Abstract
In addition to NK T cells expressing invariant Vα14 or Vα24 T cell receptors (TCRs), the CD1d-restricted T cell repertoire is comprised of T cells with diverse TCRs that mediate inflammation during autoimmune and infectious disease. Here we describe the isolation of human Vα24– T cells that are activated by antigen and CD1d. Mass spectrometric and NMR studies revealed that the stimulatory compounds were neither peptidic nor lipidic but instead were composed of sulfur and aromatic hydrocarbon rings, corresponding to the general structure of phenyl pentamethyldihydrobenzofuran sulfonates. Studies of the molecular mechanism of T cell activation showed that a clonotypic Vα2/Vβ21 TCR transmitted activating signals, which were highly specific for hydroxylation and methylation patterns at the terminal structures of stimulatory compounds. These studies provide evidence for noninvariant CD1d-restricted T cells in humans and identify the complete molecular structure of a nonlipidic small molecule that activates T cells through an αβ TCR.
The discovery of CD1 antigen presentation systems has shown that the range of antigenic structures recognized by T cells is much broader than previously thought because it includes lipid, glycolipid, and lipopeptide antigens. Four human CD1 proteins, CD1a, CD1b, CD1c, and CD1d, present lipid antigens to T cells (1–4). The known antigens are diverse and include mycobacterial mycolates, diacylglycerols, sphingolipids, polyisoprenoid lipids, sulfotrehalose-containing lipids, and lipopeptides (5–15). Each of these antigens consists of a hydrophilic cap that is linked to one or two lipid anchors composed of aliphatic hydrocarbon chains with varying degrees of branching. Crystal structures of CD1 proteins bound to phosphatidylinositol, ganglioside GM2, glucose monomycolate, and sulfatides show that the lipid anchors are inserted into the hollow antigen-binding grooves of CD1, where they interact with nonpolar amino acids that line the interior of the groove (16–19). The carbohydrate, peptide, or polyalcohol moieties of lipid antigens protrude from the groove, so that they lie on the outer, α-helical surface of CD1, where they can directly contact T cell receptors (TCRs).
Antigen and TCR diversity in the CD1-restricted T cell repertoire are central issues in understanding the biological functions of lipid-reactive T cells. T cells restricted by CD1a, CD1b, and CD1c recognize many types of microbial lipids and are normally activated during the course of acute Mycobacterium tuberculosis infections in humans (13, 14, 20). Because these cells secrete IFN-γ, produce granulysin, and kill bacteria within infected cells, their natural function is thought to involve direct recognition and elimination of infected cells as part of the host response to infection (21, 22). Pathogen recognition can be achieved by TCR contact with any one of many foreign lipids produced during infection, so the use of diverse TCRs to detect many types of foreign antigens is thought to be a central feature of their function as effector T cells. In contrast, CD1d-restricted T cells with highly conserved TCRs, known as NK T cells, express TCR-α chains composed of Vα24/Jα18 segments paired with Vβ11 in humans or Vα14/Jα18 paired with Vβ8 in mice (23, 24). NK T cells are activated by CD1d-expressing antigen-presenting cells and synthetic α-galactosyl ceramide (αGalCer), which is a surrogate for as yet unidentified natural antigens (6). The natural function of NK T cells involves activation at the earliest phases of the innate immune response to secrete large amounts of IFN-γ, IL-4, and other regulatory cytokines. Thus, the normal functioning of NK T cells relies on the expression of conserved TCRs that respond to a single or a limited number of antigenic stimuli, (25, 26) in contrast to the diverse antigens and TCRs expressed by antimicrobial T cells.
Studies of murine T cells have provided growing evidence for the existence of other types of T cells within the CD1d-restricted repertoire that are diverse with respect to TCR expression, structures of antigens recognized, and effector functions. For example, Vα14+ NK T cells can be activated by phosphatidylinositol or related lipids but fail to recognize αGalCer (9, 27, 28). Other studies have shown that CD1d activates T cells expressing Vα14– TCRs and that these T cells fail to recognize αGalCer and related compounds (29–33). Increasingly, in vivo studies show that Vα14– CD1-restricted T cells have immunoregulatory functions that are distinct from Vα14+ NK T cells. For example, in a murine model of myocarditis caused by Coxsackie virus, the inflammatory response depended on CD1d expression but was not affected by the deletion of T cells expressing the Vα14Jα18 gene (34). Likewise, hepatic inflammation in a mouse model of hepatitis B was shown to depend on CD1, but the effector T cells did not express Vα14 TCRs and did not stain with CD1d-αGalCer tetramers (35). Also, T cells that stain with CD1d-sulfatide tetramers, but not with CD1d-αGalCer tetramers, promote experimental autoimmune encephalomyelitis in mice (32). It is not yet clear whether humans have equivalent, Vα24– CD1d-restricted T cell populations. Human CD1d proteins may present antigens other than αGalCer, or the human system may rely on CD1a, CD1b, and CD1c for presentation of more structurally diverse antigens.
Here we describe the isolation of Vα24– human T cells that were activated by CD1d and small organic compounds but did not respond to any known antigen for NK T cells. Unexpectedly, detailed molecular characterization of the stimulatory compounds by MS and NMR showed that they were neither peptides nor lipids but instead were sulfur-containing small molecules, including phenyl 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonate (PPBF). PPBF did not function as a T cell mitogen but activated the T cells by means of their clonotypic Vα2/Vβ21 TCR by using a mechanism that was highly specific for methylation and hydroxylation patterns on the antigens. These studies provide evidence for Vα24– CD1d-restricted T cells in humans and identify a new type of nonpeptidic, nonlipidic molecule that activates αβ T cells.
Methods
Derivation of Human T Cell Lines and Clones. Polyclonal human T cells were derived with many antigens, including a mixture of synthetic lipopeptides (AnaSpec, San Jose, CA). The lead compound in the antigenic mixture corresponded to a sequence from human IL-1α, LKKRRL, which was chemically acylated on the εC of the third amino acid, a lysine, but this mixture also contained PPBF. To isolate CD1-restricted T cells, polyclonal human lymphocytes were stimulated three times with synthetic antigen mixtures at a concentration of 1 μg/ml and autologous dendritic cells (DC) at 2-week intervals, followed by stimulation with heterologous DC and antigens. T cells (1.25 × 105) and DC (0.25 × 105) were added per well in round-bottom, 96-well plates. T cell medium was made by supplementing 500 ml of RPMI medium 1640 with 50 ml of FCS (HyClone), penicillin, and streptomycin (GIBCO); 20 mM Hepes (GIBCO); and 4 ml 1 N NaOH solution. The IL-2 concentration was initially 0.1 nM and gradually increased to 1 nM during subsequent rounds of stimulation. T cell clones were derived by limiting dilution by using 0.6 × 105 Epstein–Barr virus-transformed B cells (100 Gray) and 1.3 × 105 heterologous peripheral blood mononuclear cells (33 Gray) as feeder cells and 1 μg/ml phytohemagglutinin (Difco) in medium containing 2 nM IL-2.
Antigens. Antigen mixtures were separated by reversed-phase HPLC with electrospray ionization MS (LCQ Advantage quadrupole ion trap mass spectrometer) and UV detection with a split interface allowing peak collection. Compounds were resolved with a C18 column (25 cm, i.d. 4.6 mm, Vydac, Hesperia, CA) by a gradient from 60% to 5% solvent A (20% acetonitrile/80% water/0.02% trifluoroacetic acid) and 40% to 95% solvent B (50% methanol/30% acetonitrile/20% water/0.02% TFA). MSn and deuterium exchange experiments were carried out by using nanoelectrospray ionization. Proton NMR spectra were acquired in CDCl3 on a Varian Inova 500-MHz spectrometer, and chemical shifts were calibrated with respect to tetramethylsilane (0.00 ppm). PPBF and structural analogs were synthesized by incubating a mixture of 10 mg of phenol or cresol, 10 mg of f luorenylmethoxycarbonyl-l-Arg(2,2,4,6,7-pentamethyldihydrobenzofuransulfonate)-OH (Glycopep, Chicago), 100 μl of trifluoroacetic acid, and 100 μl of chloroform overnight at room temperature, followed by reversed-phase HPLC purification. Didehydroxymycobactin (DDM) was purified from M. tuberculosis H37Rv as described in ref. 15.
T Cell Activation Assays. DC were prepared from peripheral blood monocytes by centrifugation over Ficoll–Hypaque, adherence of the cells to plastic flasks, culture of adherent cells with 300 units/ml granulocyte/macrophage colony-stimulating factor and 200 units/ml IL-4 for 3 days, followed by irradiation (33 Gray). T cell activation was measured by incubating 5 × 104 T cells with 5 × 104 DC or CD1-transfected C1R cells. Proliferation was measured after coculture for 3 days with antigen, followed by a 6-h pulse of 1 μCi (1 Ci = 37 GBq) of [3H]thymidine before harvesting and counting β emissions. Alternatively, 105 J.RT3-T3.5 transfectants and 5 × 104 DC were incubated in the presence of antigen and 10 nM phorbol 12-myristate 13-acetate for 24 h before the supernatants were tested for the presence of IL-2 by using the HT-2 bioassay (36). Competition assays were carried out with recombinant hCD1d proteins bound to plastic 96-well plates by incubating 10 ng/ml solutions of αGalCer with 0.8- to 500-fold excess of competitors for 1 h (37). Plates were washed, and the NK T cell hybridoma DN3A4/1-2 was added for 14 h before the supernatants were collected and IL-2 was measured by capture ELISA.
TCR Cloning. mRNA was isolated from 106 clonal or polyclonal T cells by using an Oligotex Direct mRNA kit (Qiagen, Valencia, CA), followed by cDNA synthesis with SuperScript RT (Invitrogen). PCR primer sets covering most of the TCR Vα and Vβ families (gifts from S. Gagnon and W. Biddison, National Institutes of Health, Bethesda) were used to determine the Vα and Vβ usage (38). Primers that cover the full-length Vα2 (forward, GGGCAGAAAAGAATGATGAAATC; reverse, TCAGCTGGACCACAGCC) and Vβ21 chains (forward, CCCTGCCATGGGCACCAGGCTCC; reverse, GGAGCTAGCCTCTGGAATCCTTTCTC) were used to obtain a PCR product that was cloned into the pCR-4-TOPO vector (Invitrogen). The full-length chains were cloned in the NotI/BamH1 restriction sites in pRep7 and pRep9 and transfected in J.RT3-T3.5 cells (36).
Abs. Anti-αβ TCR (clone WT31-FITC, Becton Dickinson), anti-CD4 (clone RPA-T4-FITC, Pharmingen), and anti-CD3 (OKT3) were used for flow cytometric analysis. Anti-CD1a (OKT6), -CD1b (BCD1b.3), -CD1c (F10/21A3.1), -CD1d (CD1d42), and IgG1 control (P3) were used for blocking studies at 20 μg/ml.
Experimental Results
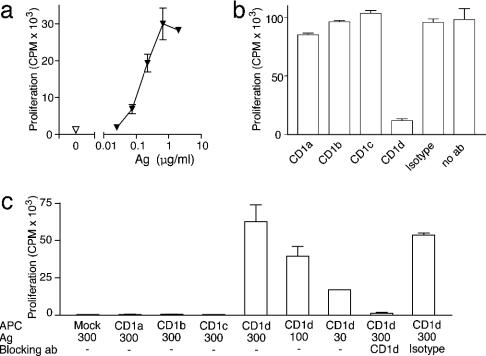
Isolation of Antigen-Dependent CD1d-Restricted T Cells. To investigate the chemical diversity of human CD1-presented antigens, we derived T cell lines by stimulation with monocyte-derived DC and a variety of antigens other than those known to be recognized by NK T cells. One of these, a mixture of synthetic lipopeptides made by acylating a synthetic peptide sequence (LKKRRL) that normally occurs in the IL-1α protein, generated a T cell line, which depended on antigen for its proliferation (Fig. 1a). Its proliferative response to DC also depended on CD1d, as blocking mAb specific for CD1d inhibited the response (Fig. 1b). Further studies for which we used CD1– lymphoblastoid cells transfected with human CD1 proteins confirmed that antigen recognition absolutely depended on CD1d expression and could not be carried out by CD1a, CD1b or CD1c (Fig. 1c).
Fig. 1.
T cell activation is CD1d-restricted and antigen (Ag)-dependent. (a and b) Proliferation of ABd T cells stimulated with DC and antigen mixtures was measured in the presence of mAb against CD1a, CD1b, CD1c, CD1d, or isotype-matched control (P3) (20 μg/ml). (c) C1R cells transfected with CD1a, CD1b, CD1c, CD1d, or vector alone (mock) were treated with the antigen mixture at the indicated concentration (in μg/ml). Blocking mAb and polyclonal ABd T cells were added, and proliferation was measured.
Flow cytometric characterization of T cell clones derived from the polyclonal cell line showed that they stained uniformly with mAb against the TCRαβ but lacked CD4 and CD8. This TCRαβ+, CD1d-restricted T cell line was named ABd. The TCR structure was further investigated by using a panel of PCR primer sets that specifically amplified human TCR variable segments (38). Only sequences corresponding to Vα2 and Vβ21 were amplified, which suggested that the ABd TCR was distinct from Vα24 TCRs found on invariant NK T cells (data not shown). This possibility was confirmed by cloning and DNA sequencing of the full-length TCR chains from the polyclonal T cell line ABd and two antigen reactive clones. All α and all β chain sequences obtained were identical, and corresponded to Vα2 (TRAV12-1*01/TRAJ6*01) and Vβ21 (TRBV11-3*01/TRBD1*01/TRBJ2-5*01) (GenBank accession nos. AY532913 and AY532914, respectively). Thus, ABd was a CD1d-restricted human T cell clone that did not express an invariant TCR found on NK T cells, prompting an investigation into the structure of the antigenic compounds.
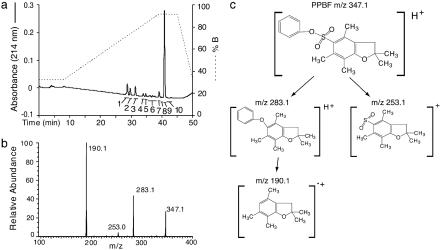
Structural Determination of Antigenic Compounds. ABd was originally derived as part of an effort to identify antigens by stimulation with synthetic lipopeptides that might have structural similarities to a mycobacterial antigen presented by CD1a (15). To identify the precise structure of the stimulatory compound from the crude mixture of synthetic compounds used to stimulate ABd, we carried out HPLC-MS separation of antigen mixtures. At least 10 separable compounds were detected, but only one of these, which eluted in HPLC fraction 9, stimulated ABd (Fig. 2a and data not shown). Electrospray ionization mass spectrometric analysis of this fraction detected only one singly charged positive ion of m/z 347.1, hereafter referred to as 347. This result was unexpected because this unknown compound had a lower molecular weight than the lipopeptides in the mixture (m/z ≈1,100).
Fig. 2.
Isolation of PPBF as T cell antigen. (a) Mixtures of stimulating compounds were resolved by using reversed-phase HPLC and monitored by UV (214 nm) absorbance (solid line). Each fraction was normalized for area under the UV absorbance curve and tested for its ability to stimulate ABd in the presence of DC. (b) Fraction 9, the only fraction that stimulated ABd, was subjected to electrospray ionization-tandem MS. The experiment showed an ion at m/z 347.1, which with collisional activation gave rise to prominent product ions that are interpreted as shown in c.
The first insight into a possible structure came from the observation that 347 showed a strong isotope peak at m/z 349, which suggested that 347 contained an atom with an abundant M + 2 isotope, such as sulfur. There was one reactant used to prepare synthetic antigens that contained sulfur, 9-fluorenyl methyloxycarbonyl-Nγ-2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl-l-arginine. This observation prompted us to consider PPBF as a candidate structure that corresponded to the known mass of 347. The tandem MS spectrum yielded product ions of m/z 253 and 283, which corresponded to the expected products resulting from the loss of phenol and sulfur dioxide, respectively (Fig. 2 b and c). The product at m/z 190 corresponded to a fragment resulting from the loss of both the phenyl group and the sulfonyl group with hydrogen transfer yielding a radical ion.
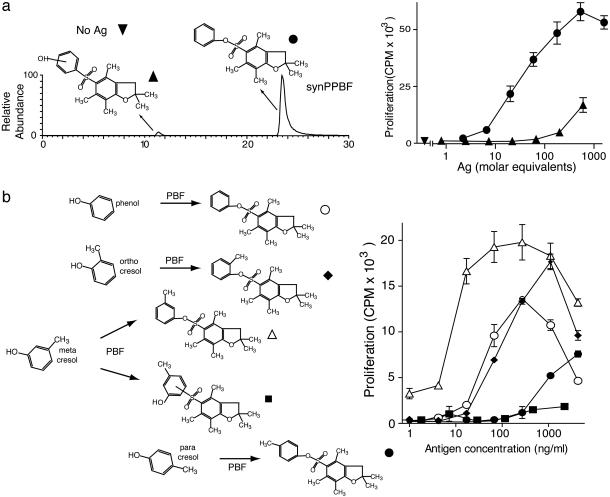
Because there is no precedent for small nonlipidic molecules with multiple aromatic rings to stimulate T cells, we sought to independently confirm this result by synthesizing PPBF from phenol and fluorenylmethoxycarbonyl-l-Arg(2,2,4,6,7-pentamethyldihydrobenzofuransulfonate)-OH. Liquid chromatography MS analysis of the reaction products detected two isomers, each with a nominal mass of 347 (Fig. 3a). The late eluting isomer showed that its elution time and tandem mass spectrum were virtually identical to that of 347 derived from fraction 9, indicating that this newly synthesized molecule (synPPBF) and 347 were chemically identical (data not shown). The structure of synPPBF was further confirmed by the one-dimensional proton NMR spectrum (Table 1). SynPPBF was able to stimulate the CD1d-restricted T cell response of ABd and its dose–response was similar to that seen with 347 (Fig. 3a Right and data not shown), providing conclusive evidence that the stimulatory factor corresponded to the structure of PPBF.
Fig. 3.
T cell fine specificity for synthetic structural analogs of PPBF. (a) By using reversed-phase HPLC and mass spectrometric ion monitoring at m/z 347, two products of the PPBF synthesis mixture with a m/z 347 were recovered. The later eluting compound showed nearly identical retention to fraction 9 from the stimulatory compounds (347) and an identical collisional mass spectrum. The early eluting compound was assigned as containing a C–S linkage as indicated. Both compounds were normalized for absorbance at 290 nm and tested for their ability to induce proliferation of ABd. Ag, antigen. (b) PPBF analogs were synthesized by coupling phenol or any of three forms of methyl phenol (cresol) with 2,2,4,6,7-pentamethyldihydrobenzofuransulfonate (PBF). After normalization for absorbance at 290 nm, the ability of PPBF analogs to induce IL-2 release by ABd was tested in the presence of C1R cells transfected with CD1d.
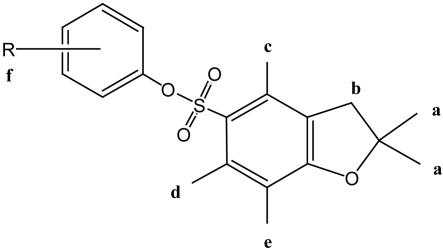
Table 1. Proton NMR data for PPBF and methylated analogs.
1H chemical shifts (ppm) for PPBF analogs were measured in deuterated chlorofom at 24.8°C. The spectrum of PPBF was characterized by the appearance of signals consistent with the 2,2,4,6,7-pentamethyl-dihydrobenzofuran nucleus linked to an isolated and otherwise unsubstituted phenyl ring. In particular, there was a resonance for the pair of 2,2 methyl groups (1.49 ppm; singlet; 6H), three additional resonances for the 4-, 6-, and 7-methyl groups (2.56, 2.33, 2.14 ppm; singlets; 3 × 3H) and a complex multiplet of aromatic signals (7.02-7.35 ppm) partially obscured by the residual CHCl3 resonance at 7.26 ppm, consistent with the isolated phenyl ring. NA, not applicable. R refers to the position at any methyl substitutions on the phenyl ring.
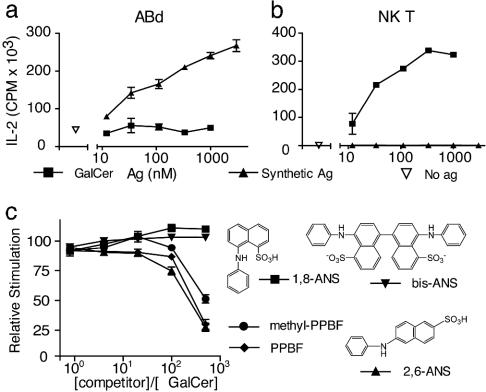
Influence of Antigen Structure on T Cell Activation. In addition to the potent response to synPPBF, ABd showed a weaker response to the early eluting isomer of m/z 347 in the reaction mixture, suggesting crossreactivity of ABd with an isomer of synPPBF (Fig. 3a). To investigate this possibility we first analyzed the early eluting antigen by using tandem MS, which revealed a spectrum similar to that of the late eluting antigen, except that the fragment resulting from the loss of S02 (m/z 283) was not seen. This result, along with deuterium exchange experiments (data not shown) suggested that the early eluting isomer was a form of PPBF in which the phenol was linked to the dihydrobenzofuran by an S–C linkage rather than an S–O linkage, as depicted in Fig. 3a. In addition, we synthesized a series of methylated PPBF isomers by substituting phenol with cresol (methyl phenol) containing a methyl branch in the ortho, meta, or para position (Fig. 3b). All of the synthetic antigens yielded NMR spectra consistent with the ortho-, meta-, or paracresol methyl group (Table 1). In most cases, the site of linkage of the phenol groups and the presence and position of the methyl groups reduced the potency of the CD1d-mediated T cell response, except for 3-methyl PPBF, which stimulated ABd more potently than PPBF. Taken together, these results indicate that human CD1d-restricted T cells can be activated by small sulfated polyaromatic hydrocarbon structures and that the response is specific for the hydroxylation and methylation patterns of these antigens.
PPBF Does Not Activate Conventional or NK T Cells. Further studies were aimed at understanding the mechanism of action and the identity of activating receptors on PPBF-reactive T cells. First, we considered the possibility that PPBFs might function as mitogenic stimuli by means of nonclonally distributed receptors on T cells. However, fresh polyclonal T cells did not proliferate in response to PPBF under the same conditions that led to strong proliferative responses to a known T cell mitogen, phytohemagglutinin (data not shown). Next, we considered the possibility that ABd T cells, although they did not express a TCR that is typical for human NK T cells, might nevertheless also be activated by αGalCer. However, ABd was not stimulated by αGalCer and PPBF did not activate an αGalCer-reactive NK T cell clone (Fig. 4 a and b).
Fig. 4.
PPBFs do not stimulate NK T cells. (a and b) The T cell line ABd and the Vα24+ T cell clone J3N.5 were stimulated with DC and a PPBF-containing antigen (Ag) mixture or αGalCer, and IL-2 release was measured after 24 h. (c) PPBF, 3-methyl PPBF and the indicated anilinonapthalene sulfonic acid (ANS) compounds were incubated in molar excess with αGalCer and washed, and then NK T cells were added for 14 h before measuring IL-2 release.
Next, to determine whether PPBF could compete with αGalCer-mediated NK T cell responses, we used an in vitro assay of PPBF added to recombinant CD1d proteins bound to a plate. When added in excess, PPBF and 3-methyl PPBF were both found to substantially block αGalCer-mediated NK T cell activation in a dose-dependent manner (Fig. 4c). The degree and titer of the inhibition by the PPBF analogs was comparable with that of 2,6-anilonapthalene sulfonic acid, a polyaromatic hydrocarbon that has been previously shown to bind to CD1d (37). Taken together, these results showed that NK T cells and ABd have differing antigen specificities and that PPBFs and αGalCer can compete for CD1d-mediated T cell activation.
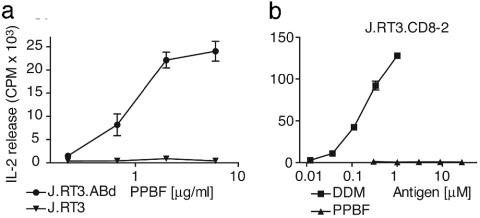
T Cell Activation by PPBF Is Mediated by the Clonotypic Vα2Vβ21 TCR. Because PPBF did not activate polyclonal T cells and NK T cells, it was unlikely to act by means of a receptor that is expressed on all T cells. In addition, previous studies had shown that CD1-lipid complexes generally activate T cells by contacting clonotypic TCRs (6, 36). To directly determine whether the response was TCR-mediated, we transfected the full-length TCR α and β chains from ABd into J.RT3 cells to generate a TCR-expressing reporter cell line (J.RT3.ABd). Transfection of both TCR α and β chains from ABd reconstituted surface-CD3 complexes and conferred the ability to produce IL-2 upon stimulation with PPBF (Fig. 5a).
Fig. 5.
PPBF recognition is mediated by variable regions of the TCR α and β chains. (a) J.RT3-T3.5 cells were transfected with the ABd TCR Vα2 and Vβ21 chains (J.RT3.ABd) or with empty vectors (J.RT3.mock) and tested for the ability to release IL-2 upon stimulation with PPBF. (b) To determine whether PPBF could activate through intact CD3 complexes that were reconstituted by transfection of a TCR with irrelevant antigen specificity, J.RT3-T3.5 cells transfected with the TCR α and β chains of the CD8-2 cell line (J.RT3.CD8-2) that recognizes DDM were tested for IL-2 release upon stimulation with DDM or PPBF.
Although these studies showed that the TCR α and β chains from ABd were sufficient to confer PPBF recognition, it remained possible that antigen recognition was mediated by invariant components of CD3 signaling complexes that are reconstituted by transfecting any TCR α and β chain. Therefore, we transfected J.RT3 cells with irrelevant Vα16 and Vβ9 TCR chains that were known to mediate recognition of the mycobacterial lipopeptide DDM. Transfection of the Vα16 and Vβ9 chains reconstituted both anti-CD3 signaling and DDM recognition but did not confer activation in response to PPBF. Taken together, these studies show that the recognition of PPBF was mediated by the clonotypic TCR of ABd.
Discussion
After decades of research on peptide antigens, the discovery of the CD1 antigen presentation has shown how lipids also function as the targets of T cell responses. Altered self-lipids or lipids from intracellular pathogens serve as natural targets of the cellular immune system. In addition, synthetic lipid antigens, like αGalCer, which are not normally encountered in the environment, can be used as drugs to modulate CD1-restricted T cell responses in an antigen-specific fashion. This new perspective has led to the use of αGalCer and other synthetic lipids as treatments in experimental models of cancer, autoimmune disease, and infection (39–43).
All of the previously known lipid antigens presented by the CD1 system contain alkyl chains. The crystal structures of CD1b and CD1a show how the fatty acids must pass through narrow channels within the CD1 groove (17, 18). Although human CD1d proteins have not been crystallized, the structure of murine CD1d shows that the groove is not organized into narrow channels found in CD1a and CD1b but instead has two pockets that are broadly contiguous, providing a more open, rounded cavity (16). The open conformation of CD1d might allow presentation of small molecules that lack alkyl chains and are instead polycyclic compounds. Supporting this concept, in vitro binding studies of human CD1 proteins have shown that whereas CD1b and CD1c preferentially bind to ligands with alkyl chains, CD1d binds to fluorescent probes composed of small, ringed hydrocarbon structures, including anilinonapthalene sulfonic acid compounds (37, 44). ANS compounds and PPBF block the presentation of αGalCer in cell-free assays, suggesting that these molecules bind to CD1d in or near the antigen binding groove (Fig. 4). Thus, the spectrum of antigens presented by the CD1 system may extend beyond conventional glycolipids with linear alkyl chains to include small molecules, such as sterols, polyketide antibiotics, or commonly used sulfa drugs that have structural homologies with ANS compounds. Our study shows that such nonlipidic small molecules can be recognized in association with CD1d proteins by describing human αβ T cells that are selectively activated by PPBF and structurally related compounds. PPBFs are synthetic rather than environmental antigens, but they share chemical features with commonly used sulfa drugs that generate strong hypersensitivity reactions in humans, including sulfisomidine, sulfadiazine, sulfasalazine, and celecoxib (45). Current studies are looking into the possibility that these antibiotics and antiinflammatory drugs may also be CD1-restricted antigens.
Because PPBFs differ structurally from all known classes of T cell antigens, we initially considered the possibility that the mechanism of action might involve activation pathways other than the clonotypic TCR. However, PPBF did not stimulate polyclonal T cells, an NK T cell clone, or T cell transformants with functional CD3 signaling complexes, ruling out ubiquitous receptors on T cells in the activation mechanism. Also, transfection of the clonotypic TCR was sufficient to confer PPBF responsiveness. This TCR-mediated response showed an absolute requirement for CD1d and PPBF, and PPBF was able to competitively inhibit formation of antigenic αGalCer-CD1d complexes in vitro (Figs. 1 and 4). Thus, the data are most consistent with a trimolecular interaction of the clonotypic TCR with CD1d and PPBF as the molecular mechanism of T cell activation. This hypothesis might be formally tested by staining T cells with PPBF-loaded CD1d tetramers, but initial experiments with such reagents have not detected an interaction, which may relate to lower affinity binding, cellular antigen processing, or other factors.
Conventionally, CD1d-restricted T cells are classified according to their TCRs, which can be invariant or diverse. Interest in the possibility that CD1d could mediate responses to a broader range of antigens has been stimulated by the discovery of diverse CD1d-restricted T cells in mice and their role in the response to infection and in autoimmune diseases in the mouse. Here, we provide evidence for an example of Vα24– CD1d-restricted T cells in humans. In this case, the ability to immunize T cells in vitro with PPBF provides proof of concept that human CD1d-restricted T cells can be activated by a compound with no structural relationship to αGalCer. This finding argues against the idea that human CD1d-restricted T cell repertoire is invariant. Whether CD1 can give rise to adaptive T cell responses, which show a diverse antigenic repertoire comparable to those of MHC class I and class II, is unknown and worthy of further study.
Acknowledgments
We thank William Biddison and Susan Gagnon for advice and PCR primers for TCR-variable regions. This work was supported by funding from the American Lung Association, the Pew Foundation Scholars in the Biomedical Sciences Program, the Cancer Research Institute, the American College of Rheumatology Research and Education Foundation, and by National Institute of Arthritis, Musculoskeletal, and Skin Diseases Grant AR 48632 and National Institute of Allergy and Infectious Diseases Grant AI 49313. G.S.B. is a Lister Jenner Research Fellow and acknowledges support from the Medical Research Council and the Wellcome Trust.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCR, T cell receptor; αGalCer, α-galactosyl ceramide; PPBF, phenyl 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonate; synPPBF, PPBF synthesized from phenol and fluorenylmethoxycarbonyl-L-Arg(2,2,4,6,7-pentamethyldihydrobenzofuransulfonate)-OH; DDM, didehydroxymycobactin; DC, dendritic cell(s).
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY532913 and AY532914).
References
- 1.Porcelli, S., Morita, C. T. & Brenner, M. B. (1992) Nature 360, 593–597. [DOI] [PubMed] [Google Scholar]
- 2.Spada, F. M., Koezuka, Y. & Porcelli, S. A. (1998) J. Exp. Med. 188, 1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckman, E. M., Melian, A., Behar, S. M., Sieling, P. A., Chatterjee, D., Furlong, S. T., Matsumoto, R., Rosat, J. P., Modlin, R. L. & Porcelli, S. A. (1996) J. Immunol. 157, 2795–2803. [PubMed] [Google Scholar]
- 4.Rosat, J. P., Grant, E. P., Beckman, E. M., Dascher, C. C., Sieling, P. A., Frederique, D., Modlin, R. L., Porcelli, S. A., Furlong, S. T. & Brenner, M. B. (1999) J. Immunol. 162, 366–371. [PubMed] [Google Scholar]
- 5.Beckman, E. M., Porcelli, S. A., Morita, C. T., Behar, S. M., Furlong, S. T. & Brenner, M. B. (1994) Nature 372, 691–694. [DOI] [PubMed] [Google Scholar]
- 6.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Motoki, K., Ueno, H., Nakagawa, R., Sato, H., Kondo, E., et al. (1997) Science 278, 1626–1629. [DOI] [PubMed] [Google Scholar]
- 7.Sieling, P. A., Chatterjee, D., Porcelli, S. A., Prigozy, T. I., Mazzaccaro, R. J., Soriano, T., Bloom, B. R., Brenner, M. B., Kronenberg, M., Brennan, P. J., et al. (1995) Science 269, 227–230. [DOI] [PubMed] [Google Scholar]
- 8.Joyce, S., Woods, A. S., Yewdell, J. W., Bennink, J. R., De Silva, A. D., Boesteanu, A., Balk, S. P., Cotter, R. J. & Brutkiewicz, R. R. (1998) Science 279, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 9.Schofield, L., McConville, M. J., Hansen, D., Campbell, A. S., Fraser-Reid, B., Grusby, M. J. & Tachado, S. D. (1999) Science 283, 225–229. [DOI] [PubMed] [Google Scholar]
- 10.Moody, D. B., Reinhold, B. B., Guy, M. R., Beckman, E. M., Frederique, D. E., Furlong, S. T., Ye, S., Reinhold, V. N., Sieling, P. A., Modlin, R. L., et al. (1997) Science 278, 283–286. [DOI] [PubMed] [Google Scholar]
- 11.Shamshiev, A., Donda, A., Carena, I., Mori, L., Kappos, L. & De Libero, G. (1999) Eur. J. Immunol. 29, 1667–1675. [DOI] [PubMed] [Google Scholar]
- 12.Shamshiev, A., Donda, A., Prigozy, T. I., Mori, L., Chigorno, V., Benedict, C. A., Kappos, L., Sonnino, S., Kronenberg, M. & De Libero, G. (2000) Immunity 13, 255–264. [DOI] [PubMed] [Google Scholar]
- 13.Moody, D. B., Ulrichs, T., Muhlecker, W., Young, D. C., Gurcha, S. S., Grant, E., Rosat, J. P., Brenner, M. B., Costello, C. E., Besra, G. S., et al. (2000) Nature 404, 884–888. [DOI] [PubMed] [Google Scholar]
- 14.Gilleron, M., Stenger, S., Mazorra, Z., Wittke, F., Mariotti, S., Bohmer, G., Prandi, J., Mori, L., Puzo, G. & De Libero, G. (2004) J. Exp. Med. 199, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moody, D. B., Young, D. C., Cheng, T. Y., Rosat, J. P., Roura-Mir, C., O'Connor, P. B., Zajonc, D. M., Walz, A., Miller, M. J., Levery, S. B., et al. (2004) Science 303, 527–531. [DOI] [PubMed] [Google Scholar]
- 16.Zeng, Z., Castano, A. R., Segelke, B. W., Stura, E. A., Peterson, P. A. & Wilson, I. A. (1997) Science 277, 339–345. [DOI] [PubMed] [Google Scholar]
- 17.Zajonc, D. M., Elsliger, M. A., Teyton, L. & Wilson, I. A. (2003) Nat. Immunol. 4, 808–815. [DOI] [PubMed] [Google Scholar]
- 18.Gadola, S. D., Zaccai, N. R., Harlos, K., Shepherd, D., Castro-Palomino, J. C., Ritter, G., Schmidt, R. R., Jones, E. Y. & Cerundolo, V. (2002) Nat. Immunol. 3, 721–726. [DOI] [PubMed] [Google Scholar]
- 19.Batuwangala, T., Shepherd, D., Gadola, S. D., Gibson, K. J., Zaccai, N. R., Fersht, A. R., Besra, G. S., Cerundolo, V. & Jones, E. Y. (2004) J. Immunol. 172, 2382–2388. [DOI] [PubMed] [Google Scholar]
- 20.Ulrichs, T., Moody, D. B., Grant, E., Kaufmann, S. H. & Porcelli, S. A. (2003) Infect. Immun. 71, 3076–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenger, S., Mazzaccaro, R. J., Uyemura, K., Cho, S., Barnes, P. F., Rosat, J. P., Sette, A., Brenner, M. B., Porcelli, S. A., Bloom, B. R., et al. (1997) Science 276, 1684–1687. [DOI] [PubMed] [Google Scholar]
- 22.Stenger, S., Hanson, D. A., Teitelbaum, R., Dewan, P., Niazi, K. R., Froelich, C. J., Ganz, T., Thoma-Uszynski, S., Melian, A., Bogdan, C., et al. (1998) Science 282, 121–125. [DOI] [PubMed] [Google Scholar]
- 23.Koseki, H., Imai, K., Nakayama, F., Sado, T., Moriwaki, K. & Taniguchi, M. (1990) Proc. Natl. Acad. Sci. USA 87, 5248–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lantz, O. & Bendelac, A. (1994) J. Exp. Med. 180, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benlagha, K. & Bendelac, A. (2000) Semin. Immunol. 12, 537–542. [DOI] [PubMed] [Google Scholar]
- 26.Gadola, S. D., Dulphy, N., Salio, M. & Cerundolo, V. (2002) J. Immunol. 168, 5514–5520. [DOI] [PubMed] [Google Scholar]
- 27.Gumperz, J. E., Roy, C., Makowska, A., Lum, D., Sugita, M., Podrebarac, T., Koezuka, Y., Porcelli, S. A., Cardell, S., Brenner, M. B., et al. (2000) Immunity 12, 211–221. [DOI] [PubMed] [Google Scholar]
- 28.Rauch, J., Gumperz, J., Robinson, C., Skold, M., Roy, C., Young, D. C., Lafleur, M., Moody, D. B., Brenner, M. B., Costello, C. E., et al. (2003) J. Biol. Chem. 278, 47508–47515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardell, S., Tangri, S., Chan, S., Kronenberg, M., Benoist, C. & Mathis, D. (1995) J. Exp. Med. 182, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behar, S. M., Podrebarac, T. A., Roy, C. J., Wang, C. R. & Brenner, M. B. (1999) J. Immunol. 162, 161–167. [PubMed] [Google Scholar]
- 31.Makowska, A., Kawano, T., Taniguchi, M. & Cardell, S. (2000) Scand. J. Immunol. 52, 71–79. [DOI] [PubMed] [Google Scholar]
- 32.Jahng, A., Maricic, I., Aguilera, C., Cardell, S., Halder, R. C. & Kumar, V. (2004) J. Exp. Med. 199, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu, Y. H., Jayawardena, J., Weiss, A., Lee, D., Park, S. H., Dautry-Varsat, A. & Bendelac, A. (1999) J. Exp. Med. 189, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber, S., Sartini, D. & Exley, M. (2003) J. Immunol. 170, 3147–3153. [DOI] [PubMed] [Google Scholar]
- 35.Baron, J. L., Gardiner, L., Nishimura, S., Shinkai, K., Locksley, R. & Ganem, D. (2002) Immunity 16, 583–594. [DOI] [PubMed] [Google Scholar]
- 36.Grant, E. P., Degano, M., Rosat, J. P., Stenger, S., Modlin, R. L., Wilson, I. A., Porcelli, S. A. & Brenner, M. B. (1999) J. Exp. Med. 189, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im, J. S., Yu, K. O., Illarionov, P. A., LeClair, K. P., Storey, J. R., Kennedy, M. W., Besra, G. S. & Porcelli, S. A. (2004) J. Biol. Chem. 279, 299–31038. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, Y., Gagnon, S. J., Kurane, I., Leporati, A. M. & Ennis, F. A. (1994) J. Virol. 68, 7614–7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, B., Geng, Y. B. & Wang, C. R. (2001) J. Exp. Med. 194, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi, M., Koseki, H., Tokuhisa, T., Masuda, K., Sato, H., Kondo, E., Kawano, T., Cui, J., Perkes, A., Koyasu, S., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 11025–11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naumov, Y. N., Bahjat, K. S., Gausling, R., Abraham, R., Exley, M. A., Koezuka, Y., Balk, S. B., Strominger, J. L., Clare-Salzer, M. & Wilson, S. B. (2001) Proc. Natl. Acad. Sci. USA 98, 13838–13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieuwenhuis, E. E., Matsumoto, T., Exley, M., Schleipman, R. A., Glickman, J., Bailey, D. T., Corazza, N., Colgan, S. P., Onderdonk, A. B. & Blumberg, R. S. (2002) Nat. Med. 8, 588–593. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa, R., Motoki, K., Nakamura, H., Ueno, H., Iijima, R., Yamauchi, A., Tsuyuki, S., Inamoto, T. & Koezuka, Y. (1998) Oncol. Res. 10, 561–568. [PubMed] [Google Scholar]
- 44.Ernst, W. A., Maher, J., Cho, S., Niazi, K. R., Chatterjee, D., Moody, D. B., Besra, G. S., Watanabe, Y., Jensen, P. E., Porcelli, S. A., et al. (1998) Immunity 8, 331–340. [DOI] [PubMed] [Google Scholar]
- 45.Pichler, W. J., Zanni, M., von Greyerz, S., Schnyder, B., Mauri-Hellweg, D. & Wendland, T. (1997) Int. Arch. Allergy Immunol. 113, 177–180. [DOI] [PubMed] [Google Scholar]