodules occupied by mostly nodC type δ rhizobia and nodC types β and δ rhizobia, respectively (Table 2). Nodules from plants inoculated with the Ecuadorian soil samples showed the four nodC types. The geographic distribution of the nodC alleles among these soils and in the collection of strains examined in the preceding section show a close correlation.
Table 2. Results from nodC typing of R. etli isolates from soils of Mexico Ecuador, Bolivia, and Argentina retrieved with the indicated bean accession.
| Nodule occupancy by nodC type
|
||||||
|---|---|---|---|---|---|---|
| Origin of soil | Bean accession | n | α | β | γ | δ |
| Mexico | Mesoamerican | |||||
| Negro Xamapa | 48 | 30 | 0 | 18 | 0 | |
| G2771 | 20 | 20 | 0 | 0 | 0 | |
| G12866 | - | - | - | - | - | |
| Andean | ||||||
| G19890 | 56 | 39 | 0 | 17 | 0 | |
| G4474 | 48 | 36 | 0 | 12 | 0 | |
| G21245* | 38 | 35 | 0 | 3 | 0 | |
| G21197 | 51 | 23 | 0 | 28 | 0 | |
| Wild beans NWA | 38 | 13 | 0 | 25 | 0 | |
| Ecuador | Mesoamerican | |||||
| Negro Xamapa | 73 | 35 | 0 | 38 | 0 | |
| G2771 | - | - | - | - | - | |
| G12866 | 14 | 11 | 0 | 0 | 3 | |
| Andean | ||||||
| G19890 | 27 | 7 | 0 | 12 | 8 | |
| G4474 | 65 | 21 | 6 | 36 | 2 | |
| G21245* | - | - | - | - | - | |
| G21197 | 29 | 10 | 0 | 19 | 0 | |
| Wild beans NWA | - | - | - | - | - | |
| Bolivia | Mesoamerican | |||||
| Negro Xamapa | 26 | 1 | 0 | 0 | 25 | |
| G2771 | - | - | - | - | - | |
| G12866 | - | - | - | - | - | |
| Andean | ||||||
| G19890 | - | - | - | - | - | |
| G4474 | - | - | - | - | - | |
| G21245* | - | - | - | - | - | |
| G21197 | 17 | 0 | 0 | 0 | 17 | |
| Wild beans NWA | 35 | 0 | 0 | 0 | 35 | |
| NWA | Mesoamerican | |||||
| Negro Xamapa | 45 | 0 | 11 | 0 | 34 | |
| G2771 | 32 | 0 | 4 | 0 | 28 | |
| G12866 | - | - | - | - | - | |
| Andean | ||||||
| G19890 | 49 | 0 | 13 | 0 | 36 | |
| G4474 | - | - | - | - | - | |
| G21245* | - | - | - | - | - | |
| G21197 | 56 | 0 | 11 | 0 | 45 | |
| Wild beans NWA | 22 | 0 | 0 | 0 | 22 | |
NWA, northwest Argentina; -, not determined.
Representative of the intermediate gene pool (5).
We concluded that the bean rhizobial populations retrieved from Mesoamerican soils by bean plants were clearly different from those retrieved from southern Andes (Bolivia and NWA) soils. Two of the nodC alleles, namely β and δ, appear absent only from the Mesoamerican soils. In particular, the population from Ecuador seems to be intermediate; all four alleles so far identified are present. Thus, Ecuador appears as a center of divergence (or convergence) of lineages of the species R. etli. Overall, these data indicated the following. (i) Bean plant populations of different origins are able to form nodules with rhizobia of each nodC type that are found to be predominant in each region, therefore ruling out the possibility that plants from either Mesoamerican or Andean centers restrict nodulation by rhizobia present in soils from other regions. (ii) R. etli lineages appear to be differently represented in soils from the three major BD centers. (iii) The fact that, for each soil sample, all wild accessions and cultivars were inoculated with the same soil–rhizobial suspension and grown in the same physiological conditions ensured that, in each case, the differences in nodule occupation by the four R. etli lineages arose from intrinsic differences in the interactions between both symbionts, and these were more favorable when both had the same geographic origin, suggesting some mutual symbiotic selectivity.
Distribution of Prevalence Among Wild and Cultivated Common Bean Populations from Different Centers of Diversity. To examine the above advanced proposition that in each region the native host plant and R. etli have developed selectivity for each other, we carried out coinoculation experiments with wild and cultivated beans from the three BD centers. Six and 16 wild common bean accessions from the Mesoamerican and Andean pools, respectively, were assayed. In addition, six and two cultivars of Mesoamerican and Andean origin, respectively, were included in this analysis (Table 4, which is published as supporting information on the PNAS web site). Most of these materials had been previously typed for phaseolin (1–5, 22–24). The group of Andean wild accessions comprises representatives of the intermediate and Andean gene pools, respectively. Mixtures of two R. etli strains representing the nodC types α and δ were used for plant inoculation. nodC type α strains CFNX5 (isolated in Mexico) and SC15 (isolated in NWA) and nodC type δ strains 55N1 and 90N3 (both isolated in NWA) were assayed in these experiments.
Table 3 summarizes the survey of 460 and 1,021 isolates each obtained from a bean nodule removed from plants representing the Mesoamerican and the Andean bean genetic pools, respectively, which had been inoculated with a mix of strains SC15 and 55N1. nodC type α was by far the dominant pattern in isolates from the Mesoamerican wild and cultivated beans: nodC type δ was found in only 3% of nodules (n = 13) formed on six Mesoamerican accessions and six cultivars with Mesoamerican introgressions from Mexico, El Salvador, Guatemala, Colombia, Brazil, and Argentina. In contrast, the average proportion of nodules formed individually by strain nodC type δ was comparatively much higher on the 16 wild and 2 cultivated beans from the Andean region (37%). This survey showed also that 100% of nodules sampled from the Mexican accessions G2771 (n = 61) and Negro Xamapa (n = 76) and from the Colombian accessions G24651 (n = 32) and 24683 (n = 37) displayed the nodC type α pattern. The strain nodC type α was also found occupying nodules formed in beans from the Andean region, albeit at much lower rates. Rates of occupancy by nodC type δ were found to be significantly higher than the average in some of the wild accessions from the southern Andes regions, such as G21245 (70%), G10025 (60%), G24318 (50%), G19894 (50%), G23724 (44%), G23445B (43%), G23583 (40%), G23454B (40%), and G19890 (30%). Also, nodC allele δ was found to be significantly represented in nodules formed in the Chilean cultivated beans G4474 (23%) and the Argentinean cv. Alubia Cerrillos (65%). Similar results were obtained by coinoculating with mixtures of strains CFNX5 plus 55N1 and SC15 plus 90N3, respectively (data not shown). We concluded that beans from the Mesoamerican and Andean genetic pools were different in their quantitative preference for nodulation by each of the two lineages of R. etli examined. Expression of this host–strain interaction was quite strong in the case of the Mesoamerican beans.
Table 3. Results of coinoculation of beans from Mesoamerica and southern Andes using a mixture of nodC type α and type δ strains.
| Region | N* | Nodule occupancy by nodC type δ,† % |
|---|---|---|
| Mesoamerican | ||
| Mexico, El Salvador, Guatemala, and Colombia | 460 | 3 ± 2.4 |
| Andean | ||
| Ecuador, Peru, Bolivia, Chile, and Argentina | 1,021 | 37 ± 10 |
Number of nodules examined.
Range of means at P = 0.05 by Student's t test.
Discussion
R. etli is the predominant P. vulgaris-nodulating species in the Americas. Our search for intraspecific diversity within the R. etli populations found associated to wild common beans from the southern extreme of the Andean BD center led us to identify polymorphism in the nodC gene. The use of molecular markers such as phaseolin, isozymes, RFLP, and amplified RFLP have been instrumental in identifying the major Mesoamerican and Andean groups in P. vulgaris (4, 5, 24). Similarly, here we demonstrated that the RFLP analysis of the nodC gene proved to be useful as a molecular marker for the identification of R. etli lineages associated with each center of bean domestication. In this study we analyzed the R. etli populations from various geographical origins, mainly from American BD centers, and found that each nodC type has a markedly distinct geographical distribution. The discovery that one lineage characterized by nodC type δ that was prevalent among the populations isolated from NWA was also present among isolates from Ecuador and Peru prompted us to examine the soils, as well as wild and cultivated beans, from the regions that had been proposed as centers of bean diversity and domestication. The finding that R. etli populations retrieved from Mexican soils were clearly different from those retrieved from soils from Ecuador, Bolivia, and Argentina was one of the most significant features of this research, indicating that the distribution of R. etli lineages follows that of centers of bean diversity and domestication. R etli lineages characterized by the presence of nodC alleles types α and γ were found in the Mesoamerican soil sample from Mexico, whereas lineage nodC type δ was found in soils from Ecuador and from the southern Andes in Bolivia and Argentina. It is therefore interesting to observe that nodC allele δ is found geographically restricted to the southern Andes regions and Ecuador. The area of Ecuador is considered intermediate between that of the Mesoamerican and Andean bean gene pools (5). It is noteworthy that nodC alleles β and δ were also identified as the only nodC alleles found among the very limited number of bean-retrieved isolates carrying the R. leguminosarum bv. phaseoli 16S rRNA allele and that nodC type β was found tightly associated with the 16S rRNA allele of R. leguminosarum. Whether in R. leguminosarum the allele nodC type β evolved from the allele nodC type δ after lateral transfer from R. etli or, conversely, the nodC type δ arose in R. leguminosarum from the type β and afterward transferred to R. etli remains to be investigated.
To test whether lineages that were recovered at high frequency from each soil sample are more competitive for nodulation or are just better adapted to soil and numerically dominant, assays for nodulation competitiveness were performed with representative bean plants from the different genetic pools. Although several studies have used agriculturally cultivated beans (8, 26), our study was performed mostly with wild common beans, and its resulting data were quantitative and suitable for statistical analysis. We found accessions of wild as well as cultivated common beans that are preferentially nodulated by the lineage nodC type α, whereas other accessions nodulate preferentially with the lineage nodC type δ. Overall, the occupancy of the nodC type δ strain in nodules of Andean beans was significantly higher than in those of Mesoamerican beans; the reverse was true for nodC type α strains. Moreover, this result and the distribution and abundance of the two rhizobial types in soils of both regions were in agreement. Therefore, these results seem to reveal that a particular R. etli lineage is not only more abundant in soils from a given region but also has a certain affinity for plants collected in the same region. Together, these observations may result in a synergism in which, during the growth cycle of beans, the more competitive rhizobia will turn into the soil after nodule senescence and therefore contribute to an increase in their representation in soil. Furthermore, from the point of view of the plant and the microsymbiont as well, this result suggested the presence of genes in beans and in rhizobia that determine affinity between plant and rhizobia and also suggested that geographical isolation led to some specificity. It is remarkable that in these coinoculation experiments an almost exclusive nodule occupation by nodC type α occurs in the case of Mesoamerican plants. This observation suggests that the selectivity for nodC type α is a common trait among Mesoamerican bean populations. Because most of the accessions we have examined had been characterized previously, we were able to extend comparisons to other traits, such as the phaseolin type (4, 5, 22, 24). Accessions showing affinity for lineage nodC type α have mostly S and M variants of phaseolin, whereas those accessions having significant nodulation with lineage nodC type δ were characterized by having the T-, C-, Pa-, and I-type phaseolin. This result indicated that our observations still are valid for the variability present within each genetic pool. Analysis of R. etli populations from regions that were the target of bean dissemination, such as Spain, has shown nodC allele types α and γ. Gepts and Bliss (23) and Ocampo et al. (22) have examined Spanish bean cultivars and found that phaseolin type C was detected at the highest frequency, followed by type T, type S, and type H, indicating that most Spanish beans share the south Andean gene pool, therefore suggesting that Spanish bean domestication may have developed from materials introduced from the Andean zone. Assuming that rhizobia are indeed carried by seeds, it appears that only two types among the three lineages of R. etli we found in the Americas are currently detected in Spain: α and γ. It is possible that only those two lineages survived desiccation stress on seeds (27) or, alternatively, were successful in competition and adaptation to new environments.
Our results invite speculation that each of the major genetic pools of P. vulgaris and rhizobia coevolved independently of the others after geographic separation. Arguments in favor of this hypothesis are our data on the characterization of R. etli strains isolated from different regions of the Americas that show that the nodC type of rhizobia in populations from the Andean region is different from those isolated from the Mesoamerican region; they may represent evolutionarily distinct lineages. The high prevalence of nodC type δ among the R. etli isolates from wild common beans in NWA may reflect the environmental isolation in which the Argentinean wild bean populations are found (2).
By using analysis of DNA fingerprinting, Bernal and Graham (8) examined a collection of bean rhizobial isolates from Mexico and Ecuador, some of which are also investigated in this work, and found three clusters that corresponded to isolates from Mexico and the northern and southern Andean regions, respectively, providing additional evidence of the diverse genetic structures of R. etli populations in Latin America.
Our previous results, as well as data from other investigators, indicate that R. etli is the species most commonly associated with beans in the Americas; therefore, the presumed coevolution we propose here represents an event that might have taken place after P. vulgaris and Rhizobium spp. coevolved symbiotically toward species R. etli. The distinct organization of R. etli common nodulation genes, in which, unlike in most rhizobial species, nodA is separated from the nodBC genes (28), could be considered a molecular marker (“remains”) of the early evolutionary process of “gathering” specific symbiotic genes. Afterward, diversification of species R. etli in geographically separated and isolated areas might have occurred.
Coevolution between host plant and microorganisms has been shown in pathogenic interactions in beans and in rhizobia–legume interactions. Geffroy et al. (6) performed cross-inoculations between common bean plants and the fungal pathogen Colletotrichum lindemuthianum, which was collected in the three BD centers, and found that coevolution led to differentiation of host resistance in the three centers of domestication, with each plant population showing resistance against allopatric strains. A typical example of rhizobia–legume coevolution is that described by Lie et al. (29), in which the pea cultivar from Afghanistan is nodulated by rhizobia isolated from soils of the Middle East and Central Asia but not by the European R. leguminosarum strains. These two cases are examples of coevolution due to geographical isolation, which might also have been the situation in the common bean–rhizobia interaction. But what could be the basis for this host plant–rhizobia interaction? In the present work, the diverse lineages of R. etli have been defined according to their distinct RFLP profile of the nodC gene, actually due to the presence of the HinfI polymorphism. The nodC gene product is an N-acetyl-glucosaminyltransferase, which is involved in the first step in Nod-factor assembly (30, 31). We have examined the nucleotide sequence of nodC from strains that represent each lineage of R. etli and found that the polymorphism arises from single nucleotide transversions at positions within the HinfI recognition site that do not affect the translational reading frame and represent neutral amino acid changes (data not shown). Therefore, it is unlikely that this nucleotide polymorphism in nodC causes functional changes in the NodC protein or changes in the plant–rhizobia interaction. Rather, we believe that the nodC profile is a molecular marker associated with a certain affinity for beans from each BD center, the basis of which remains to be discovered. Interestingly, R. etli strains belonging to each of the lineages are able to nodulate beans from the different centers of diversity and domestication, arguing against a situation of restricted nodulation such as is found with certain soybeans, peas, and beans. For instance, Sinorhizobium fredii strain USDA191 forms nodules on the soybean cultivars McCall and Peking, but S. fredii strain USDA257 nodulates only cultivar Peking (32). The Afghanistan pea cultivar is resistant to nodulation by European R. leguminosarum strains but is efficiently nodulated by strains isolated from Israel (29). Further investigations demonstrated that the ability to acetylate the nodulation factor introduced into the European strains lifts the restriction and makes them able to nodulate cv. Afghanistan (33). In contrast to this, in our case the absence of absolute restriction limits to cross-nodulation is clearly marked by the effective nodulation of bean accessions by allopatric R. etli strains (nodC type δ strains nodulating Mesoamerican accessions, and its reverse, type α strains forming nodules with Andean accessions; Table 2), and the existence of strong sympatric competition effects (Table 3) demonstrates nodulation preference between lineages of R. etli and beans from the same region of diversity.
Although our results provide no evidence for the basis and mechanisms underlying this nodulation preference, we find them provocative. It would be of interest to determine whether features we found in this work are encoded by the symbiotic plasmid in each lineage, because it was shown that symbiotic genes in R. etli are borne in plasmids (20). It also will be of interest to pursue investigations of the bean genetic background involved in this trait in the plant. Overall, these findings might offer clues for applied aspects of the bean–rhizobia symbiosis, such as a search for strains with better chances to compete against indigenous strains in a program of bean inoculation.
Supplementary Material
Acknowledgments
We thank all the people who provided samples of soil, seeds, and strains and Gabriel Favelukes for his critical revision of the manuscript. O.M.A. is a member of and O.R. is the recipient of a training studentship from Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina.
Abbreviations: BD center, center of bean genetic diversification; NWA, northwestern Argentina; RFLP, restriction fragment length polymorphism.
References
- 1.Becerra Velásquez, V. L. & Gepts, P. (1994) Genome 37, 256–263. [DOI] [PubMed] [Google Scholar]
- 2.Beebe, S., Rengifo, J., Gaitan, E., Duque, M. C. & Tohme, J. (2001) Crop Sci. 41, 854–862. [Google Scholar]
- 3.Singh, S. P., Gepts, P. & Debouck, D. G. (1991) Econ. Bot. 45, 379–396. [Google Scholar]
- 4.Tohme, J., Gonzalez, D. O., Beebe, S. & Duque, M. C. (1996) Crop Sci. 36, 1375–1384. [Google Scholar]
- 5.Kami, J., Becerra Velásquez, V., Debouck, D. G. & Gepts, P. (1995) Proc. Natl. Acad. Sci. USA 92, 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geffroy, V. D., Sicard, D., de Oliveira, J. C. F., Sevignac, M. S., Cohen, S., Gepts, P., Neema, C., Langin, T. & Dron, M. (1999) Mol. Plant–Microbe Interact. 12, 774–784. [DOI] [PubMed] [Google Scholar]
- 7.Amarger, N. (2001) Adv. Agron. 73, 109–168. [Google Scholar]
- 8.Bernal, G. & Graham, P. H. (2001) Can. J. Microbiol. 47, 526–534. [DOI] [PubMed] [Google Scholar]
- 9.Montealegre, C. & Kipe-Nolt, J. (1994) Arch. Microbiol. 162, 352–356. [Google Scholar]
- 10.Rodríguez-Navarro, D. M., Buendía, A. M., Camacho, M., Lucas, M. M. & Santamaría, C. (2000) Soil Biol. Biochem. 32, 1601–1613. [Google Scholar]
- 11.Sessitsch, A., Hardarson, G., Akkermans, A. D. & de Vos, W. M. (1997) Mol. Ecol. 6, 601–608. [Google Scholar]
- 12.Silva, C., Vinuesa, P., Eguiarte, L. E., Martínez-Romero, E. & Souza, V. (2003) Appl. Environ. Microbiol. 69, 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilar, O. M., López, M. V., Riccillo, P. M., González, R. A., Pagano, M., Grasso, D. H., Pühler, A. & Favelukes, G. (1998) Appl. Environ. Microbiol. 64, 3520–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilar, O. M., López, M. V. & Riccillo, P. M. (2001) J. Biotechnol. 91, 181–188. [DOI] [PubMed] [Google Scholar]
- 15.Vincent, J. M. (1970) A Manual for the Practical Study of the Root-Nodule Bacteria (IBP Handbook No. 15) (Blackwell Scientific, Oxford).
- 16.Alippi, A. M. & Aguilar, O. M. (1998) J. Inverterbr. Pathol. 72, 21–27. [DOI] [PubMed] [Google Scholar]
- 17.Laguerre, G., Nour, S. M., Macheret, V., Sanjuan, J., Drouin, P. & Amarger, A. (2001) Microbiology 147, 981–993. [DOI] [PubMed] [Google Scholar]
- 18.Steel, R. G. D. & Torrie, J. H. (1990) Principles and Procedures of Statistics: A Biometrical Approach (McGraw–Hill, New York).
- 19.Amarger, N., Macheret, V. & Laguerre, G. (1997) Int. J. Syst. Bacteriol. 47, 996–1006. [DOI] [PubMed] [Google Scholar]
- 20.Brom, S., Girard, L., García-de los Santos, A., Sanjuán-Pinilla, J. M., Olivares, J. & Sanjuán, J. (2002) Appl. Environ. Microbiol. 69, 2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michiels, J., Van Soom, T., D'hooghe, I., Dombrecht, B., Benhassine, T., de Wilde, P. & Vanderleyden, J. (1998) J. Bacteriol. 180, 1729–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ocampo, C. H., Martin, J. P., Ortiz, J. M., Sánchez-Yélamo, M. D., Toro, O. & Debouck, D. G. (2002) Annu. Rep. Bean Improv. Coop. 45, 236–238. [Google Scholar]
- 23.Gepts, P. & Bliss, F. A. (1988) Econ. Bot. 42, 86–104. [Google Scholar]
- 24.Gepts, P. (1990) Econ. Bot. 44, 28–38. [Google Scholar]
- 25.Chaverra, M. H. & Graham, P. H. (1992) Crop Sci. 32, 1432–1436. [Google Scholar]
- 26.Graham, P. H., Rosas, J. C., Estevez de Jensen, C., Peralta, E., Tlusty, B., Acosta-Gallegos, J. & Arraes Pereira, P. A. (2003) Field Crops Res. 82, 179–192. [Google Scholar]
- 27.Pérez-Ramírez, N. O., Rogel, M. A., Wang, E., Castellanos, J. Z. & Martínez-Romero, E. (1998) FEMS Microbiol. Ecol. 26, 289–296. [Google Scholar]
- 28.Vázquez, M., Dávalos, A., de las Peñas, A., Sánchez, F. & Quinto, C. (1991) J. Bacteriol. 173, 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lie, T. A., Goktan, D., Egin, M., Pijnenborg, J. & Anlarsal, E. (1987) Plant Soil 100, 171–181. [Google Scholar]
- 30.Long, S. R. (2001) Plant Physiol. 125, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perret, X., Staehelin, C. & Broughton, W. J. (2000) Microbiol. Mol. Biol. Rev. 64, 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pueppke, S. G., Bolaños-Vasques, M. C., Werner, D., Bec-Forté, M.-P., Promé, J.-C. & Krishnan, H. B. (1998) Plant Physiol. 117, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firmin, J. L., Wilson, K. E., Carlson, R. W., Davies, A. E. & Downie, J. A. (1993) Mol. Microbiol. 10, 351–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


