Abstract
The cAMP-response element-binding protein (CREB) transcription factor was initially identified as a mediator of cAMP-induced gene expression. CREB binds to a target sequence termed the cAMP-response element (CRE) found in many cellular and viral gene promoters. One of the best-characterized CREs resides in the promoter of the gene encoding the neuropeptide somatostatin, and this element has served as a model for studies of CREB function. Phosphorylation of CREB by protein kinase A allows recruitment of the coactivator CREB-binding protein (CBP). A central tenet of the CREB–CBP model is that CREB binds constitutively to the CRE and that regulation occurs through the phosphorylation-dependent recruitment of CBP. In this report, we use chromatin immunoprecipitation assays to show that CREB does not interact in vivo with the somatostatin CRE, or similar elements in several other genes, in PC12 cells, a standard model for studies of CREB function. Rather, CREB binding in vivo is regulated in a cell-specific manner, a finding that was confirmed by using in vivo genomic footprinting assays. The CREs in other genes were also found to interact differentially with CREB in PC12 cells, hepatoma cells, and cortical neurons. We conclude that the family of CREB target genes differs from one cell type to another and that the ability of CREB to bind to a particular CRE represents an important component of gene regulation.
The transcription factor cAMP-response element-binding protein (CREB) was discovered through its ability to bind to the cAMP-response element (CRE) (TGACGTCA) in the promoter of the neuropeptide gene, somatostatin (1–3). The regulation of somatostatin gene expression by the cAMP-protein kinase A pathway, in concert with the presence of a consensus protein kinase A phosphorylation site within the CREB activation domain (4), led to the suggestion that CREB mediates the cAMP-induced activation of somatostatin gene expression. A similar involvement of CREB has been proposed for a multitude of other cellular and viral genes (5, 6). In fact, recent studies have predicted that there may be as many as 10,000 CREB-binding sites in the mammalian genome (7). In part because of its simplicity, the cAMP-CREB model is regarded as one of the best-characterized transcriptional signaling pathways. CREB is phosphorylated at the same critical serine residue, Ser-133, by a variety of kinases in addition to protein kinase A (6). Phosphorylation at Ser-133 is essential for the recruitment of the transcriptional coactivator, CREB-binding protein (CBP) (8, 9), and this association, the first well characterized phosphoserine-dependent protein–protein interaction, has been described in atomic detail (10). CBP and its paralogue, p300, are believed to activate gene expression by interacting with components of the general transcriptional machinery (11–13) and by acetylating chromatin (14–16). Thus, the selection of which genes are activated is determined by CREB, and activation, per se, is determined, for the most part, by CBP. Because of its ability to serve as a target for multiple kinase pathways, CREB has been implicated in a large number of biological processes, including long-term neuronal plasticity, cell survival, circadian rhythms, adaptation to drugs, and hormonal regulation of metabolism (6).
The CREB-CBP pathway can be broken down into two distinct steps: binding of CREB to the CRE sequences in target genes and recruitment of the CBP coactivator. The first step, CREB binding, is believed to be constitutive, and the second step, gene activation, is thought to occur through the regulated recruitment of CBP (5). The idea that CREB binds constitutively stems primarily from in vitro binding assays by using purified DNA. Gel-mobility shift assays, for example, show that phosphorylation does not affect the interaction of CREB with naked DNA (17), although this point has been controversial (18). More sensitive fluorescence anisotropy binding assays show that CREB binds to the consensus CRE at high affinity (≈2 nM) and that variant CREs bind only slightly weaker (19). In these fluorescence studies, phosphorylation also did not affect CREB binding (19). In reporter assays, CREB seems to activate most CRE-containing promoters in a similar manner (4), supporting the idea that CREB binding to its targets in various gene promoters is not regulated and that the CREs in most genes are occupied by CREB under basal conditions.
Nonetheless, other studies have suggested mechanisms that could conceivably regulate CREB binding in the context of specific CREs (5). Potential regulatory factors include magnesium, which is required for CREB interaction with consensus but not variant CREs (20), and associated proteins, such as the Tax protein of the human T cell leukemia virus (21–23), which cooperates with CREB to allow binding to the Tax-responsive element in the human T cell leukemia virus-1 long-terminal repeat. Additionally, the existence of a CpG at the center of the CRE makes this element ideally suited for regulation by DNA methylation. Indeed, studies have shown that methylation of the central CpG prevents CREB binding in vitro and in vivo (24–26). X-ray crystallographic analyses suggest that methylation of the central CpG sterically hinders interaction with arginine 301 in the CREB DNA-binding domain (27) and reduces the flexibility required for optimal CREB interaction (28).
Probably most relevant, however, are studies examining whether CREB binding is regulated in vivo. In experiments performed over a decade ago, Schutz and coworkers (29) found that protein kinase A signaling regulated occupancy of a variant CRE sequence in the tyrosine aminotransferase promoter. These studies used an in vivo genomic footprinting assay, however, which by itself cannot identify the particular factor involved in binding. At least in vitro, CRE sequences can interact with many transcription factors in addition to CREB, so it is possible that the regulated binding involves a different protein. Wolfl et al. (30) showed that CREB binding to the corticotropin-releasing hormone promoter was similarly regulated by cAMP signaling. This study was also performed before the advent of chromatin immunoprecipitation (ChIP) assays, but clearly showed regulated CREB occupancy at a variant CRE site. Most recently, Strauss and coworkers (31) used ChIP assays to determine that CREB binding to the steroidogenic acute regulatory protein promoter was increased by cAMP signaling in adrenal cells.
In this study, we used both ChIP and in vivo genomic footprinting assays to explore whether CREB occupies the CRE sequences of target promoters in different cell types under basal and stimulated conditions. Our studies suggest that in contrast to the currently accepted model, CREB binding varies in a cell type-specific manner. Thus, the family of CREB targets differs from one cell type to another. Interestingly, the best-studied CREB target, the somatostatin CRE, does not interact with CREB in PC12 cells, where it has been so extensively studied.
Materials and Methods
Cell Culture. PC12 cells were grown in DMEM with 10% neonatal calf serum and 5% FBS plus 100 units/ml penicillin and 100 units/ml streptomycin (pen/strep). H4IIE rat hepatoma cells (American Type Culture Collection) were grown in DMEM (American Type Culture Collection) supplemented with 10% FBS (HyClone) and pen/strep (Invitrogen). Cortical neurons were cultured from postnatal day 1–2 rats and plated at a density of 4 × 105 cells per well onto 10-cm plates. Cells were maintained in Neurobasal A medium (Invitrogen) supplemented with 1% B27 (Invitrogen) and 0.5% FBS, 35 mM glucose, 1 mM l-glutamine, and pen/strep, 5–7 days before experimentation. PC12 and H4IIE cells were cultured under serum-free conditions overnight before treatment with 10 μM forskolin (Sigma) for 15 min.
ChIP. Cells were fixed for 20 min with 1% formaldehyde in PBS at room temperature. Cells were harvested with 100 mM Tris·HCl, pH 9.4/10 mM DTT and pelleted by centrifugation for 5 min at 2,000 × g. The pellets were washed with 1 ml of ice-cold PBS, resuspended in 0.6 ml of cell lysis buffer (0.1% SDS/0.5% Triton X-100/20 mM Tris·HCl, pH 8.1/150 mM NaCl/1 × protease inhibitor mixture; Roche Molecular Biochemicals), sonicated, and then centrifuged for 10 min at 13,000 rpm with an Eppendorf microcentrifuge at 4°C. Supernatants were transferred to fresh tubes and precleared with protein A-Sepharose [80 μl of 50% slurry in 10 mM Tris·HCl, pH 8.1/1 mM EDTA (TE)/0.5% BSA/19.2 μg of glycogen] for 1 h at 4°C. Immunoprecipitation was performed overnight at 4°C with 10 μg of anti-CREB antibody (32), 3 μg of anti-dimethyl methyl K4 histone H3 (abcam, Cambridge, MA), or anti-β-Gal (5 Prime → 3 Prime). Immune complexes were captured with 80 μl of 50% protein A-agarose slurry for 1 h at 4°C. The beads were collected by centrifugation at 7,600 rpm for 1 min and washed as follows: two times in lysis buffer for 10 min, two times in washing buffer (0.1% SDS/0.5% Triton X-100/2 mM EDTA, pH 8.0/20 mM Tris·HCl, pH 8.1/150 mM NaCl) for 10 min, one time in LiCl buffer (0.25 M LiCl/1% Nonidet P-40/1% deoxycholate/1 mM EDTA, pH 8.0/10 mM Tris·HCl, pH 8.1), and two times with TE. CREB-DNA complexes were eluted from the beads, first by 15-min incubation with 100 μl of elution buffer (1% SDS/0.1 M NaHCO3, pH 8.0) at room temperature. Beads were collected by centrifugation, and a second elution was performed by 10-min incubation with 50 μl of elution buffer. After the addition of 10 μl of 5 M NaCl, eluates were pooled and heated at 65°C overnight to reverse the formaldehyde cross-linking. DNA fragments were purified by using a Qiagen (Valencia, CA) kit and quantified by real-time PCR.
In Vivo Genomic Footprinting. Genomic footprints followed the ligation-mediated PCR technique of Mueller and Wold (33) and Riggs et al. (34) with some modifications. Briefly, PC12 and H4IIE cells (≈8 × 107) were either not treated or treated with 0.2% dimethyl sulfate at room temperature for 5 min to alkylate the guanine bases by the Maxam–Gilbert method (35). Dimethyl sulfate treatment was followed by washing and harvesting in 1× PBS. Genomic DNA was isolated, and DNA from the untreated cells was subjected to 0.5% dimethyl sulfate treatment in vitro for 4 min at room temperature and stopped by addition of 375 mM sodium acetate (pH 7.0) and 250 mM 2-mercaptoethanol. Cleavage of the modified guanosines was carried out by treatment with 1 M piperidine for 30 min at 90°C. Cleaved DNA (2.5 μg) was used in primer extension reactions at 48°C by using the program sequenase version 2.0 (United States Biochemical) with 40 nM of the following gene-specific primers designed against the bottom strand of the rat sequence: somatostatin (5′-GTCAGTCTAGACGCAGGT-3′); c-fos (5′-GGCAAAGCTCGGCGAG-3′); and phosphoenolpyruvate carboxykinase (PEPCK; 5′-CCGGATTTCCCCTGTT-3′). The primer extension products were ligated with a linker (top strand, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; and bottom strand, 5′-GAATTCAGATC-3′); which served as a fixed end for ligation-mediated PCR. The top-strand primer and a gene-specific primer (each 100 nM) were used in 20 cycles of PCR amplification with TaqDNA polymerase plus Q buffer (Qiagen). The amplification primers were somatostatin (5′-AGGTCCGCAGGCAGCAGAG-3′); c-fos (5′-AGGGGTCCAGGGGTAGACAC-3′); and PEPCK (5′-CTGTTGGCCAAGGGTGTGTTCC-3′). A final three rounds of primer extension was carried out by using γ33P-ATP end-labeled primers [(5 pmol of 1.4 μCi/pmol (1 Ci = 37 GBq) specific activity, with partial removal of free nucleotides by precipitation with ethanol with addition of 5 M ammonium sulfate]. The end-labeled primers were somatostatin (5′-GACGACTCCAGTAGCGTCTCCTTCAG-3′); c-fos (5′-ACTGGTGGGAGCTGCAGAGCAG-3′); and PEPCK (5′-GAAGGCCAACCGTGCTTGGTAGCTAG-3′). Labeled ligation-mediated PCR products were phenol-chloroform-extracted, ethanol-precipitated, and resuspended in formamide gel-loading buffer. Samples were electrophoresed on a 7% acrylamide-urea gel and the footprints were visualized by exposure to a PhosphorImager screen (Molecular Dynamics).
RT-PCR. PC12 cells (1–5 × 104), H4IIE, and cortical neurons were treated as described, and total RNA was isolated by using TRIzol (Invitrogen) or an RNeasy kit (Qiagen) according to manufacturer's instructions. RNA (from 50 ng to 3 μg) was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and 50–250 ng of random primers (Invitrogen). cDNA was amplified with platinum Taq (Invitrogen) for 28 cycles (c-fos), 29 cycles [brain-derived neurotrophic factor (BDNF)], or 33 cycles for all other genes. PCR products were resolved in 2% agarose gels. Primers will be provided on request.
Quantitative PCR. Primers surrounding CRE sites were designed by using MIT's primer3 software (http://frodo.wi.mit.edu/cgibin/primer3/primer3_www.cgi) with default parameters, except rodent and simple repeat library was on, product size was 80–200 bp, primer size was 18–27 bases, Tm was 66°–70°C, maximum self-complementarity was 4, and maximum 3′ complementarity was 1. Primer sequences are available on request. PCRs (10 μl) contained 1 μl of 10× PCR buffer (Invitrogen), 2.5 mM MgCl2, 200 μM dNTP (Roche), 0.125–0.25 μM primer (Integrated DNA Technologies, Coralville, IA), 1× SYBR green I (Invitrogen), and 1 unit of Platinum Taq (Invitrogen). PCR was performed on an Opticon OP346 (MJ Research, Cambridge, MA) for one cycle at 95°C for 35 s, 30–50 cycles at 94°C for 15 s, and 68–70°C for 40 s. Data were expressed as either nanograms of gel-purified (Qiagen) amplicon or fold-change over IgG.
Results
ChIP assays were used to examine the association of CREB with endogenous promoters in PC12 cells, a frequently used model for studies of CREB action. To determine whether CREB interacts constitutively with all CREs in their native context, we tested elements in three well studied promoters, the consensus CREs in the somatostatin (3) and inducible cAMP early repressor (ICER) (36) genes and the variant CRE in the c-fos promoter (37). PC12 cells, derived from a rat pheochromocytoma, express c-fos and ICER but not somatostatin. Of note, although c-fos gene expression is induced strongly by various stimuli, a significant proportion of the induction is due to the release of a block of transcriptional elongation (38, 39). Thus, although considered to be an immediate early gene, c-fos expresses a 5′-truncated transcript in the absence of stimulation. The GAPDH gene was used as a negative control because its promoter does not bind CREB and it is not induced transcriptionally by cAMP. As depicted in Fig. 1A, a high level of CREB binding is detected over the c-fos and ICER CREs but not over that in somatostatin. By using in vitro fluorescence anisotropy measurements, we have shown that CREB interacts better with the somatostatin CRE than it does with the CRE in the c-fos promoter (20). Thus, the ability of CREB to interact with the CRE in vitro does not predict its binding in vivo. No significant stimulation of CREB binding at c-fos and ICER promoters was observed with cells treated with forskolin, an activator of adenylyl cyclase.
Fig. 1.
Differential binding of CREB to CRE-containing promoters. (A) PC12 cells were treated without (gray bar) or with (white bar) 10 μM forskolin for 15 min. ChIP assays were performed by using 10 μg of anti-CREB or anti-β-Gal (IgG control, black bar) antibodies, and the levels of immunoprecipitated c-fos, ICER, and somatostatin promoter regions were measured by real-time PCR. The data represent 15 individual ChIPs from four experiments. Error is SEM. (B) In vivo genomic footprinting was performed on PC12 cells as described in Materials and Methods. In vitro denotes dimethyl sulfate-cleaved naked genomic DNA. In vivo denotes cleaved DNA from cells that were treated with dimethyl sulfate in vivo. Arrows denote footprinted or hypersensitive bands, including several over the CRE and immediately 3′ of the TATA box in the c-fos promoter. No footprints were detected over the somatostatin promoter.
The epitope recognized by the anti-CREB antibody could conceivably be masked in specific contexts and thereby cause an artifactually negative ChIP result. No binding of CREB to the somatostatin promoter was observed when we used another anti-CREB antiserum, however (data not shown). Because activation of the somatostatin CRE in PC12 cells has served as a model for CREB function, we sought to examine CREB occupancy by using an independent method, in vivo genomic footprinting. This assay cannot determine the identity of factors interacting with a DNA element, but it can assess whether a binding site is occupied. In vivo genomic footprinting assays were performed to examine the c-fos and somatostatin promoters (Fig. 1B). Consistent with the ChIP studies, the c-fos CRE showed a clear and reproducible footprint, represented by three bands of diminished intensity and one hypersensitive site. A weak but reproducible footprint was also detected immediately 3′ to the c-fos TATA sequence as a band of diminished intensity. In contrast, no footprints were detected over the CRE or TATA sequences of the somatostatin gene. This experiment confirms the results of the ChIP assays.
To determine whether the promoter-specific occupancy of CREB in PC12 cells was a general phenomenon or unique to somatostatin, we performed a similar analysis for other promoters known to interact with CREB in vitro and to be induced by cAMP in transient transfection assays (Fig. 2A). We found that CREB binding was undetectable at a variety of other CRE-containing promoters in PC12 cells, including those in the BDNF, gonadotropin-releasing hormone receptor, PEPCK, or insulin. CREB binding appeared to correlate with the potential for gene expression (Fig. 2B), as transcripts for BDNF, gonadotropin-releasing hormone receptor, PEPCK, and insulin are undetectable in PC12 cells, whereas c-fos is expressed at high levels, at least after induction by forskolin.
Fig. 2.
CREB does not interact with the promoters of transcriptionally silent genes. (A) PC12 cells were treated without (gray bar) or with (white bar) 10 μM forskolin for 15 min. Black bar denotes IgG signal. ChIP assays were performed to measure the level of CREB binding on BDNF, gonadotropin-releasing hormone receptor, PEPCK, and insulin promoters by employing real-time PCR. The data represent 10 individual ChIPs from four experiments. Error is SEM. (B) PC12 cells were treated without (lanes, 2, 5, 8, 11, and 14) or with (lanes, 3, 6, 9, 12, and 15) 10 μM forskolin for 1 h. mRNAs for CREB target genes indicated were detected by reverse transcription and PCR. The molecular weight marker (MW) is a 1-kb DNA ladder, and lanes 1, 4, 7, 10, and 13 are no reverse transcriptase controls.
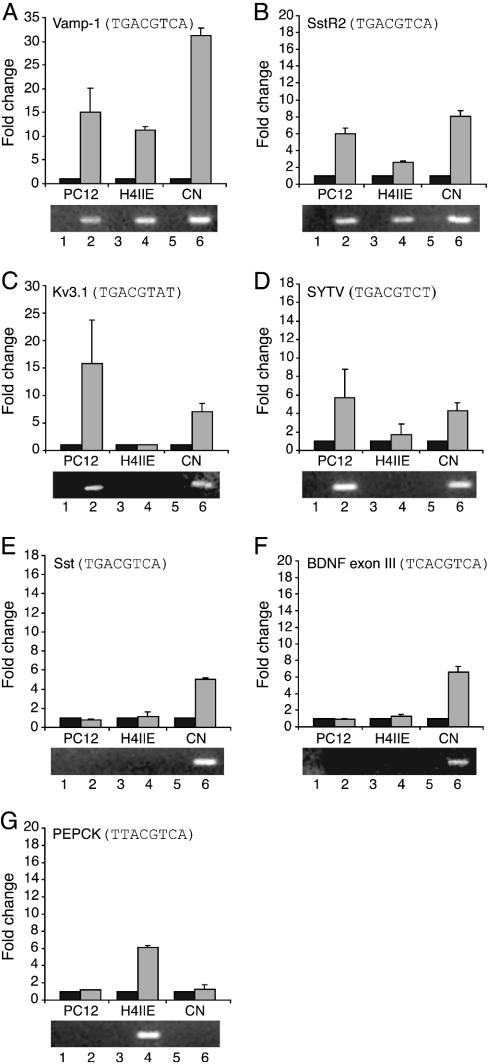
We next performed ChIP assays on a variety of additional genes, comparing the results in PC12 cells, H4IIE hepatoma cells, and primary cortical neurons. Because of the difficulty comparing absolute ChIP signals in separate immunoprecipitations, we used real-time PCR to compare the fold-increase in signal obtained by using anti-CREB antiserum versus nonspecific IgG. As shown in Fig. 3, some CREs bind CREB in all three cell types [Vamp-1 and type II somatostatin receptor (SstR2) (Fig. 3 A and B), whereas others (the potassium channel, Kv 3.1, and synatotagmin V) associate with CREB only in PC12 cells and cortical neurons (Fig. 3 C and D). CREB binding to the somatostatin and BDNF CREs was evident only in cortical neurons (Fig. 3 E and F). In contrast, the PEPCK CRE interacted with CREB in H4IIE cells, but not in PC12 cells or cortical neurons (Fig. 3G). CREB occupancy of the CRE correlates tightly with gene expression (or the potential for gene expression). To confirm our hypothesis that CREB binds to CREs in a cell type-specific manner, we performed additional in vivo genomic footprinting assays, this time examining the PEPCK promoter. A clear footprint over the CRE can be observed in H4IIE cells, but not in PC12 cells (Fig. 4). We conclude from these experiments that CREB binds to its target CREs in a cell-type-specific manner, which is not consistent with the idea that the CRE is occupied constitutively.
Fig. 3.
CRE occupancy by CREB is cell type-specific. ChIPs were performed by using PC12 and H4IIE cells, and primary cortical neurons (CN). The level of immunoprecipitated promoter regions was measured by real-time PCR. The black bar indicates IgG control, and the gray bar indicates CREB. Lanes 1, 3, and 5 are no reverse transcriptase controls. Lanes 2, 4, and 6 are reverse transcriptase-PCR products from PC12 cells, H4IIE cells, and cortical neurons, respectively. For study of BDNF expression (F, lane 6), cortical neurons were stimulated with K+ for 1 h. Data are presented as fold changes over IgG (average of 12 individual ChIPs from at least two different experiments). Error is SD.
Fig. 4.
The PEPCK CRE is occupied in H4IIE but not in PC12 cells. In vivo genomic footprinting was performed as described in Fig. 1. Footprints or hypersensitive sites were visualized only in the H4IIE sample. Locations of the CRE and NF-1 sites are indicated.
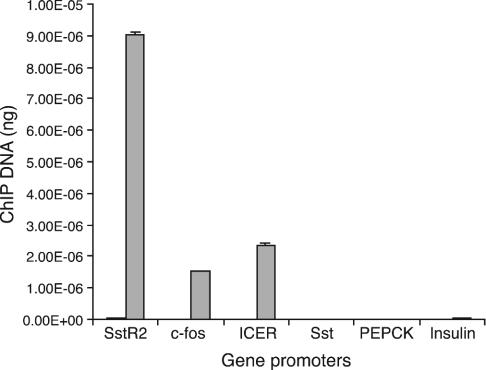
To test whether CREB binding to DNA correlated with nucleosomal modifications that reflect the activation state of promoters, we examined the methylation status of histone H3 at Lys-4. This modification is believed to inhibit the binding of the repressive Nurd complex and seems to be a marker for chromatin capable of transcriptional activity (40). Fig. 5 shows that three genes capable of being activated in PC12 cells, SstR2, c-fos, and ICER, bind CREB and are heavily methylated at Lys-4. The high level of Lys-4 methylation is evident even in the absence of stimulation for c-fos and ICER, which either reflects their potential for transcriptional activation or the basal transcriptional activity of these immediate early genes. Genes that are inactive, including somatostatin, PEPCK, and insulin, do not bind CREB and are not associated with methylated Lys-4.
Fig. 5.
Correlation of histone H3 Lys-4 methylation and CREB binding. ChIP was performed by using an antibody against dimethylated histone H3 Lys-4 in PC12 cells. The level of methylation at the indicated promoters was measured by real-time PCR. Black bars represent the IgG control. Histone methylation was detected in SstR2, c-fos, and ICER promoters but not in the Sst, PEPCK, or insulin promoters. Data from two experiments are shown.
Discussion
In this study, we demonstrate that CREB binding is highly tissue-specific. Binding is apparent at genes that are either transcriptionally active or potentially active but not at promoters that are not expressed. Therefore, the repertoire of genes that are potentially regulated by CREB (the CREB regulon) is distinct in different cell types. The presence of the epigenetic marker, histone H3 dimethyl-Lys-4, distinguishes active versus inactive genes and correlates with CREB binding. This finding suggests that CREB binding is regulated through an epigenetic mechanism (or that binding of CREB determines an active chromatin conformation).
Occupancy of many genetic regulatory elements is determined by the tissue-specific expression of particular transcription factors. CREB expression seems to be ubiquitous, however. The finding that CREB does not bind to all CREs was unexpected and is entirely contrary to the generally accepted model of CREB function. Most importantly, our results suggest that the presence of a CRE sequence in a gene promoter does not necessarily predict that CREB participates in its regulation. Thus, our findings may require the reevaluation of many genes proposed to be regulated through the CREB pathway, especially those characterized by using reporter genes and transient transfection assays. Indeed, the origin of the idea that CREB binds constitutively to CRE sequences is somewhat obscure. Support for this model include the findings that CREB is highly expressed, that it has a high affinity for the CRE, and that CREB binding to DNA in vitro does not seem to be regulated. The dogma that CREB binds constitutively to all CRE sequences is not entirely vestigial, however. Euskirchen et al. (7) recently used a human chromosome 22 microarray in ChIP-on-ChIP assays to examine CREB binding to the CREs of expressed and nonexpressed promoters in human choriocarcinoma placental cells. Because CREB seemed to interact with several targets that are generally classified as “neuronal” rather than “placental,” these workers concluded that CREB binding was constitutive. There are several possible explanations for our discordant interpretation. First, it is well known that many neuronal genes are expressed in nonneuronal tissues; for example, BDNF and its receptor, trkB, are expressed in immune cells (41). Second, genes are often aberrantly regulated in transformed cell lines, which underscores the need to combine ChIP data with more direct measures of transcription. The difference could also relate to the quality of the antibodies used or other aspects of the ChIP assays. The idea that CREB binds to CRE sequences in a tissue-specific manner is not unprecedented, however. For example, CREB was proposed to bind to a CRE-like element in the tyrosine aminotransferase gene in hepatoma cells, which express tyrosine aminotransferase, but not in fibrosarcoma cells, where the gene is inactive (42). These studies were performed before the advent of ChIP assays, however, so it is not certain that they actually monitor CREB binding, as opposed to some other transcription factor. Our results clearly show that CREB does not bind to genes that are inactive in PC12 cells, H4IIE cells, and primary cortical neurons. Therefore, CRE occupancy by CREB is not constitutive.
Because CREB was believed to bind its target genes constitutively, the mechanisms underlying its tissue-specific binding have received little attention. It is possible, in fact, that CREB could control the tissue-specific expression of some genes, rather than the converse. For example, BDNF is highly expressed in hippocampal neurons but not in some other neuronal subtypes, in response to activity (43–45). Because CREB activation occurs in both the expressing and nonexpressing neurons, how CREB drives BDNF expression in distinct neuronal populations has been in question. Tao et al. (46) reported that in response to membrane depolarization, both CREB and a transcription factor termed CaRF are required for BDNF exon III expression in rat embryonic cortical neurons. Interestingly, these authors pointed out that BDNF expression does not occur in PC12 cells, even though these cells contain both CREB and CaRF. It is possible that the failure of CREB to occupy the BDNF exon III CRE accounts for the lack of BDNF expression.
What controls whether CREB binds to a specific CRE? The binding of transcription factors to their recognition sites in DNA can be prevented through multiple mechanisms, including steric occlusion, sequestration by association with other proteins, or competition by other transcription factors. The most likely mechanism for the CREB block is steric occlusion. Our genomic footprinting data support this idea because of the complete absence of footprints over the CREs in nonexpressing cells. Steric occlusion in this case is likely the result of chromatin, as opposed to other competing transcription factors, because nucleosomes are invisible to dimethyl sulfate treatment. Thus, we believe that chromatin structure precludes CREB from binding to its target sites in certain situations. This conclusion is consistent with the findings of Ji et al. (47), who used in vivo genomic footprinting to show that occupancy of the bcl-2 CRE occurs only after its translocation to a transcriptionally active Ig heavy-chain locus.
In conclusion, the correlation of gene expression, histone H3 Lys-4 methylation, and CRE occupancy supports the idea that CREB binding is determined by the methylation status of histones at specific promoters. We found no detectable histone H3 Lys-4 methylation signal at the somatostatin, PEPCK, or insulin promoters in PC12 cells, and high levels of dimethylated Lys-4 at active genes, such as SstR2, c-fos, and ICER. Of interest, the c-fos promoter binds TFIIB and RNA polymerase II under basal conditions (48), but the ICER promoter does not and is only minimally active. This result indicates, as has been suggested by others, that Lys-4 methylation correlates with the potential for activation, rather than by the absolute levels of gene expression (49). However, in all cases where the promoter contained the dimethyl-Lys-4 epigenetic modification, gene expression could be detected by RT-PCR, and no transcription was detected from genes lacking dimethylated Lys-4. These findings suggest that epigenetic differences in chromatin from one cell type to another determine the population of genes capable of binding CREB and, in turn, the pattern of gene responses to the wide variety of pathways that activate CREB function. Although the mechanism that attenuates binding of CREB to transcriptionally silent target genes is not known, this process likely participates in specifying the transcriptional programs that underlie development and differentiation. Future work will be required to determine whether epigenetic modifications regulate occupancy by CREB or vice versa.
Acknowledgments
We thank Jeremy Boss (Emory University School of Medicine, Atlanta) for providing anti-CREB antisera and Gail Mandel for helpful comments. This work was supported by grants from the National Institutes of Health.
Abbreviations: CRE, cAMP-response element; CREB, CRE-binding protein; CBP, CREB-binding protein; ChIP, chromatin immunoprecipitation; BDNF, brain-derived neurotrophic factor; PEPCK, phosphoenolpyruvate carboxykinase; ICER, inducible cAMP early repressor; SstR2, type II somatostatin receptor.
References
- 1.Gonzalez, G. A., Yamamoto, K. K., Fischer, W. H., Karr, D., Menzel, P., Biggs, W., III, Vale, W. W. & Montminy, M. R. (1989) Nature 337, 749–752. [DOI] [PubMed] [Google Scholar]
- 2.Montminy, M. R. & Bilezikjian, L. M. (1987) Nature 328, 175–178. [DOI] [PubMed] [Google Scholar]
- 3.Montminy, M. R., Sevarino, K. A., Wagner, J. A., Mandel, G. & Goodman, R. H. (1986) Proc. Natl. Acad. Sci. USA 83, 6682–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez, G. A. & Montminy, M. R. (1989) Cell 59, 675–680. [DOI] [PubMed] [Google Scholar]
- 5.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 6.Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- 7.Euskirchen, G., Royce, T. E., Bertone, P., Martone, R., Rinn, J. L., Nelson, F. K., Sayward, F., Luscombe, N. M., Miller, P., Gerstein, M., et al. (2004) Mol. Cell. Biol. 24, 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chrivia, J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365, 855–859. [DOI] [PubMed] [Google Scholar]
- 9.Kwok, R. P., Lundblad, J. R., Chrivia, J. C., Richards, J. P., Bachinger, H. P., Brennan, R. G., Roberts, S. G., Green, M. R. & Goodman, R. H. (1994) Nature 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 10.Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. R. & Wright, P. E. (1997) Cell 91, 741–752. [DOI] [PubMed] [Google Scholar]
- 11.Felzien, L. K., Farrell, S., Betts, J. C., Mosavin, R. & Nabel, G. J. (1999) Mol. Cell. Biol. 19, 4241–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima, T., Uchida, C., Anderson, S. F., Lee, C. G., Hurwitz, J., Parvin, J. D. & Montminy, M. (1997) Cell 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 13.Swope, D. L., Mueller, C. L. & Chrivia, J. C. (1996) J. Biol. Chem. 271, 28138–28145. [DOI] [PubMed] [Google Scholar]
- 14.Bannister, A. J. & Kouzarides, T. (1996) Nature 384, 641–643. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., Lin, R. J., Schiltz, R. L., Chakravarti, D., Nash, A., Nagy, L., Privalsky, M. L., Nakatani, Y. & Evans, R. M. (1997) Cell 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 16.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. (1996) Cell 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 17.Hagiwara, M., Brindle, P., Harootunian, A., Armstrong, R., Rivier, J., Vale, W., Tsien, R. & Montminy, M. R. (1993) Mol. Cell. Biol. 13, 4852–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols, M., Weih, F., Schmid, W., DeVack, C., Kowenz-Leutz, E., Luckow, B., Boshart, M. & Schutz, G. (1992) EMBO J. 11, 3337–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards, J. P., Bachinger, H. P., Goodman, R. H. & Brennan, R. G. (1996) J. Biol. Chem. 271, 13716–13723. [DOI] [PubMed] [Google Scholar]
- 20.Craig, J. C., Schumacher, M. A., Mansoor, S. E., Farrens, D. L., Brennan, R. G. & Goodman, R. H. (2001) J. Biol. Chem. 276, 11719–11728. [DOI] [PubMed] [Google Scholar]
- 21.Kwok, R. P., Laurance, M. E., Lundblad, J. R., Goldman, P. S., Shih, H., Connor, L. M., Marriott, S. J. & Goodman, R. H. (1996) Nature 380, 642–646. [DOI] [PubMed] [Google Scholar]
- 22.Cox, J. M., Sloan, L. S. & Schepartz, A. (1995) Chem. Biol. 2, 819–826. [DOI] [PubMed] [Google Scholar]
- 23.Yin, M. J., Paulssen, E. J., Seeler, J. S. & Gaynor, R. B. (1995) J. Virol. 69, 3420–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iguchi-Ariga, S. M. & Schaffner, W. (1989) Genes Dev. 3, 612–619. [DOI] [PubMed] [Google Scholar]
- 25.Martinowich, K., Hattori, D., Wu, H., Fouse, S., He, F., Hu, Y., Fan, G. & Sun, Y. E. (2003) Science 302, 890–893. [DOI] [PubMed] [Google Scholar]
- 26.Mancini, D. N., Singh, S. M., Archer, T. K. & Rodenhiser, D. I. (1999) Oncogene 18, 4108–4119. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher, M. A., Goodman, R. H. & Brennan, R. G. (2000) J. Biol. Chem. 275, 35242–35247. [DOI] [PubMed] [Google Scholar]
- 28.Konig, P. & Richmond, T. J. (1993) J. Mol. Biol. 233, 139–154. [DOI] [PubMed] [Google Scholar]
- 29.Weih, F., Stewart, A. F., Boshart, M., Nitsch, D. & Schutz, G. (1990) Genes Dev. 4, 1437–1449. [DOI] [PubMed] [Google Scholar]
- 30.Wolfl, S., Martinez, C. & Majzoub, J. A. (1999) Mol. Endocrinol. 13, 659–669. [DOI] [PubMed] [Google Scholar]
- 31.Hiroi, H., Christenson, L. K., Chang, L., Sammel, M. D., Berger, S. L. & Strauss, J. F., III (2004) Mol. Endocrinol. 18, 791–806. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, C. S., Beresford, G. W., Louis-Plence, P., Morris, A. C. & Boss, J. M. (1999) Immunity 10, 143–151. [DOI] [PubMed] [Google Scholar]
- 33.Mueller, P. R. & Wold, B. (1989) Science 246, 780–786. [DOI] [PubMed] [Google Scholar]
- 34.Riggs, A. D., Singer-Sam, J. & Pfeifer, G. P. (1998) in Chromatin: A Practical Approach, ed. Gould, H. (Oxford Univ. Press, Oxford), pp. 79–109.
- 35.Maxam, A. M. & Gilbert, W. (1980) Methods Enzymol. 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 36.Molina, C. A., Foulkes, N. S., Lalli, E. & Sassone-Corsi, P. (1993) Cell 75, 875–886. [DOI] [PubMed] [Google Scholar]
- 37.Sheng, M., Dougan, S. T., McFadden, G. & Greenberg, M. E. (1988) Mol. Cell. Biol. 8, 2787–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mechti, N., Piechaczyk, M., Blanchard, J. M., Jeanteur, P. & Lebleu, B. (1991) Mol. Cell. Biol. 11, 2832–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinaud, S. & Mirkovitch, J. (1998) J. Mol. Biol. 280, 785–798. [DOI] [PubMed] [Google Scholar]
- 40.Zegerman, P., Canas, B., Pappin, D. & Kouzarides, T. (2002) J. Biol. Chem. 277, 11621–11624. [DOI] [PubMed] [Google Scholar]
- 41.Besser, M. & Wank, R. (1999) J. Immunol. 162, 6303–6306. [PubMed] [Google Scholar]
- 42.Weih, F., Nitsch, D., Reik, A., Schutz, G. & Becker, P. B. (1991) EMBO J. 10, 2559–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson, S. L., Grover, L. M., Schwartzkroin, P. A. & Bothwell, M. (1992) Neuron 9, 1081–1088. [DOI] [PubMed] [Google Scholar]
- 44.Tao, X., Finkbeiner, S., Arnold, D. B., Shaywitz, A. J. & Greenberg, M. E. (1998) Neuron 20, 709–726. [DOI] [PubMed] [Google Scholar]
- 45.Shieh, P. B., Hu, S. C., Bobb, K., Timmusk, T. & Ghosh, A. (1998) Neuron 20, 727–740. [DOI] [PubMed] [Google Scholar]
- 46.Tao, X., West, A. E., Chen, W. G., Corfas, G. & Greenberg, M. E. (2002) Neuron 33, 383–395. [DOI] [PubMed] [Google Scholar]
- 47.Ji, L., Mochon, E., Arcinas, M. & Boxer, L. M. (1996) J. Biol. Chem. 271, 22687–22691. [DOI] [PubMed] [Google Scholar]
- 48.Fass, D. M., Butler, J. E. & Goodman, R. H. (2003) J. Biol. Chem. 278, 43014–43019. [DOI] [PubMed] [Google Scholar]
- 49.Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C. & Kouzarides, T. (2004) Nat. Cell Biol. 6, 73–77. [DOI] [PubMed] [Google Scholar]