Abstract
Study of the maize (Zea mays L.) somatic chromosomes (2n = 20) has been difficult because of a lack of distinguishing characteristics. To identify all maize chromosomes, a multicolor fluorescence in situ hybridization procedure was developed. The procedure uses tandemly repeated DNA sequences to generate a distinctive banding pattern for each of the 10 chromosomes. Fluorescence in situ hybridization screening trials of nonsubtracted or subtracted PCR libraries resulted in the isolation of microsatellite 1-26-2, subtelomeric 4-12-1, and 5S rRNA 2-3-3 clones. These three probes, plus centromeric satellite 4 (Cent4), centromeric satellite C (CentC), knob, nucleolus-organizing region (NOR), pMTY9ER telomere-associated sequence, and tandemly repeated DNA sequence 1 (TR-1) were used as a mixture for hybridization to root-tip chromosomes. All 10 chromosomes were identified by the banding and color patterns in the 14 examined lines. There was significant quantitative variation among lines for the knob, microsatellite, TR-1, and CentC signals. The same probe mixture identifies meiotic pachytene, late prophase I, and metaphase I chromosomes. The procedure could facilitate the study of chromosomal structure and behavior and be adapted for other plant species.
In maize (Zea mays L., 2n = 20) pachytene chromosomes have been used extensively for karyotyping and cytogenetic analyses. The first procedure to identify maize meiotic chromosomes was developed by McClintock (1), and the method was refined and detailed by Longley (2) and Rhoades (3) in the middle of the 20th century. Pachytene-stage karyotyping has contributed to maize genetics in numerous ways [i.e., constructing chromosome maps (4), examining the structure and behavior of chromosomal aberrations (5), discovering transposable elements (6), and developing A–A and B–A translocation series (7, 8)]. A detailed morphological pachytene chromosome map is available (4). However, the pachytene stage is a relatively short period, and only a small percentage of anthers carry this stage in the tassel as a whole. In this sense, the procedure is limited by the availability of the appropriate cell type. Thus, there would be advantages for the study of maize chromosomes if each of the 10 members of the karyotype could be identified in somatic cells. Such a system would permit the screening of many individuals in a short period. A root tip contains many dividing cells and the tissue is readily available. However, the identification of somatic chromosomes has been difficult because the highly condensed chromatin structure conceals the fine details that are used for chromosome identification at the pachytene stage, such as cytologically observable knobs, heterochromatic regions, arm ratios, and total chromosome length (5).
Recent development of fluorescence in situ hybridization (FISH) technology has provided improved karyotyping on both meiotic (9) and mitotic (10) cells in maize. However, because of the paucity of landmarks and the polymorphism of knobs among varieties (11), these procedures are effective only for specific lines. To build on this procedure for general use with different maize varieties, an increase in the number of probes was necessary. To this end, we attempted bacterial artificial chromosome (BAC) detection on maize chromosomes. However, without the precise level of blocking with unlabeled repetitive DNA, the FISH procedure tends to label all chromosomes nonspecifically because of the presence of retrotransposons in the probe. Comparative genomic hybridization (CGH) has proved to be effective in distinguishing mammalian chromosomes (12); however, our CGH trials using chromosome 2 and 4 trisomics failed, even after computerized data processing of the images. Of the maize genome, ≈85% consists of repeated sequences (13), making BAC and CGH detection difficult because of the low unique-gene density. Therefore, we sought repetitive sequences that are located at specific chromosomal regions that could be used for karyotyping. Screening was carried out on random PCR libraries to recover sequences that were used as FISH probes to identify the somatic chromosomes. These probes, coupled with changes in the chromosome-preparation procedure that improve fluorescent signal detection, allowed the development of a FISH karyotyping method that is effective on all tested maize lines.
Materials and Methods
Construction of a Random PCR Library. The FISH screen using a random PCR library. Genomic DNA was extracted from immature ears of maize inbred line Mo17 by using a urea-based extraction protocol. DNA was partially digested by DNase I (catalog no. 104–132, Roche Applied Science), and fragments in the size range of 0.5–1.5 kb were collected by using a repeated gel-shift procedure. An adapter sequence (EBH1F, 5′-AGAATTCGGATCCAAGCTTCTGGTTTGT-3′; and EBH1R+p, 5′-pACAAACCAGAAGCTTGGATCCGAA-3′) was ligated to the fragments, and the DNA was gel purified again to eliminate the low-molecular-weight DNA and adapter dimers. We suspended 1 μl of the DNA solution in 499 μl of molecular-grade DMSO (stored at room temperature; D-4818; Sigma), and the number of PCR amplifiable DNA fragments was determined by using the EBH1F primer. After dilution, an average of 48 PCR-amplifiable fragments were added to a 300-μl PCR solution (Qiagen, Valencia, CA; with use of nuclease-free water, Ambion, Austin, TX), and 3 μl of this solution was added to 96 PCR tubes (average of 0.5 fragments per tube). The fragments were then amplified by using a GeneAmp 9700 PCR machine (Applied Biosystems), and the PCR products that showed a single band on gel electrophoresis were reamplified by adding 20 μl of PCR solution to the respective PCR tube. The PCR product was then ethanol-precipitated, and the DNA was labeled with biotin-14-dATP (Invitrogen) by the nick-translation procedure (14). Without further purification, the probes were hybridized to maize Oh43 root-tip chromosome spreads, and FISH signals were detected by dichlorotriazinyl aminofluorescein (DTAF)–streptavidin system (Jackson ImmunoResearch) with or without the tyramide amplification (PerkinElmer Life Science), according to the manufacturer's instructions.
The FISH screen using a subtracted PCR library. During the initial FISH screening, we found that most of the PCR products that showed signals were homologous to previously identified highly repetitive sequences, such as knob (15), centromeric satellite C (CentC) (16), nucleolus-organizing region (NOR) (17), and retroelements heavily distributed over the maize genome. To eliminate these repeated sequences, the mixture of PCR products of the first screening and B repeat (18) were biotin-labeled by nick translation. Then, a 10-times-larger amount of biotin labeled DNA was added to the partially digested and adapter ligated B73 genomic DNA (containing four B chromosomes). A different adapter was used to avoid amplification of contaminants from the first screening (i.e., BEH2F, 5′-AGGATCCGAATTCAAGCTTGTCTTTG-3′; and BEH2R+p, 5′-pCAAAGACAAGCTTGAATTCGGA3-′). After denaturing and annealing, the DNA fragments were passed through a Vectrex Avidin D column (Vector Laboratories) according to the manufacturer's instructions. The column selectively binds DNA sequences that are annealed to the biotin labeled repeated DNA sequences. This subtractive process was repeated, and the second PCR library was constructed by using the same procedures as described above. The DNA sequences were screened by the tyramide-amplified FISH procedure on root-tip chromosome spreads of a B73 × Mo17 hybrid (containing B chromosomes). Among the DNA sequences exhibiting FISH signals, 50 fragments were selected for cloning into the pGem-T vector (Promega) based on potential usefulness for karyotyping and future work.
Preparation of Chromosome Spreads. For analysis, we used 12 commonly used maize inbred lines (A188, A632, B37, B55, B73, KYS, M14, Mo17, Oh43, stock6, W22, and W23) and two maize varieties [Black Mexican Sweet and an abnormal chromosome 10 line (K10) (19)]. Kernels were germinated at 30°C for 2–3 days. Excised root tips were treated with nitrous oxide gas (20) for 2 h. Treated root tips were fixed in ice-cold 90% acetic acid for 10 min and stored in 70% ethanol at –20°C until use. After washing in water on ice, the section containing dividing cells was dissected and digested in 1% pectolyase Y23 (ICN) and 2% cellulase Onozuka R-10 (Yakult Pharmaceutical, Tokyo) solution for 65 min at 37C (one section per tube with 20 μlofenzyme solution). After digestion, the root sections were washed in ice-cold distilled water and then washed in 100% ethanol two times briefly. The root sections were carefully broken by using a needle and vortexed at maximum speed in 100% ethanol for 30 sec at room temperature to separate cells from one another. The cells were collected at the bottom of the tube by centrifugation and resuspended in acetic acid/ethanol (9:1 dilution) solution. The cell suspension was dropped onto glass slides in a box lined with wet paper towels and dried slowly.
For meiotic chromosome preparations, immature tassels of inbred line Oh43 were fixed in ethanol/acetic acid (3:1 dilution) and stored at –20°C in 70% ethanol. Anthers at the pachytene, late prophase I, or metaphase I stages were selected under light microscopy by examining one anther stained with iron acetocarmine. The remaining anthers were digested in the enzymatic solution, and air-drying was performed in the same manner as described above for root-tip slides.
Probe-Mixture Constitution. The following nine repeated DNA sequences were used for karyotyping: coumarin-5-dUTP-labeled knob 180-bp sequence (40 ng/μl) (15), Oregon green-488–5-dUTP-labeled NOR-173 clone (0.2 ng/μl), fluorescein-12-dATP, fluorescein-12-dGTP, fluorescein-12-dCTP, fluorescein-12-dUTP-labeled subtelomeric 4-12-1 clone (40 ng/μl) (Table 1; GenBank accession no. CL569186), fluorescein-12-dUTP-labeled CentC (2 ng/μl) (16), fluorescein-12-dUTP and Texas red-5-dUTP-labeled 5S 2-3-3 clone (Table 1; GenBank accession no. CL569181) (18 ng/μl), Texas red-5-dUTP-labeled microsatellite 1-26-2 clone (Table 1) (6 ng/μl), Texas red-5-dUTP labeled centromeric satellite 4 (Cent4) (10 ng/μl) (21), Texas red-5-dATP, Texas red-5-dGTP, Texas red-5-dCTP, Texas red-5-dUTP-labeled pMTY9ER telomere-associated sequence (40 ng/μl) (22), and Cy5-dUTP-labeled tandemly repeated DNA sequence 1 (TR-1) (20 ng/μl) (23). These sequences were all labeled by the nick-translation procedure. The NOR-173 clone was obtained from a separate plasmid-based random cloning of maize genomic DNA (A.K., unpublished data, GenBank accession no. CL569243); sequencing results indicate that this clone is a 17S maize rRNA sequence. The TR-1 clone was obtained from TA cloning (pGem-T vector; Promega) of PCR products amplified by MR77-specific primers (24). The sequence MR77 (GenBank accession no. AF020266) reported by Chen et al. (24) is a TR-1 sequence (88% homology; GenBank accession no. AF071123) that was reported by Ananiev et al. (23).
Table 1. Cloned FISH positive DNA sequences separated by root-tip screening.
| FISH signals | Clone no. | Homology to known sequences |
|---|---|---|
| Chromosomes 1, 2, and 4 | 1-26-2, 4-12-4, 4-12-6, and 4-12-12 | Microsatellite TAG repeat |
| Chromosome 2 | 2-3-3 | 5S |
| Chromosomes 2, 4, and 6 | 4-12-7 | TR-1 |
| Centromere | 1-26-68, 4-12-16, 4-12-22, 6-9-26, 6-9-34, 12-29-1, and 12-29-6 | CRM or Cinful1 |
| Centromere diffuse | 4-12-18, 4-12-19, 6-9-1, 6-9-2, 6-9-3, 6-9-4, 6-9-5, 6-9-6, 6-9-7, 6-9-9, 6-9-10, 6-9-11, 6-9-12, 6-9-13, 6-9-14, 6-9-16, 6-9-17, 6-9-18, 6-9-19, 6-9-20, 6-9-21, 6-9-22, 6-9-23, 6-9-24, 6-9-25, 6-9-27, 6-9-28, 6-9-29, 6-9-30, 6-9-31, 6-9-36, and 12-2-7 | Various sequences, most are homologous to partial sequences of known maize BACs |
| Centromere diffuse, B chromosome long arm enriched | 4-12-20 and 4-12-21 | 4-12-20 is homologous to the maize BAC ZM16H10. 4-12-21 shows no homology to known sequences. |
| Uniform in A chromosome and less in B chromosome | 4-12-9 and 4-12-10 | Rire-2 and Huck-2 |
| Chromosome-specific subtelomeric | 4-12-1 | Gardiner's telomere associated sequence (22), Burr's subtelomere (28) |
Clones that showed signals at the positions of 180-bp knob, CentC, and NOR are not included.
After labeling, these probes were purified by column chromatography (BioGel P-60; Bio-Rad) to eliminate unincorporated dNTPs and then coprecipitated with autoclaved (20 min) salmon-sperm DNA (50 μg) and dried. The pellets were resuspended in 2× SSC (containing 1 mM EDTA; 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) solution and stored at –20°C. Each probe concentration for the karyotyping mixture was adjusted by increasing the amount of probes that show weaker signals or decreasing the probes that show stronger signals to capture the weakest signals with the more intense ones without interference.
FISH Procedure. After the cell spreads were dried on slides, they were UV-crosslinked for 2 min (total energy, 120 mJ/cm2) and fixed in 10% formaldehyde solution for 5 min. Slides were washed sequentially in water and 100% ethanol and then dried. At the center of the cell spreads, 3 μl of 2× SSC solution containing autoclaved salmon-sperm DNA (1 μg/μl) were dropped. After application of a mineral-oil-coated plastic coverslip, the slide preparation was denatured by being placed on a wet paper towel in an aluminum tray floating in boiling water (100°C) for 5 min. The slides were cooled immediately on a metal plate placed on ice. After removing the plastic coverslip, denatured (100°C, 5 min) and rapidly cooled probe mixture (5 μl, in 2× SSC/1 mM EDTA) was applied. After reapplication of the plastic coverslip, the slides were incubated at 55°C overnight in a humidity chamber containing 2× SSC soaked paper toweling. Slides were washed in 2× SSC for 20 min at 55°C. After a brief wash with PI buffer (0.2 M NaH2PO4, pH 7.8/0.1% Igepal CA-630; I-3021; Sigma), the slides were mounted with Vectashield mounting medium (Vector Laboratories) without counter stain.
Image Capture and Data Processing of FISH Images. Chromosome spreads were identified by using an oil lens (×25 magnification) and a triple band-pass filter of a Universal microscope (Zeiss). FISH images were captured by an Optronics MagnaFire charge-coupled device (CCD) camera and plan apo oil lenses (×100 objective for mitotic cells, and ×63 objective for meiotic cells). Four single channel (blue, green, red, and infrared) images were captured in 8-bit depth black and white and were later superimposed in photoshop 7.0 (Adobe Systems, San Jose, CA). Assignments for color of each channel after being superimposed were the same as the fluorochrome color used (blue, coumarin; green, fluorescein or Oregon green; and red, Texas red), except for Cy5, for which a white color was assigned. Computerized background subtraction and image-feature intensification were conducted by using the “curve” and “layer overlay” command in photoshop 7.0 to make the weak signals recognizable. Aligned- and paired-chromosome figures were generated by using the “cut and paste” and “rotation” commands in photoshop 7.0.
Results
A total of 1,000 DNA fragments in the initial PCR library were examined by the FISH procedure applied to root tips of the inbred line Oh43. Among them, 10% of the fragments showed signals at knobs, 2% showed signals at CentC regions (16), 2% showed signals at the NOR on chromosome 6 (17), 1.6% showed signals at positions on chromosomes 1, 2, and 4 (a microsatellite cluster), and 0.5% showed centromere-specific retrotransposon of maize-like (25) signals. Of the fragments, 75% labeled most of the length of all chromosomes after tyramide amplification or had a distinct enhancement in the regions around the centromeres in what we term a “centromere-diffuse” pattern. The remaining 10% did not show any signal. For the subtracted PCR library, 1,100 fragments were analyzed on the B73 × Mo17 hybrid (with one to two B chromosomes). Among them, 3.5% were present at knob positions, 1.6% showed signals at the NOR, 0.7% showed signals at CentC regions, 0.4% showed centromere patterns different from CentC, 0.3% were present at microsatellite positions, and 0.3% corresponded to TR-1 sites (chromosomes 4 and 6). In addition, one fragment was specific to chromosome 2 (2-3-3), one fragment showed subtelomeric signals (4-12-1), two fragments were more intense on the B than A chromosomes, and two fragments showed labeling along the length of the A chromosomes but with a diminished hybridization on the B; 65% showed nonspecific or centromere diffuse signals, and ≈30% were not detectable.
Among these FISH-positive fragments, 50 were cloned and sequenced for the potential use in karyotyping (Table 1) (GenBank accession nos. CL569181–CL569242). The four clones that showed chromosome 1-, 2-, and 4-specific patterns proved to be difficult to sequence; however, the partial results indicated that they are (TAG)n simple sequence repeats. Sequencing results of clone 2-3-3, which showed a chromosome 2-specific signal, determined this fragment to be part of a 5S rRNA gene (26). Most centromere signals that showed patterns different from those of CentC were homologous to centromere-specific retrotransposon of maize (25, 27) or Cinful1 retrotransposons. The 32 cloned centromere-diffuse sequences were related to various DNA sequences, including segments of maize BACs or retrotransposable elements. The chromosome-specific subtelomere sequence 4-12-1 was homologous to the maize subtelomere sequence (GenBank accession no. S46925) (28) or the pMTY7SC2 maize telomere-associated sequence (GenBank accession no. U39641) (22). We selected the microsatellite 1-26-2 clone, the 5S 2-3-3 clone, and the subtelomeric 4-12-1 clone for FISH karyotyping in combination with the following other repeated sequences: knob, TR-1, CentC, pMTY9ER telomere-associated sequence (22), Cent4, and NOR-173.
Distribution of Each Sequence. The chromosomes of the inbred lines Oh43 (29) and KYS (9, 10) have been characterized for the position of heterochromatic knobs, 5S sequence, size, arm ratio, and the presence of the secondary constriction (chromosome 6). Based on these and other articles that describe the distribution of the sequences of Cent4 (21) and TR-1 (23), the FISH signal distributions on Oh43 of the repeated sequences used were determined as follows (Fig. 1). Knob 180-bp repeat: 1S (small), 2L, 4L, 5L, 6SL, 7L, 8S (very small), and 9S; TR-1: 2L, 4L, and 6S; microsatellite 1-26-2 clone: 1L, 2SL, and 4S; CentC: present at all centromeres, and chromosomes 7 and 8 have the strongest signals; 5S 2-3-3 clone: 2L; Cent4: chromosome 4 primary constriction; NOR-173 clone: 6S; subtelomeric 4-12-1 clone: 2SL, 4SL, 5S, and 8L; and pMTY9ER telomere-associated sequence: 2SL, 3SL, 4SL, 5S, 7S, and 8L.
Fig. 1.
Probe localization on maize Oh43 root-tip chromosomes. (a) Microsatellite 1-26-2 clone (red), CentC (green), TR-1 (white), and knob 180-bp (blue) signals. (b) The 5S 2-3-3 clone (yellow), Cent4 (red), NOR-173 clone (green), and knob180-bp (blue) signals. (c) pMTY9ER telomere-associated sequence (red), subtelomeric 4-12-1 clone (green), and knob 180-bp (blue) signals. (d) The signals of all nine probes. (Scale bar, 10 μm.)
All somatic chromosomes showed distinctive staining patterns, and chromosome numbers were identified (Figs. 1 and 2). In meiotic cells of Oh43, all 10 chromosomes are identifiable by using the same hybridization mixture, which helped confirm the location of the hybridization sites. In the late prophase I stage, the chromosomes are well separated and all 10 chromosome pairs can be recognized (Fig. 3). At metaphase I, identification of all chromosome pairs is possible (Fig. 3).
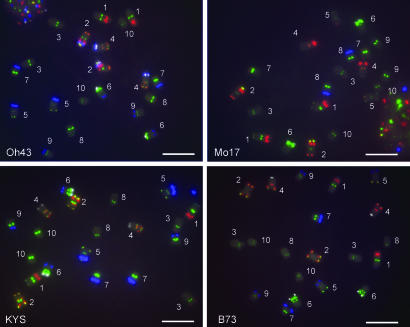
Fig. 2.
Somatic-chromosome identification in four maize inbred lines probed with the FISH mixture described in the text. (Scale bar, 10 μm.)
Fig. 3.
FISH signals on maize Oh43 meiotic cells. (Upper) Late prophase I. All 10 chromosome pairs are identifiable. (Lower) Metaphase I. Chromosomes are identifiable from the signal combinations. (Scale bar, 10 μm.)
To test the applicability of this system for identifying each chromosome in other varieties, many of the commonly used inbred lines were examined. In the root-tip chromosome spreads of 14 tested lines, distinguishing all 10 chromosomes was possible by using this multicolor FISH procedure (Fig. 4). The features of the each chromosome are as follows.
Fig. 4.
Somatic chromosome karyotyping of 14 maize lines probed with the FISH mixture. Knob 180-bp repeat (blue), 5S 2-3-3 (yellow, 2L), NOR-173 clone (green, 6S), CentC (green), subtelomeric 4-12-1 clone (green), Cent4 (red, 4C), microsatellite 1-26-2 clone (red), pMTY9ER telomere-associated sequence (red), and TR-1 (white). (Scale bar, 10 μm.)
Chromosome 1. The large microsatellite signals are at the middle of the long arm (red in Fig. 4). Very small knob signals (blue in Fig. 4) are at the tip of the short arm. It is the longest chromosome.
Chromosome 2. The 5S signals (yellow in Fig. 4) are at the long-arm tip. Microsatellite signals (red in Fig. 4) are present at the short-arm tip and middle of the long arm. TR-1 (white in Fig. 4) or knob (blue in Fig. 4) signals are sometimes present in the long arm.
Chromosome 3. The telomere-associated pMTY9ER signal (red in Fig. 4) is present at the ends of both chromosome arms, and 4-12-1 subtelomeric signals are absent.
Chromosome 4. The Cent4 signals (red in Fig. 4) are present at the primary constriction. Microsatellite signals (red in Fig. 4) are present in the short arm. Some lines carry knob (blue in Fig. 4) or TR-1 signals (white in Fig. 4) in the long arm.
Chromosome 5. The subtelomeric 4-12-1 signals (green in Fig. 4) are present on the tip of the short arm and knob (blue in Fig. 4) in the long arm. The arm ratio is ≈1:1.
Chromosome 6. The NOR signal (green in Fig. 4) is present in the short arm, and knob signals (blue in Fig. 4) are present at both chromosome ends, which are sometimes obscured by adjacent signals. TR-1 signals (white in Fig. 4) are present at the tip of short arm.
Chromosome 7. The telomere-associated pMTY9ER signals (red in Fig. 4) are present at the tip of the short arm. This chromosome tends to have larger CentC signals (green in Fig. 4) and large knob signals (blue in Fig. 4) in the long arm.
Chromosome 8. The 4-12-1 subtelomeric signals (green in Fig. 4) are at the tip of the long arm. This signal is invariant in the lines examined. The arm ratio is 1:3.
Chromosome 9. Knob signals (blue in Fig. 4) are always present at the tip of the short arm but vary in size.
Chromosome 10. This chromosome is the smallest. There are no landmarks on this chromosome, except for abnormal 10 in the K10 line.
There is significant variation for the presence and size of the repeated sequences among lines. The presence of knobs (blue in Fig. 4) is variable on chromosomes 2, 4, 5, and 8 among lines. TR-1 signals (white in Fig. 4) are variable on chromosomes 2, 4, and 6L. Chromosome 9 in stock 6 has a large TR-1 and knob hybridization in the short arm. Microsatellite signals (red in Fig. 4) are variable in the chromosome 6 centromere region and on the long or short arms of chromosome 5. The size of the CentC signals (green in Fig. 4) is different among chromosomes as well as among lines. All chromosomes of the 14 lines tested have a chromosome 9 short-arm knob, although the size of this knob in B37, BMS, K10, Mo17, and W23 is small. Chromosome 1 short-arm knobs are found consistently, except in BMS and Mo17. The 4-12-1 subtelomeric signals on the chromosome 8 long-arm tip are detected consistently in all of the tested lines. The telomere-associated pMTY9ER signals in the short arm of chromosome 7 are weak and variable. In A188, A632, B37, B73, W22, and W23, the 7S subtelomeric signals are undetectable. Also, these signals on chromosome 3 are weak and variable. Despite these variations, all somatic chromosomes are identifiable in the examined lines based on the conserved signals by using the current multicolor FISH technique.
Discussion
The FISH screening of the random PCR library proved to be effective for separating repeated sequences from the maize genome. Most of the major repeated sequences, knob, CentC, NOR, 5S, TR-1, centromere-specific retrotransposon of maize, microsatellite, and 4-12-1 subtelomeric repeats were separated during the screening. However, Cent4, telomere (30), and pMTY9ER telomere-associated sequences were not found. This result implies that there might be other remaining unknown repeated sequences that could be useful for maize karyotyping. Subtraction was effective in reducing the known repeated sequences from the population of the original PCR library. Judging from the number of PCR fragments that showed knob signals, the process eliminated 70–80% of these sequences.
The current system contains a sufficient number of probes to distinguish all chromosomes in commonly used lines of maize. It provides a baseline to which additional probes can be added. One possibility involves BACs, which are used as probes in other organisms, such as Arabidopsis thaliana (31), rice (Oryza sativa L.) (32), and sorghum [Sorghum bicolor (L.) Moench] (33), as well as sorghum BACs onto maize chromosomes (34). However, detection of maize BACs on chromosome spreads is difficult because of the low gene density (13). Other repeated sequences or tandem gene arrays may provide additional features that could aid in distinguishing chromosomes for the further refinement of the probe collection. The procedure could be adapted to other species by the isolation of a respective collection of repetitive DNA sequences.
The morphologies of chromosomes 2–4 resemble each other, as do chromosomes 7–9 (Fig. 4). Distinguishing these chromosomes cannot be achieved reliably without the use of unique banding patterns. Changes in chromosome arm length can be affected 5–20% by the polymorphism of knobs (Fig. 4) as well as chromosome-preparation procedures. The multicolor FISH procedure described here permits the distinction of these chromosomes.
Examination of the various inbred lines revealed significant variation for many of the repetitive gene arrays examined. The copy number of the respective sequences is apparently subject to change by mechanisms that are currently unknown. The probe collection described here provides the tools to examine this variability throughout other maize lines and the mechanism by which this variation arises.
For detailed structural analysis of chromosomes, the pachytene stage of meiosis is unparalleled because of the less condensed chromatin that permits high resolution of chromosomal features; however, the ease of observation at this stage is genotype-dependent (5). The technique described here is effective for identifying all maize somatic chromosomes in a wide variety of lines and allows such analysis in a genotype-independent manner. Somatic chromosomes are typically better spread, permitting greater ease of identification. Also, it is reasonable to design experiments involving hundreds of individuals that can be analyzed in a relatively short period. The use of root tips for karyotyping has great advantages because studies can be completed shortly after germination and individuals of interest can be retained and grown to maturity for future analysis. This system can be used for detection of chromosomal aberrations, determination of specific chromosomes involved in aneuploidy, detection of variation of repetitive sequences in the genome, analysis of chromosomal behavior in mitosis and meiosis, localization of large transgenes to chromosomal region, and many other applications for which chromosomal identification is useful.
Acknowledgments
We thank R. L. Phillips (University of Minnesota, Saint Paul) for providing the CentC clone; L. Dennis (CSIRO Plant Industry, Canberra, Australia) for the knob clone; J. Gardiner (University of Arizona, Tucson) for the pMTY9ER telomere-associated sequence. We also thank J. M. Vega (University of Missouri, Columbia) for providing B73 + 4 B plant tissue; E. H. Coe (University of Missouri, Columbia) for stock 6; S. Melia-Hancock (University of Missouri, Columbia) for inbred lines A632, B37, and W23; and J. Eta-ndu (University of Minnesota, Saint Paul) for A188; T. L. Phelps-Durr (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and J. L. Cooper (University of Washington, Seattle) for comments on the cloning procedure; T. Wako (National Institute of Agrobiological Sciences, Tsukuba, Japan) for information about the tyramide amplification system; and D. L. Auger (South Dakota State University, Brookings) and T. Ream (Washington University, Saint Louis) for discussions. This work was supported by the Monsanto Company and in part by National Science Foundation Grant DBI 9975827.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FISH, fluorescence in situ hybridization; BAC, bacterial artificial chromosome; Cent4, centromeric satellite 4; CentC, centromeric satellite C; NOR, nucleolus-organizing region; TR-1, tandemly repeated DNA sequence 1.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CL569181 and CL569243).
References
- 1.McClintock, B. (1930) Proc. Natl. Acad. Sci. USA 16, 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longley, A. E. (1939) J. Agric. Res. 59, 475–490. [Google Scholar]
- 3.Rhoades, M. M. (1950) J. Hered. 41, 58–67. [DOI] [PubMed] [Google Scholar]
- 4.Neuffer, G. M., Coe, E. H. & Wessler, S. (1997) in Mutants of Maize (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 32–53.
- 5.Carlson, W. R. (1988) in Corn and Corn Improvement, eds. Sprague, G. F. & Dudley, J. W. (Am. Soc. Agronomy, Crop Science Soc. America, and Soil Science Soc. America, Madison, WI), 3rd Ed., pp. 259–331.
- 6.McClintock, B. (1950) Proc. Natl. Acad. Sci. USA 36, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coe, E. H. (1993) in The Maize Handbook, eds. Freeling, M. & Walbot, V. (Springer, New York), pp. 364–376.
- 8.Beckett, J. B. (1978) J. Hered. 69, 27–36. [Google Scholar]
- 9.Chen, C. C., Chen, C. M., Hsu, F. C., Wang, C. J., Yang, J. T. & Kao, Y. Y. (2000) Theor. Appl. Genet. 101, 30–36. [Google Scholar]
- 10.Sadder, M. T. & Weber, G. (2001) Plant Mol. Biol. Rep. 19, 117–123. [Google Scholar]
- 11.Longley, A. E. & Kato, Y. T. A. (1965) Res. Bull. CIMMYT (Chapingo, Mexico) 1, 1–112. [Google Scholar]
- 12.Wells, D. & Levy, B. (2003) BioEssays 25, 289–300. [DOI] [PubMed] [Google Scholar]
- 13.Meyers, B. C., Tingey, S. V. & Morgante, M. (2001) Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegant, J., Verwoerd, N., Mascheretti, S., Bolk, M., Tanke, H. J. & Raap, A. K. (1996) J. Histochem. Cytochem. 44, 525–529. [DOI] [PubMed] [Google Scholar]
- 15.Peacock, W. J., Dennis, E. S., Rhoades, M. M. & Pryor, A. J. (1981) Proc. Natl. Acad. Sci. USA 78, 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananiev, E. V., Phillips, R. L. & Rines, H. W. (1998) Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein, N., Ponelies, N., Musket, T., McMullen, M. & Weber, G. (1998) Plant J. 13, 281–289. [Google Scholar]
- 18.Alfenito, M. R. & Birchler, J. A. (1993) Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhoades, M. M. & Dempsey, E. (1966) Genetics 53, 989–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, A. (1999) Biotech. Histochem. 74, 160–166. [DOI] [PubMed] [Google Scholar]
- 21.Page, B. T., Wanous, M. K. & Birchler, J. A. (2001) Genetics 159, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardiner, J. M., Coe, E. H. & Chao, S. (1996) Genome 39, 736–748. [DOI] [PubMed] [Google Scholar]
- 23.Ananiev, E. V., Phillips, R. L. & Rines, H. W. (1998) Proc. Natl. Acad. Sci. USA 95, 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, C., Yan, H., Zhai, W., Zhu, L. & Sun, J. (2000) Genome 43, 181–184. [PubMed] [Google Scholar]
- 25.Zhong, C. X., Marshall, J. B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J. A., Jiang, J. M. & Dawe, R. K. (2002) Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, Y. B., Burner, D. M. & Legendre, B. L. (2000) Genetica 108, 285–295. [DOI] [PubMed] [Google Scholar]
- 27.Nagaki, K., Song, J., Stupar, R. M., Parokonny, A. S., Yuan, Q., Ouyang, S., Liu, J., Hsiao, J., Jones, K. M., Dawe, R. K., Buell, C. R. & Jiang, J. (2003) Genetics 163, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burr, B., Burr, F. A., Matz, E. C. & Romero-Severson, J. (1992) Plant Cell 4, 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakeda, K., Yamagata, H., Fukui, K., Ohno, M., Fukui, K., Wei, Z. Z. & Zhu, F. S. (1990) Theor. Appl. Genet. 80, 265–272. [DOI] [PubMed] [Google Scholar]
- 30.Cox, A. V., Bennett, S. T., Parokonny, A. S., Kenton, A., Callimassia, M. A. & Bennett, M. D. (1993) Ann. Bot. 72, 239–247. [Google Scholar]
- 31.Lysak, M. A., Pecinka, A. & Schubert, I. (2003) Chromosome Res. 11, 195–204. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, J., Gill, B. S., Wang, G. L., Ronald, P. C. & Ward, D. C. (1995) Proc. Natl. Acad. Sci. USA 92, 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Islam-Faridi, M. N., Childs, K. L., Klein, P. E., Hodnett, G., Menz, M. A., Klein, R. R., Rooney, W. L., Mullet, J. E., Stelly, D. M. & Price, H. J. (2002) Genetics 161, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koumbaris, G. L. & Bass, H. W. (2003) Plant J. 35, 647–659. [DOI] [PubMed] [Google Scholar]