Abstract
MicroRNAs (miRNAs) are endogenous, ~22 nucleotide, non-coding RNA molecules that function as post-transcriptional regulators of gene expression. miRNA dysregulation has been observed in cancer and in neurodegenerative disorders such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS) and the neurological disorder, epilepsy. Neuronal degradation and death are important hallmarks of neurodegenerative disorders. Additionally, abnormalities in metabolism, synapsis and axonal transport have been associated with AD, PD and frontotemporal dementia (FTD). A number of recently published studies have demonstrated the importance of miRNAs in the nervous system and have contributed to the growing body of evidence on miRNA dysregulation in neurological disorders. Knowledge of the expressions and activities of such miRNAs may aid in the development of novel therapeutics. In this review, we discuss the significance of miRNA dysregulation in the development of neurodegenerative disorders and the use of miRNAs as targets for therapeutic intervention.
Keywords: miRNAs, Neurodegenerative diseases (NDDs), Alzheimer’s disease (AD), Parkinson’s disease (PD), Epilepsy, Huntington’s disease (HD), Amyotrophic lateral sclerosis (ALS), Epilepsy
1. miRNA Biogenesis
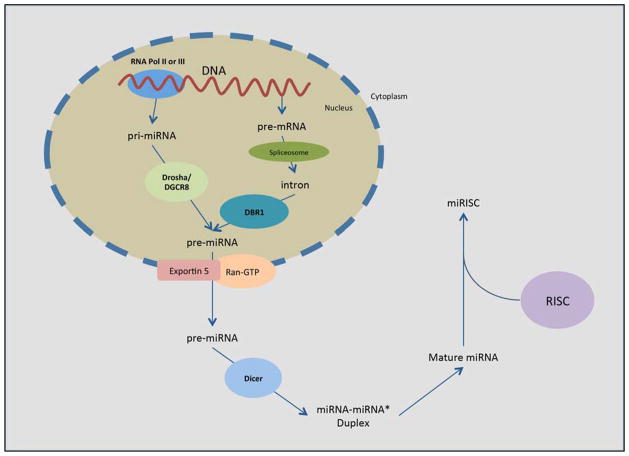
miRNAs are one of the most well-studied classes of non-coding RNA molecules. They are found in animals, plants and DNA viruses that regulate gene expression at the post-transcriptional level. miRNAs influence gene expression through degradation and translational repression of messenger RNA (mRNA) transcripts. An individual miRNA can have hundreds of targets, while a single target transcript may be regulated by many different miRNAs. miRNA biogenesis begins in the nucleus when 1–3 kilobase (kb) precursor miRNAs (pre-miRNAs) are generated either by microprocessor complex (Drosha and DGCR8) RNase III enzyme processing of primary miRNA (pri-miRNAs) transcripts or by spliceosome processing of precursor mRNA (pre-mRNA) transcripts followed by Lariat Debranching Enzyme (DBR1) processing of the resulting introns [1, 2]. pre-miRNAs can be produced from two separate processes because miRNA-encoding loci are present in both intragenic and intergenic chromosomal DNA regions [3, 4]. After they are transported from the nucleus into the cytoplasm by the karyopherin Exportin 5 and Ran-GTP [5–7], the 70–100 nucleotide pre-miRNAs are processed by the RNase III enzyme Dicer [8]. The resulting 20–25 nucleotide mature miRNAs are subsequently incorporated into the RNA-induced silencing complex (RISC) in order to pair with the 3′ untranslated regions (3′-UTR) of target mRNAs and cleave them using the Argonaute (AGO) family proteins (AGO1–4), which are the catalytic components of RISC [9]. The active RISC/miRNA complex is called a miRISC. miRISC induces downregulation of target transcript expression by either degradation of the mRNA or translational repression (Fig. 1).
Fig 1. Biogenesis pathway of miRNA, and miRISC.
pre-miRNA is produced in the nucleus via either transcription of pri-miRNA and Drosha/DGCR8 processing, or via DBR1 processing of pre-mRNA introns. pre-miRNA is exported from the nucleus to the cytoplasm via Exportin 5 and Ran-GTP and processed by Dicer to produce the miRNA-miRNA* duplex. The two strands comprising the duplex disassociate to form two mature miRNAs. One mature miRNA is incorporated into RISC to produce miRISC and enact translational repression of target transcripts.
2. Neurodegenerative diseases
Neurodegenerative disorders (NDDs) are characterized by the progressive loss of neuronal function and structure, which eventually leads to neuronal death in the nervous system. NDDs include Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS). Pathological cellular and molecular events in neurodegeneration include oxidative stress, protein oligomerization and aggregation, axonal transport deficiency, calcium deregulation, mitochondrial dysfunction, neuroinflammation, neuron–glial interactions, aberrant RNA processing and DNA damage. The greatest risk factor for neurodegeneration is age in combination with mitochondrial DNA mutations and neuronal damage from oxidative stress. Other possible risk factors include gender, endocrine conditions, poor education, inflammation, stroke, hypertension and diabetes. Due in part to the gains in knowledge of the human genome, made possible by next-generation sequencing and similar techniques, there is increasing evidence of the involvement of miRNAs in the development of the central nervous system and in neurodegeneration[10]. Recent reports have shown that aberrant miRNA expression contributes to neurodegeneration, and that NDDs may be considered as a class of RNA disorders [11, 12]. The failures of a number of surgical and pharmacological treatments for NDDs highlight the need for novel therapeutic approaches to regulate several targets and molecular pathways. The inherent ability of miRNAs to regulate many transcripts makes them ideal candidates for investigation towards the development of therapeutics for NDDs.
2.1 Alzheimer’s disease
Alzheimer’s disease (AD) is characterized by neuronal inflammation and death in areas of the brain that affect memory, behavior, language, and cognition. AD brain contains several pathological structures: intracellular Hirano bodies [13] and neurofibrillary tangles (NFTs), as well as extracellular amyloid-β (Aβ) plaques. These irregular structures are believed to be the cause of the extensive neuronal degeneration observed in AD. Hirano bodies are rod-shaped actin aggregates that contain cofilin, a protein that negatively affects the cytoskeleton in AD and plays a dynamic role in the pathogenesis of the disease [14, 15]. NFTs are intracellular, insoluble aggregates of hyperphosphorylated microtubule-binding protein tau. Amyloid-β (Aβ) deposition is also one of the hallmarks of neuronal degeneration in AD brain. Aβ is generated from the sequential cleavage of amyloid precursor protein (APP) by beta-site APP cleaving enzyme 1 (BACE1) and γ-secretase.
Many miRNAs have been found to be dysregulated in the brains of AD patients [16, 17]. Decreased brain levels of miR-29a, miR-29b-1, and miR-9 are associated with AD [16]. One review by Maes et al. demonstrated the potential diagnostic role of the AD-specific let-7 miRNAs (let-7f, let-7b, and let-7i), as well as miR-9, miR-181, and miR-29 [18]. miR-26b expression was found to be elevated in the substantia nigra of AD brain during disease progression [19]. Dysregulation of miR-146 is also observed in AD brains. Small-scale studies in the hippocampus of fetal, adult, and AD brains [20], as well as large-scale analyses profiling AD brain, peripheral blood, and CSF [16, 17, 21–24] have demonstrated the use of miRNAs as potential biomarkers of disease. Decreased expressions of miR-298, miR-328 and miR-195 have been observed in AD model mouse brain [25, 26].
miRNA dysregulation is also observed in the blood and cerebrospinal fluid (CSF) of AD patients. The levels of 60 miRNAs were found to be significantly altered in the CSF of AD patients as compared to normal volunteers in a recent large-scale miRNA expression profile study [22]. Quantitative real-time PCR (qRT-PCR) showed significant alterations in the expressions of miR-34a, miR-181b and let-7f in blood from late-onset AD patients when compared to age-matched controls (n =17 per group) [21]. The same study used a miRNA microarray in blood mononuclear cells (BMC) from patients with sporadic AD to show significant upregulation of miR-34a and miR-181b. A fluorescent miRNA array study in AD CSF and short post-mortem interval, brain tissue-derived extracellular fluid (ECF) showed significant increases in the expression levels of miR-9, miR-146a, miR-125b, and miR-155 [20].
Recent laboratory and clinical research studies have used a standard workflow of assays [27] to provide evidence of causal relationships between brain miRNA dysregulation and Aβ production leading to neurodegeneration [16, 23, 28]. Many of these studies have identified miRNAs that influence the expression of APP, and thus effect Aβ production [29]. One such study indicated that the downregulation of miR-29b in peripheral blood mononuclear cells (PBMC) might be involved in the pathogenesis of AD [30] due to the fact that miR-29b targets transcription factor Sp1, which in turn promotes the transcription of APP [31] and tau [32]. miR-375 has also been shown to target Sp1, though it’s downregulation did not influence Sp1 expression in the PBMC study [33]. miR-106a and miR-520c, both members of the miR-20a family that includes miR-20a, miR-106a/b, miR-17, have been found to inhibit the expression of APP in vitro [34]. miR-17 and miR-20a, along with miR-147, -153, -323-3p, -644 and -655 were subsequently found to directly target APP [35], as was miR-16 in 2012 [36]. Finally, Long and colleagues released two studies in 2011 and 2012 showing that miR-101 and miR-153 both inhibit APP expression, and that miR-153 is downregulated in the brains of advanced AD patients [37, 38].
A number of studies have also identified dysregulated miRNAs that influence Aβ production through their regulation of BACE1, the APP-cleaving enzyme that initiates the generation of Aβ. BACE1 appears to be one of the many miRNA target transcripts that are targeted by multiple miRNAs, a number of which are dysregulated in AD. For example, downregulation of miR-107 was associated with increased expression of BACE1 [39]. miR-107, however, has not been shown to directly target BACE1. Studies of miR-29c over-expression in HEK-293T and SH-SY5Y cells, as well as in transgenic mouse models of AD, have shown decreases in BACE1 proteins levels [40]. BACE1 expression was also shown to increase in response to inhibition of miR-124 in cultured rat PC12 cell lines and primary cultured hippocampal neurons [41]. This result is particularly significant, considering that the expression of miR-124 was observed to be downregulated in the brains of AD patients [42]. miR-124 has also been shown to regulate polypyrimidine tract binding protein 1 (PTBP1), an RNA-binding protein that inhibits neuronal splice variants and increases AD-related splice variants of APP [43]. Another study found that miR-339-5p downregulates BACE1 expression in human primary cortical neurons and that it is expressed at significantly lower levels in brain tissues of AD patients as compared to age-matched controls [44]. Additionally, miR-195, -298 and -328 have all been shown to regulate BACE1 in mouse models of AD [25, 26].
Additional studies have identified dysregulated miRNAs that contribute to AD pathogenesis through aberrant regulation of various proteins involved in the progression of the disease. Downregulated expression of miR-103 or miR-107 can lead to increased cofilin 1 (CFL1) levels in AD brains, which may contribute to the formation of intracellular Hirano bodies [45]. Another study showed that changes in neuronal miR-802 expression in the CSF of AD patients (n=14) were correlated with decreased caveolin-1 expression as compared to that of normal controls (n =8) [46]. Increased expression of miR-98 is known to be involved in the negative regulation of insulin-like growth factor 1 (IGF-1), which results in increases in Aβ expression, and in tau phosphorylation [47]. Decreased expressions of miR-137, miR-181c, miR-9, miR-29a and miR-29b-1 in AD brain lead to upregulation of serine palmitoyltransferase (SPT) long chain subunit 1 (SPTLC1) and 2 (SPTLC2). These proteins are the essential components of SPT, the rate-limiting enzyme in the synthesis of ceramides, which are known to be upregulated in AD [48, 49]. miR-34a-mediated inhibition of tau protein was identified using multiple prediction algorithms [50]. One recent study showed that miR-15a regulates extracellular signal-regulated kinase 1 (ERK1), which is involved in tau hyperphosphorylation [51], while another found that miR-15a was correlated with neuritic plaque score in AD patients [52]. Increased expression of miR-26b, in addition to decreased expression of retinoblastoma Rb, causes enhanced tau phosphorylation and leads to apoptosis and neurodegeneration in primary cortical neurons [19]. One study has shown that PSEN2 may regulate microglia activity via miR-146 [53]. Finally, a number of proteins and pathways involved in both AD and cancer have been found to be modulated by dysregulated miRNAs [54].
2.2 Parkinson’s Disease
Parkinson’s disease (PD) is an age-associated neurodegenerative disorder. This disease is quite heterogeneous [55] and characterized by the loss of dopaminergic neurons and/or presence of Lewy bodies in the midbrain. Clinical symptoms associated with the disease include rigidity, bradykinesia and tremors [56]. This disease can also spread to other regions of brain, such as the cingulate gyrus, amygdala and higher cortical regions, thus leading to dementia and psychosis. Although there is a substantial body of research on PD, the currently available methods for diagnosis and treatment are at preliminary stages, particularly in relation to early disease. The emerging field of miRNA research in relation to PD has the potential to generate novel diagnostic and therapeutic strategies.
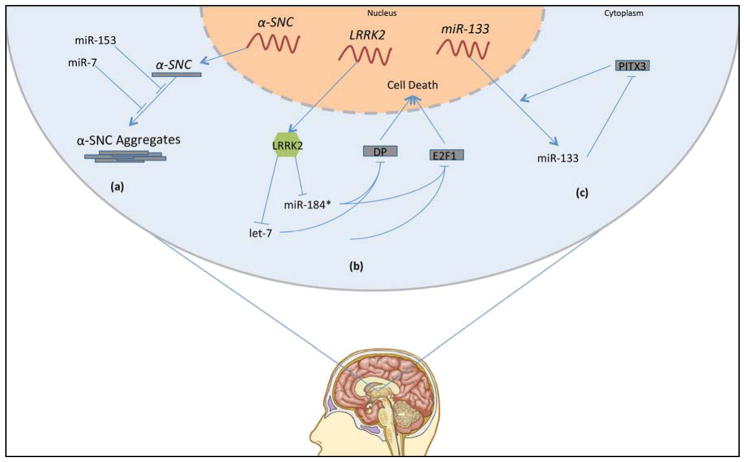
Comparative studies of miRNA profiles in postmortem brain tissue from PD patients and healthy controls show that aberrant miRNA levels are an important component of this neurodegenerative disease. For example, the expression of the PD-associated protein α-synuclein is targeted by miR-7 and miR-153. Single-nucleotide polymorphisms in fibroblast growth factor 20 (FGF20), a protein whose overexpression is considered a risk factor for PD, prevented its translational repression by miR-433 and led to increased levels of α-synuclein in Neuro2A cells[57, 58] (Fig. 2a). Mitochondrial dysfunction has also been reported to be a result of miRNA dysregulation in PD [59].
Fig 2. Parkinson disease molecular mechanisms.
(a) miR-7 and -153 suppress translation of α-synuclein (α-SNC). (b) LRRK2 antagonizes let-7 and miR-184, both of which inhibit translation of DP and E2F1. DP and E2F1 overexpression is toxic to neurons. (c) miR-133 inhibits translation of PITX3. PITX3 is a transcription factor that induces transcription of miR-133 in addition other genes relevant to dopaminergic neuron differentiation and maintenance.
Several miRNAs found to be dysregulated in PD brain have been studied in depth, particularly miR-133 and miR-34. miR-133b was abnormally expressed in an analysis of 224 different miRNAs isolated from the midbrain of PD and control subjects [60]. miR-133b expression was also found to be decreased in PD patients and to be mainly involved in the regulation of dopaminergic neuron development. The exact role of miR-133b in dopaminergic neuron maturation, however, has yet to be thoroughly evaluated. One of the physiological targets of miR-133b is Pitx3, a transcription factor that plays a key role in dopaminergic neuron differentiation [61–63] (Fig. 2c). Interestingly, Pitx3 promotes the transcription of miR-133b, which in turn decreases Pitx3 expression in a negative-feedback loop mechanism.
A considerable amount of research has been devoted to miR-34 dysregulation in PD, as well. Decreased levels of miR-34b and c were observed in the frontal cortex, cerebellum, and amygdala of Area 17 brain. Downregulation of miR-34b and c was also observed in the substantia nigra of PD brains. miR-133b expression was not found to be altered in any of the areas in which miR-34b and c downregulation was observed [64]. miR-34b and c were also suggested to indirectly reduce the expressions of both Parkin and Parkinson protein 7 (DJ1), as well as increase the rate of cell death. For example, cell death, altered mitochondrial activity and oxidative stress were observed in differentiated SH-SY5Y neuroblastoma cells in which miR-34b and c were silenced. Further investigation is required into the roles of the aforementioned miRNAs in PD-relevant regulatory cellular pathways, as well as the potential physiological targets of these miRNAs.
Two additional studies demonstrated the involvement of dysregulated miRNAs in the modulations of systems relevant to PD pathology. Gehrke and colleagues showed that leucine-rich repeat kinase 2 (LRRK2), a protein to which gain-of-function mutations induce familial and sporadic PD, reduces the levels of let-7 and miR-184*, two microRNAs that normally repress the translation of the transcription factors E2F1 and DP [65]. Overexpression of E2F1 and DP in multiple models of let-7 and miR-184* downregulation led to increased cell death and pathogenicity similar to that observed in LRRK2 gain-of-function models (Fig. 2b). This study suggests that the pathogenicity of LRRK2 mutation in PD may be mediated by the subsequent downregulation of let-7 and miR-184* and resulting upregulation of the toxic transcription factors E2F1 and DP.
2.3 Huntington’s disease
Huntington’s disease (HD) is an incapacitating, hereditary neurodegenerative disorder distinguished by progressive impairment of motor and cognitive functions, and is induced by a mutation in exon 1 of the huntingtin (Htt) gene. Present on chromosome 4 in humans, this mutation takes the form of a polyglutamine tract CAG trinucleotide repeat expansion in the N-terminus of ubiquitously expressed Htt protein [66]. Whereas Htt polyglutamine tracts within the normal population have up to 35 CAG repeats, those of HD patients are expanded to 36 or more CAG repeats [67]. Although the exact molecular function of endogenous normal (wild type) Htt has yet to be fully characterized, genetic data in both transgenic animal HD models and humans imply that excess polyglutamine expansions confer a deleterious gain-of-function on target proteins [68], although loss-of-function effects also have been described [69]. Htt is present within the nucleus and appears to impact the transcription of the numerous genes that are dysregulated in HD; in particular those mediated via the cAMP response element binding (CREB) protein, retinoic acid receptor (RAR), and specificity protein-1 (Sp1) [70–72]. Of key relevance, Htt appears to be involved with modulation of brain-derived neurotrophic factor (BDNF) expression, vesicle trafficking, clathrin associated endocytic pathways and cytoskeletal configuration [69, 73–75]. The mutated Htt form, consequent to its polyglutamine expansion, possesses an aberrant protein structure that leads to its aggregation in brain that, in addition to its transcription-induced actions, invariably results in neuronal degeneration [76].
A next-generation sequence analysis in 26 HD and 36 control subjects suggests that among the 938 miRNAs present in quantifiable amounts in both HD and control brain, 75 are differentially regulated [77]. Across a number of studies, miR-129a, miR-132, miR-29a, miR-29b, miR-330, miR-17, miR-196, miR-222, miR-485, and miR-486 have been reported to be dysregulated in HD due, in large part, to the disrupted miRNA transcriptome [78, 79]. Downregulation of miR-22, miR-29c and miR-128 has been reported in mice, while miR-29b, and miR-124a have been found to be downregulated in humans [80, 81].
A number of miRNAs have been observed to be upregulated in HD brain. One study detected increases in the expressions of miR-30b, miR-30c and miR-30e in HD-FC (frontal cortex) and HD-ST (striatum). The same study found decreased expressions of let-7a, let-7c, let-7d, let-7e, miR-221 and miR-222 in HD [78]. Another study demonstrated upregulation of five miRNAs in human HD brain: miR-10b-5p, miR-196a-5p, miR-196b-5p, miR-615-3p and miR-1247-5p, all of which are located intergenica to Hox gene clusters except for miR-1247-5p [82]. An additional study suggested that miR-34b could be used as a potential diagnostic biomarker for HD since its expression was increased in HD before the onset of symptoms [83, 84].
Several miRNAs that target transcripts relevant to HD pathology have been investigated. Downregulation of miR-132 and increased expression of p250GAP have been observed in the brains of mouse models of HD and in the postmortem brains of HD patients [79]. The p250GAP gene encodes a protein that is a member of the Rac/Rho family of GTPase-activating proteins and that is involved in the inhibition of neurite growth [85]. Exogenous expression of miR-214, miR-150, miR-146a and miR-125b reduced HTT aggregate formation in mouse cells [86]. A comparative analysis of networks has identified two upregulated miRNAs: miR-200a and miR-200c, which regulate genes involved in mutant HTT-induced neuronal dysfunction, including abnormal synaptic transmission and perturbed neurogenesis [87].
One particularly well-characterized system of miRNA dysregulation in HD is that of miR-9/9*, and its regulation of REST/CoREST. Decreased expressions of miR-9 and 9* were observed in the cerebral cortex of HD-affected subjects and correlated with disease progression [84]. miR-9 regulates REST, while miR-9* regulates CoREST. REST and CoREST are both components of the repressor element 1 (RE1) silencing transcription factor (REST) complex, which is known to suppress the expression of neuronal genes in non-neuronal cells. Overexpression of REST in neurons is pathological. In healthy neurons, REST is bound to Htt and partly sequestered in the cytoplasm [88]. In diseased neurons, due to pathogenic polyglutamine expansions in Htt, REST disassociates from Htt and translocates to the nucleus, where it decreases neuronal gene expression and eventually causes neuronal death. Decreased levels of miR-9/miR-9* in HD brain lead to increased expression of REST/CoREST and facilitate translocation of REST from the cytoplasm to the nucleus. Due to the fact that miR-9 and 9* are regulated by REST, their dysregulation leads to a pathogenic positive feedback loop that continually downregulates their own transcription, which, in turn, upregulates REST.
Another consequence of REST overexpression is downregulation of BDNF. BDNF is a REST-modulated neuronal gene. Decreased expression of BDNF leads to loss of medium spiny neurons [69]. BDNF is also related to miRNA dysregulation due to the fact that BDNF protein levels in HD-FC and HD-ST appear to be downregulated in response to increased expression of members of the miR-30 family of miRNAs, particularly miR-30a-5p [89].
2.4 Amyotrophic lateral sclerosis (ALS)
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, is an NDD characterized by abnormal function in muscle tissue, such as atrophy and paralysis. The disease leads to death within a span of 3 to 5 years from symptom onset. ALS affects nerve cells within the spinal cord brainstem and cortex, causing gradual degeneration and death of motor neurons [90, 91]. A genetic etiology has been recognized in up to 20% of seemingly sporadic and 60% of familial ALS cases, with the involvement of at least 16 genes and genetic loci being associated with pathogenesis [92]. Key amongst these are mutations within the superoxide dismutase 1 (SOD1), TARDBP, which encodes the transcriptional repressor TAR DNA-binding protein 43 (TDP-43), and FUS genes. The most common genetic cause of ALS relates to the C9orf72 gene, identified from expanded repeats in a noncoding region of chromosome 9 open reading frame 72 [93]. Rare mutations in additional genes, including, ANG, VAPB, DAO, OPTN, VCP, and UBQLN2, have also been reported [91]. Defective RNA metabolism and neuronal cytoplasmic protein aggregation are common pathogenic mechanism for ALS. Mitochondrial dysfunction, excitotoxicity, and oxidative stress are evident and ultimately lead to a loss of neuromuscular junction integrity, retrograde axonal degeneration, and motor neuronal cell dysfunction and death [92]. The currently available drug Riluzole, that blocks tetrodotoxin-sensitive sodium channels, modestly slows the progression of ALS but does not prevent the life-ending consequences of the disease. A significant portion of recent ALS research has investigated the involvement of miRNAs in the pathogenesis of the disease.
Involvement of miRNAs in ALS was suggested by a Dicer deletion study performed in a mouse model of ALS. Deletion of Dicer (the RNase responsible for converting pre-miRNAs into mature miRNAs [94]) in post-mitotic motor neurons led to motor neuron degeneration, which is similar to the neuromuscular phenotype observed in mouse models of spinal muscular atrophy (SMA) [95]. A number of studies have since demonstrated miRNA dysregulation in ALS. For example, increased expression of the skeletal-muscle-specific miR-206 was observed in the muscle of a mouse model of ALS, which carries a mutation for SOD1 [96]. Increased expression of miR-338-3p, as well as down-regulation of several other miRNAs was observed in the leukocytes of ALS patients in a miRNA profiling study of 14 ALS patients and 14 controls [97]. One study by Haramati et al. in 2010 identified miR-9 and 9* dysregulation in mutant embryonic-stem-cell-derived motor neurons.
A number of other studies have identified ALS-relevant transcripts and pathways that are targeted by dysregulated miRNAs. Recent findings have shown that increased expressions of miR-23a, miR-29b, and miR-455 in skeletal muscle of ALS patients leads to mitochondrial gene expression [98]. MiR-132, miR-134, and miR-9 are known to be involved in synaptic plasticity and neuronal development [99]. miR-132 regulates genes such as acetyl cholinesterase and polypyrimidine tract binding protein 2 (PTBP2), which influences alternative splicing of tau [100–104], while miR-134 is involved in the regulation of neuronal development and dendritogenesis [105].
Two particularly well-studied and ALS-relevant systems that have been connected to miRNA dysregulation are those of TDP-43 and FUS/TSL. Dominant mutations have been observed in the trans-activating response region (TAR) DNA-binding protein 43 (TDP-43) in ALS patients [106–109]. TDP-43 is an important component of ubiquitinated protein aggregates observed in the motor neurons of sporadic ALS patients. Mutations are also found in the gene encoding fused in sarcoma/translocated in sarcoma (FUS/TLS) in ALS patients [110, 111]. The pathological effects of FUS/TLS mutations are similar to those of TDP-43. Ling et al. showed that TDP-43 associates with the Drosha/Pasha microprocessor complex, as well as proteins in the heterogeneous nuclear ribonucleoproteins family [112]. Normal, and particularly mutant TDP-43, interact with FUS/TSL [113]. Dysregulation of miRNA biogenesis was caused by misallocation of TDP-43 and FUS/TLS. The mutation in TDP-43 led to differential expressions of miR-132, miR-143 and miR-558, all of which are involved in ALS [114].
3. Epilepsy
Epilepsy is a chronic neurological disorder that is characterized by seizures, which are episodes of abnormal, hypersynchronous neuronal activity within the brain. Affecting about 1–2% of the population, it is now recognized that a tally of in excess of 500 genes and genetic loci are linked to seizures [115], and these possesses a broad biological diversity – covering almost every facet of neurotransmission.
There is a considerable amount of research on miRNA dysregulation in epilepsy. A recent study used TaqMan low-density arrays (TLDAs) to show that 51% of miRNAs detected in control hippocampus were downregulated in epileptic hippocampus. An additional 24% of miRNAs found in control brain were not found in epileptic brain [116]. Several miRNAs have shown aberrant regulation in a rat model of temporal lobe epilepsy as well. miR-23a, -27a, -31, -33, -34a, 146a, -152, -203, -210 and -211 were up-regulated, while miR-19a, -135b, -136, -138*, -144, -153, -190, -296*, -301a, -325-5p, -380, -542-3p, -542-5p and -543 were down-regulated [117]. A genome-wide miRNA profiling study revealed a number of miRNAs involved in human mesial temporal lobe epilepsy (MTLE) pathogenesis. This study also suggested that altered miRNA levels influence the expression of immunomodulatory proteins in MTLE. One recent study showed that 21 miRNAs were upregulated, whereas 12 miRNAs were down-regulated in focal-onset status epilepticus brain [118].
Many of the miRNAs that have been shown to be dysregulated in epileptic brain are involved in cellular processes relevant to the disease, such as cell proliferation and migration, neuroinflammation, and neuronal apoptosis. Several dysregulated miRNAs in epileptic brain appear to have significant effects on cell proliferation and migration. For example, Dicer depletion studies using the Nestin-Cre system have demonstrated the role of Dicer, and thus miRNAs, in cortical neuron migration [119]. Dicer depletion increases the expression of Doublecortin (Dcx), a protein that regulates tangential and radial neuron migration. Increased Dcx expression leads to premature maturation of neurons in the cortex. Studies have also shown that miR-134 may be involved in the regulation of cell migration through an interaction with Dcx [120, 121]. Reduction of cortical layers and disruption of interneuron tangential migration from basal forebrain were reported in miR-9 knockout mice [122]. Decreased miR-9 expression is correlated with reduced proliferation, as well as enhanced migration of human embryonic neural progenitors due to miR-9’s targeting of stathmin, which causes microtubule instability in migrating neuroblasts [123]. Finally, the role of miR-137 in the induction of premature differentiation and outward migration via its regulation of lysine-specific histone demethylase 1A (KDM1A) has been demonstrated in embryonic neural stem cells [124]. Neural stem/progenitor cell migration also increased upon introduction of exogenous miR-125b [125].
Dysregulated miRNAs connected to inflammation have been identified in a number of studies. For example, significant upregulation of miR-146a has been found in MTLE patients [126, 127]. This miRNA has been implicated in the astrocyte-mediated inflammatory response in the brain [128]. A follow-up study showed significant upregulation of miR-146a in astrocytes exposed to interleukin-1 beta (IL-1b) [129]. Additionally, miR-221 and miR-222 regulate ICAM1 expression and were shown to be expressed concurrently with ICAM1 in astrocytes of MTLE patients [130]. The involvement of dysregulated miRNAs in MTLE inflammation has been reported elsewhere [131]. miR-155 has also been implicated in inflammatory pathways related to MTLE. One study demonstrated upregulation of miR-155 in an immature rat epilepsy model [132]. The same study showed a positive correlation between miR-155 expression and TNF-α expression in nervous tissue. Dysreuglated miRNAs have been implicated in the inflammatory response of focal cortical dysplasia (FCD) a common cause of epilepsy characterized by the presence of giant/dysmorphic neurons, balloon cells, and cell abnormalities [133–136]. In situ hybridization studies have shown that miR-146a is upregulated in reactive astrocytes of dysplastic cortex in FCD type IIb, suggesting that miR-146a may play a role in FCD-related inflammation.
miRNAs have also been implicated in the overexpression of adenosine kinase (ADK) observed during seizures and in the neuronal apoptosis that occurs in epileptic brain. For example, lentiviral-mediated miRNA expression targeting ADK transduction in human mesenchymal stem cells led to an 80% reduction in ADK expression [137, 138]. As for the effects of miRNAs on neuronal apoptosis, recent evidence suggests that miR-34a has direct pro-apoptotic effects in cells and plays a role in the regulation of p53 [139]. In non-neuronal cells, upregulation of miR-34a has been reported to lead to apoptosis [140]. The aforementioned studies suggest that miR-34a may play an important role in the mechanisms underlying neuronal death induced by seizures [117, 141]. These studies have shown miRNAs to be key players in seizure-induced neuronal death and novel therapeutic targets for epilepsy.
In addition to the many studies on dysregulated miRNAs related to cell proliferation and migration, inflammation and apoptosis, a number of studies have investigated individual dysregulated miRNAs that are involved in processes more specific to epileptic pathology. miR-134, in particular, has been the subject of multiple studies. miR-134 is brain-specific and known to be actively involved in the regulation of neuronal microstructure. miR-134 regulates LIM domain kinase 1 (Limk1), which inactivates cofilin via phosphorylation [142, 143]. Over-expression of miR-134 in neurons of the postnatal mouse brain was also shown to reduce dendritic length. One qPCR study investigated the gene expression of brain-specific, inflammation-related miRNAs in the hippocampal tissues of rats at three different stages of MTLE, as well as in those of healthy control rats [144]. Whereas miR-132 and miR-21 have been shown to be upregulated in acute and chronic stages of MTLE, miR-124 and miR-134 were shown to have similar expression patterns in all three stages of disease. miR-132 was upregulated and miR-21 was down-regulated in the latent stage. The upregulation of miR-124 and miR-134 in MTLE could provide potential targets for the treatment of developing epileptic brains.
4. Therapeutic role of miRNAs in Neurodegenerative diseases and Epilepsy
Evidence of the involvement of miRNAs in signaling cascades that regulate cell fate, brain development and, most importantly, neurodegeneration has been provided by a multitude of recent studies. miRNA expression profile studies in several neurodegenerative disorders have provided important information on the role miRNAs in the progression of neurodegeneration. These studies have also indicated potential roles for miRNAs as biomarkers used in the diagnosis and prognostication of NDDs, and as targets for novel therapeutic strategies. Ectopic expression and silencing studies on targeted miRNAs both in vitro and in vivo have been shown to alter miRNA expression during the development of NDDs.
miRNAs can be classified as tumor-inducing oncogenes or as tumor suppressor genes due to their diverse regulatory functions. Both suppression of upregulated tumor-inducing miRNAs and replacement of down-regulated miRNAs by re-introduction of tumor-suppressor miRNAs have been proven to be functional therapeutic methods for combating cancer. Anti-miRNAs have been synthesized to silence or block a target miRNA that acts as an oncogene. Anti-miRNAs include cholesterol-conjugated “antagomirs”, polylysine-conjugated peptide nucleic acids (PNAs) and locked nucleic acid (LNA) oligonucleotides, all of which have demonstrated effectiveness in vivo [145].
Several obstacles are associated with the therapeutic use of both miRNA agonists and antagonists in vivo. These obstacles include endothelial barriers, glomerular filtration, and hepatic metabolism. Chemical modifications to miRNAs, as well as non-viral vectors have been developed to overcome such barriers [146–150]. Delivery of miRNA across the blood-brain barrier (BBB), however, remains to be the most challenging aspect of the treatment of NDDs with miRNA-based therapeutics. Potassium channel agonists and minoxidil sulfates could help facilitate the transport of miRNA molecules across the BBB [151, 152]. Banks and colleagues have also demonstrated the ability of APP and lipoprotein receptor related protein-1 (LRP-1) antisense phosphorothioate oligodeoxynucleotides to cross the BBB, and reduce or enhance AD pathology, respectively [153, 154]. Recent studies have shown that exosomes can serve as good delivery agents for miRNAs across the BBB because they can protect miRNAs from RNAase agents in the cell and deliver them to specific targets [155–158]. One study showed successful knockdown of BACE1 expression when exosomes with siRNAs targeting BACE1 were delivered intravenously [159]. These exosomes were derived from dendritic cells and bioengineered to express Lamp 2b fused to the acetylcholine receptor.
LNA-modified anti-miRNAs could enhance the specificities and reduce the effective doses of anti-miRNA molecules [160]. The endonucleolytic DNAzyme motif substation in LNA-based anti-miRNAs and in siRNA could enhance their silencing ability, in addition to their stability [161, 162]. Several factors, such as dosage, duration of activity and clearance of anti-miRNA molecules, need to be evaluated before successful miRNA delivery to target sites in in vivo models of neurodegeneration is achieved. Synthetic sponge miRNAs have been developed to inhibit endogenous miRNAs in recent years. These sponge miRNAs have complementary binding sites to a miRNA of interest and inhibit specific miRNA sequences rather than non-specific sequences that affect many miRNAs and other genes [163–169]. Expression of the sponge sequences with cytomegalovirus (CMV) promoters can enhance the efficiency of miRNA sponges. Even though sponge technology has shown high specificity towards target miRNAs, in addition to having several other advantages, further studies are required to develop the technology into a clinical strategy.
5. Conclusion
Increasing knowledge of the roles that miRNAs play in complex diseases such as NDDs and epilepsy will help to improve our understanding of basic cellular disease mechanisms, and our ability to modulate miRNA expression for therapeutic ends. miRNAs have been shown to be involved in the regulation of many signaling pathways that are key to the progression of NDDs and epilepsy (Table 1). It is important to note that several microRNAs target more than one protein related to the diseases discussed in this review. The miR-29 family, for example, targets BACE1, SPTLC2 and SP1, all of which encode proteins relevant to AD. Similarly, miR-124 influences AD pathology by targeting both BACE1 and PTBP1. let-7 and miR-184 both target two transcription factors that are pathologically dysregulated in PD, while the miR-9 family targets two essential components of a HD-relevant transcription factor, miR-132 targets two ALS-relevant proteins, and miR-134 targets two protein relevant to epilepsy.
Table 1.
Summary of miRNAs Regulating Disease-Relevant Transcripts
| Target mRNA (Protein) | Protein Expression in Disease Brain | miRNA Regulator(s) | miRNA Expression in Disease Brain |
|---|---|---|---|
| Alzheimer’s Disease (AD) | |||
| APP (Amyloid Precursor Protein) | - | miR-16 [36] | Down [36, 176] |
| miR-17 [35] | Normal [176] | ||
| miR-20a [35] | Normal [176] | ||
| miR-101 [177] | Up [176]; Down [178, 179] | ||
| miR-106a [172] | Normal [176] | ||
| miR-147 [35] | Normal [176] | ||
| miR-153 [35, 180] | Up [176]; Down [180] | ||
| miR-323-3p [35] | Normal [176] | ||
| miR-520c [172] | Normal [176] | ||
| miR-644 [35] | - | ||
| miR-655 [35] | - | ||
| BACE1 (β-site amyloid precursor protein cleaving enzyme 1) | - | miR-29c [40] | Normal [178]; Down [179] |
| miR-124 [181] | Down [182] | ||
| miR-195 [26] | Up [182]; Normal [176] | ||
| miR-298 [25] | - | ||
| miR-328 [25] | Down [176] | ||
| miR-339-5p [44] | Up [176] | ||
| SPTLC2 (Serine palmitoyltransferase, long chain base subunit 2) | Up [183, 184] | miR-9 [184] | Up [185]; Down [176, 178] |
| miR-29a [184] | Up [176] | ||
| miR-29b-1 [184] | Up [176]; Down [178, 179] | ||
| CFL1 (Cofilin 1) | - | miR-103 [186] | Down [176] |
| miR-107 [186] | Normal [176]; Down[187] | ||
| SP1 (Specificity protein 1) | Up [188] | miR-29b [189] | Up [176]; Down [178, 179] |
| miR-375 [33] | Normal [176] | ||
| SPTLC1 (Serine palmitoyltransferase, long chain base subunit 1) | Up [184] | miR-137 [184] | Normal [176] |
| miR-181c [184] | Down [176, 178, 179] | ||
| CAV1 (Caveolin 1) | Up [190] | miR-802 [191] | - |
| ERK1 (Extracellular signal-regulated kinase 1) | Normal [192, 193] | miR-15a [194] | Up [176]; Down [178, 179] |
| IGF1 (Insulin-like growth factor 1) | Up [195] | miR-98 [196] | Down [176] |
| PTBP1 (Polypyrimidine tract-binding protein 1) | - | miR-124 [43] | Down [182] |
| MAPT (Tau) | - | miR-34a [197] | Up [176] |
| Parkinson’s Disease (PD) | |||
| SNCA (α-synuclein) | - | miR-7 [57, 198] | Normal [199] |
| miR-153 [57] | - | ||
| TFDP1 (Transcription factor Dp-1) | - | let-7 [65] | - |
| miR-184* [65] | Normal [199] | ||
| E2F1 (Transcription factor E2F1) | Up [200] | let-7 [65] | - |
| miR-184* [65] | Normal [199] | ||
| FGF20 (Fibroblast growth factor 20) | - | miR-433 [201] | Normal [199] |
| PITX3 (Pituitary homeobox 3) | - | miR-133b [202] | Normal [199]; Down [202] |
| Huntington’s Disease (HD) | |||
| HTT (Huntingtin) | - | miR-125b [203] | - |
| miR-146a [203] | Normal [204] | ||
| miR-150 [203] | - | ||
| miR-214 [203] | - | ||
| BDNF (Brain-derived neurotrophic factor) | Down [205] | miR-30a-5p [206] | Normal [204] |
| RCOR1 (REST corepressor 1 [CoREST]) | - | miR-9* [207] | Down [207] |
| RICS (p250GAP) | - | miR-132 [85] | Up [204, 207]; Normal [204] |
| REST (RE1-silencing transcription factor) | - | miR-9 [207] | Up [204]; Down [204, 207] |
| TRIM2 (Tripartite motif-containing 2) | - | miR-200a [208] | Up [208] |
| Amyotrophic Lateral Sclerosis (ALS) | |||
| ACHE (Acetylcholinesterase) | - | miR-132 [209] | - |
| PTBP2 (Polypyrimidine tract-binding protein 2) | - | miR-132 [101] | - |
| Epilepsy | |||
| ICAM1 (Intercellular adhesion molecule 1) | - | miR-221 [210] | Down [210] |
| miR-222 [210] | Down [210] | ||
| DCX (Doublecortin) | Up [211] | miR-134 [212] | Up [213] |
| LIMK1 (LIM domain kinase 1) | Up [214] | miR-134 [143] | Up [213] |
| KDM1A (Lysine-specific histone demethylase 1A) | - | miR-137 [124] | - |
| TP53 (p53) | Up [215] | miR-34a [139] | Up [117] |
| STMN1 (Stathmin 1) | Up [216] | miR-9 [123] | Down [210] |
Only direct target/regulator pairs were considered for this table. Only articles that measured protein concentrations in human brain tissues were considered for column 2, “Protein Expression in Disease Brain,” and only articles that measured microRNA concentration in human brain tissues were considered for column 4, “miRNA Expression in Disease Brain.” Only microRNAs confirmed to regulate target transcripts experimentally were included in column 3, “miRNA Regulator(s).” The dash (-) symbols denote a lack of available information.
Interestingly, a number of miRNAs are also involved in more than one the four diseases discussed: miR-153 targets proteins involved in AD and PD, the miR-9 family targets proteins involved in AD, HD and epilepsy, miR-137 targets proteins involved in AD and epilepsy, as does miR-34a, while miR-132 targets proteins involved in HD and ALS. These multifaceted miRNAs, particularly those that target multiple proteins involved in a single disease, are particularly intriguing and should be further investigated in order to explore their ability to regulate multiple pathological systems simultaneously.
Researchers have great interest in understanding the involvement of miRNAs in these complex disorders. Recent studies have shown that dysregulated miRNAs may function as useful biomarkers of disease. Studies have shown that individual miRNAs can regulate more than 100 different mRNAs, and also that one miRNA could be target of multiple other miRNAs. This regulatory complexity reflects a significant biological network of functionally associated molecules in human cells. Comprehensive analysis of miRNA dysregulation in NDDs could aid in the development of novel therapeutic strategies [170, 171]. Finally, advances in miRNA-based therapeutics could help in understanding the early causative factors in NDD and epilepsy pathogenesis and lead to vast improvements in the treatment of these debilitating diseases.
Acknowledgments
This work was supported in part by Gland Pharma Limited (HKK) and the Krishna Institute of Medical Sciences (KIMS), Hyderabad, India, and by the Intramural Research Program of the National Institute on Aging (NHG, IAT), Baltimore, MD, USA
Footnotes
6. Conflict of Interests
Authors declare no conflict of interests.
References
- 1.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 2.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 7.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szafranski K, Abraham KJ, Mekhail K. Non-coding RNA in neural function, disease, and aging. Front Genet. 2015:6. doi: 10.3389/fgene.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilen J, Liu N, Bonini NM. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R, Noble W, Tartaglia GG, Buckley NJ. Neurodegeneration as an RNA disorder. Prog Neurobiol. 2012;99:293–315. doi: 10.1016/j.pneurobio.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano A. Hirano bodies and related neuronal inclusions. Neuropathol Appl Neurobiol. 1994;20:3–11. doi: 10.1111/j.1365-2990.1994.tb00951.x. [DOI] [PubMed] [Google Scholar]
- 14.Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628–636. doi: 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- 15.Maloney MT, Bamburg JR. Cofilin-mediated neurodegeneration in Alzheimer’s disease and other amyloidopathies. Mol Neurobiol. 2007;35:21–44. doi: 10.1007/BF02700622. [DOI] [PubMed] [Google Scholar]
- 16.Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013;33:14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 21.Schipper HM. Biomarker potential of heme oxygenase-1 in Alzheimer’s disease and mild cognitive impairment. Biomark Med. 2007;1:375–385. doi: 10.2217/17520363.1.3.375. [DOI] [PubMed] [Google Scholar]
- 22.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 23.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 25.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, Duan DX. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res Bull. 2012;88:596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Long JM, Lahiri DK. Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp Neurol. 2012;235:402–418. doi: 10.1016/j.expneurol.2011.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hébert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 29.Long JM, Lahiri DK. Current drug targets for modulating Alzheimer’s amyloid precursor protein: role of specific micro-RNA species. Curr Med Chem. 2011;18:3314–3321. doi: 10.2174/092986711796504592. [DOI] [PubMed] [Google Scholar]
- 30.Villa C, Ridolfi E, Fenoglio C, Ghezzi L, Vimercati R, Clerici F, Marcone A, Gallone S, Serpente M, Cantoni C, Bonsi R, Cioffi S, Cappa S, Franceschi M, Rainero I, Mariani C, Scarpini E, Galimberti D. Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J Alzheimers Dis. 2013;35:487–494. doi: 10.3233/JAD-122263. [DOI] [PubMed] [Google Scholar]
- 31.La Fauci G, Lahiri DK, Salton SR, Robakis NK. Characterization of the 5′-end region and the first two exons of the beta-protein precursor gene. Biochem Biophys Res Commun. 1989;159:297–304. doi: 10.1016/0006-291x(89)92437-6. [DOI] [PubMed] [Google Scholar]
- 32.Heicklen-Klein A, Ginzburg I. Tau promoter confers neuronal specificity and binds Sp1 and AP-2. J Neurochem. 2000;75:1408–1418. doi: 10.1046/j.1471-4159.2000.0751408.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Delay C, Calon F, Mathews P, Hebert SS. Alzheimer-specific variants in the 3′UTR of Amyloid precursor protein affect microRNA function. Mol Neurodegener. 2011:6. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Liu C, Zhu J, Shu P, Yin B, Gong Y, Qiang B, Yuan J, Peng X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol Aging. 2012;33:522–534. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 37.Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long JM, Ray B, Lahiri DK. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012;287:31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer’s disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zong Y, Wang H, Dong W, Quan X, Zhu H, Xu Y, Huang L, Ma C, Qin C. miR-29c regulates BACE1 protein expression. Brain Res. 2011;1395:108–115. doi: 10.1016/j.brainres.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 41.Fang M, Wang J, Zhang X, Geng Y, Hu Z, Rudd JA, Ling S, Chen W, Han S. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Smith P, Al Hashimi A, Girard J, Delay C, Hébert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 43.Smith P, Al Hashimi A, Girard J, Delay C, Hébert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 44.Long JM, Ray B, Lahiri DK. MicroRNA-339–5p down-regulates protein expression of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem. 2014;289:5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT. MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One. 2010;5:e15546. doi: 10.1371/journal.pone.0015546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol. 2011;22:1087–1098. doi: 10.1681/ASN.2010090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu YK, Wang X, Li L, Du YH, Ye HT, Li CY. MicroRNA-98 induces an Alzheimer’s disease-like disturbance by targeting insulin-like growth factor 1. Neurosci Bull. 2013;29:745–751. doi: 10.1007/s12264-013-1348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 50.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem. 2013;127:739–749. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 52.Bekris LM, Lutz F, Montine TJ, Yu CE, Tsuang D, Peskind ER, Leverenz JB. MicroRNA in Alzheimer’s disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013;18:455–466. doi: 10.3109/1354750X.2013.814073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jayadev S, Case A, Alajajian B, Eastman AJ, Möller T, Garden GA. Presenilin 2 influences miR146 level and activity in microglia. J Neurochem. 2013;127:592–599. doi: 10.1111/jnc.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holohan KN, Lahiri DK, Schneider BP, Foroud T, Saykin AJ. Functional microRNAs in Alzheimer’s disease and cancer: differential regulation of common mechanisms and pathways. Front Genet. 2013:3. doi: 10.3389/fgene.2012.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis. 2012;46:279–284. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 56.Shtilbans A, Henchcliffe C. Biomarkers in Parkinson’s disease: an update. Curr Opin Neurol. 2012;25:460–465. doi: 10.1097/WCO.0b013e3283550c0d. [DOI] [PubMed] [Google Scholar]
- 57.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, Navarro F, Dykxhoorn D, Lieberman J, Gorospe M. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci U S A. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res. 2003;114:123–131. doi: 10.1016/s0169-328x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 63.Martinat C, Bacci JJ, Leete T, Kim J, Vanti WB, Newman AH, Cha JH, Gether U, Wang H, Abeliovich A. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc Natl Acad Sci U S A. 2006;103:2874–2879. doi: 10.1073/pnas.0511153103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Martí E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 65.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 67.Myers RH. Huntington’s disease genetics. NeuroRx. 2004;1:255–262. doi: 10.1602/neurorx.1.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ross CA. When more is less: pathogenesis of glutamine repeat neurodegenerative diseases. Neuron. 1995;15:493–496. doi: 10.1016/0896-6273(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 69.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 70.Kita H, Carmichael J, Swartz J, Muro S, Wyttenbach A, Matsubara K, Rubinsztein DC, Kato K. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum Mol Genet. 2002;11:2279–2287. doi: 10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 71.Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 72.Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft TF, Kato K, Rubinsztein DC. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 73.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 74.Velier J, Kim M, Schwarz C, Kim TW, Sapp E, Chase K, Aronin N, DiFiglia M. Wild-type and mutant huntingtins function in vesicle trafficking in the secretory and endocytic pathways. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 75.Hoffner G, Kahlem P, Djian P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: relevance to Huntington’s disease. J Cell Sci. 2002;115:941–948. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- 76.Labbadia J, Morimoto RI. Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38:378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoss AG, Labadorf A, Latourelle JC, Kartha VK, Hadzi TC, Gusella JF, MacDonald ME, Chen JF, Akbarian S, Weng Z, Vonsattel JP, Myers RH. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genomics. 2015;8:10. doi: 10.1186/s12920-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martí E, Pantano L, Bañez-Coronel M, Llorens F, Miñones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Johnson R, Buckley NJ. Gene dysregulation in Huntington’s disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 81.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, Roh JK. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Hoss AG, Kartha VK, Dong X, Latourelle JC, Dumitriu A, Hadzi TC, Macdonald ME, Gusella JF, Akbarian S, Chen JF, Weng Z, Myers RH. MicroRNAs located in the Hox gene clusters are implicated in huntington’s disease pathogenesis. PLoS Genet. 2014:10. doi: 10.1371/journal.pgen.1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinha M, Ghose J, Bhattarcharyya NP. Micro RNA -214,-150,-146a and-125b target Huntingtin gene. RNA Biol. 2011;8:1005–1021. doi: 10.4161/rna.8.6.16035. [DOI] [PubMed] [Google Scholar]
- 87.Jin J, Cheng Y, Zhang Y, Wood W, Peng Q, Hutchison E, Mattson MP, Becker KG, Duan W. Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J Neurochem. 2012;123:477–490. doi: 10.1111/j.1471-4159.2012.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 89.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomsen GM, Gowing G, Svendsen S, Svendsen CN. The past, present and future of stem cell clinical trials for ALS. Exp Neurol. 2014;262(Pt B):127–137. doi: 10.1016/j.expneurol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 91.Bucchia M, Ramirez A, Parente V, Simone C, Nizzardo M, Magri F, Dametti S, Corti S. Therapeutic Development in Amyotrophic Lateral Sclerosis. Clin Ther. 2015;37:668–680. doi: 10.1016/j.clinthera.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 92.Vucic S, Rothstein JD, Kiernan MC. Advances in treating amyotrophic lateral sclerosis: insights from pathophysiological studies. Trends Neurosci. 2014;37:433–442. doi: 10.1016/j.tins.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 93.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 95.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I, Rubin LL, Misawa H, Tabin CJ, Brown R, Chen A, Hornstein E. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci U S A. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Felice B, Guida M, Coppola C, De Mieri G, Cotrufo R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508:35–40. doi: 10.1016/j.gene.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 98.Russell AP, Wada S, Vergani L, Hock MB, Lamon S, Leger B, Ushida T, Cartoni R, Wadley GD, Hespel P, Kralli A, Soraru G, Angelini C, Akimoto T. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol Dis. 2013;49:107–117. doi: 10.1016/j.nbd.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 99.Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31:4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 102.Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012 doi: 10.1038/nn.3082. [DOI] [PubMed] [Google Scholar]
- 103.Hébert SS, Sergeant N, Buée L. MicroRNAs and the Regulation of Tau Metabolism. Int J Alzheimers Dis. 2012;2012:406561. doi: 10.1155/2012/406561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shaltiel G, Hanan M, Wolf Y, Barbash S, Kovalev E, Shoham S, Soreq H. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct Funct. 2013;218:59–72. doi: 10.1007/s00429-011-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 109.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 111.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 114.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noebels J. Pathway-driven discovery of epilepsy genes. Nat Neurosci. 2015;18:344–350. doi: 10.1038/nn.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, Sano T, Michalak Z, Moran C, Delanty N, Farrell M, O’Brien D, Meller R, Simon RP, Stallings RL, Henshall DC. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS One. 2012;7:e35921. doi: 10.1371/journal.pone.0035921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu K, Xie YY, Zhang C, Ouyang DS, Long HY, Sun DN, Long LL, Feng L, Li Y, Xiao B. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012;13:115. doi: 10.1186/1471-2202-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McLoughlin HS, Fineberg SK, Ghosh LL, Tecedor L, Davidson BL. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012;223:285–295. doi: 10.1016/j.neuroscience.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 121.Reiner O, Coquelle FM, Peter B, Levy T, Kaplan A, Sapir T, Orr I, Barkai N, Eichele G, Bergmann S. The evolving doublecortin (DCX) superfamily. BMC Genomics. 2006;7:188. doi: 10.1186/1471-2164-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, Chen B, Li X, Dai J. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci. 2012;13:116. doi: 10.1186/1471-2202-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Omran A, Peng J, Zhang C, Xiang QL, Xue J, Gan N, Kong H, Yin F. Interleukin-1β and microRNA-146a in an immature rat model and children with mesial temporal lobe epilepsy. Epilepsia. 2012;53:1215–1224. doi: 10.1111/j.1528-1167.2012.03540.x. [DOI] [PubMed] [Google Scholar]
- 127.Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 128.Iyer A, Zurolo E, Prabowo A, Fluiter K, Spliet WG, van Rijen PC, Gorter JA, Aronica E. MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS One. 2012;7:e44789. doi: 10.1371/journal.pone.0044789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aronica E, Crino PB. Inflammation in epilepsy: clinical observations. Epilepsia. 2011;52:26–32. doi: 10.1111/j.1528-1167.2011.03033.x. [DOI] [PubMed] [Google Scholar]
- 130.Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, O’Duibhir E, de Jager W, Van Rijen PC, Gosselaar PH, de Graan PN, Pasterkamp RJ. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci. 2012;69:3127–3145. doi: 10.1007/s00018-012-0992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vezzani A, Friedman A, Dingledine RJ. The role of inflammation in epileptogenesis. Neuropharmacology. 2013;69:16–24. doi: 10.1016/j.neuropharm.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ashhab MU, Omran A, Kong H, Gan N, He F, Peng J, Yin F. Expressions of tumor necrosis factor alpha and microRNA-155 in immature rat model of status epilepticus and children with mesial temporal lobe epilepsy. J Mol Neurosci. 2013;51:950–958. doi: 10.1007/s12031-013-0013-9. [DOI] [PubMed] [Google Scholar]
- 133.Guerrini R, Dobyns WB, Barkovich AJ. Abnormal development of the human cerebral cortex: genetics, functional consequences and treatment options. Trends Neurosci. 2008;31:154–162. doi: 10.1016/j.tins.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 134.Palmini A, Najm I, Avanzini G, Babb T, Guerrini R, Foldvary-Schaefer N, Jackson G, Lüders HO, Prayson R, Spreafico R, Vinters HV. Terminology and classification of the cortical dysplasias. Neurology. 2004;62:S2–8. doi: 10.1212/01.wnl.0000114507.30388.7e. [DOI] [PubMed] [Google Scholar]
- 135.Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, Palmini A, Jacques TS, Avanzini G, Barkovich AJ, Battaglia G, Becker A, Cepeda C, Cendes F, Colombo N, Crino P, Cross JH, Delalande O, Dubeau F, Duncan J, Guerrini R, Kahane P, Mathern G, Najm I, Ozkara C, Raybaud C, Represa A, Roper SN, Salamon N, Schulze-Bonhage A, Tassi L, Vezzani A, Spreafico R. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52:158–174. doi: 10.1111/j.1528-1167.2010.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sisodiya SM, Fauser S, Cross JH, Thom M. Focal cortical dysplasia type II: biological features and clinical perspectives. Lancet Neurol. 2009;8:830–843. doi: 10.1016/S1474-4422(09)70201-7. [DOI] [PubMed] [Google Scholar]
- 137.Boison D. Engineered adenosine-releasing cells for epilepsy therapy: human mesenchymal stem cells and human embryonic stem cells. Neurotherapeutics. 2009;6:278–283. doi: 10.1016/j.nurt.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ren G, Boison D. Engineering human mesenchymal stem cells to release adenosine using miRNA technology. Methods Mol Biol. 2010;650:225–240. doi: 10.1007/978-1-60761-769-3_17. [DOI] [PubMed] [Google Scholar]
- 139.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 140.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012:3. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant Adeno-Associated Virus-Mediated microRNA Delivery into the Postnatal Mouse Brain Reveals a Role for miR-134 in Dendritogenesis in Vivo. Front Neural Circuits. 2010;3:16. doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 144.Peng J, Omran A, Ashhab MU, Kong H, Gan N, He F, Yin F. Expression patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. J Mol Neurosci. 2013;50:291–297. doi: 10.1007/s12031-013-9953-3. [DOI] [PubMed] [Google Scholar]
- 145.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 146.Borchard G. Chitosans for gene delivery. Adv Drug Deliv Rev. 2001;52:145–150. doi: 10.1016/s0169-409x(01)00198-3. [DOI] [PubMed] [Google Scholar]
- 147.Wang J, Zhang PC, Lu HF, Ma N, Wang S, Mao HQ, Leong KW. New polyphosphoramidate with a spermidine side chain as a gene carrier. J Control Release. 2002;83:157–168. doi: 10.1016/s0168-3659(02)00180-3. [DOI] [PubMed] [Google Scholar]
- 148.Juliano RL. Peptide-oligonucleotide conjugates for the delivery of antisense and siRNA. Curr Opin Mol Ther. 2005;7:132–136. [PubMed] [Google Scholar]
- 149.Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 150.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ningaraj NS, Salimath BP, Sankpal UT, Perera R, Vats T. Targeted brain tumor treatment-current perspectives. Drug Target Insights. 2007;2:197–207. [PMC free article] [PubMed] [Google Scholar]
- 152.Bidros DS, Liu JK, Vogelbaum MA. Future of convection-enhanced delivery in the treatment of brain tumors. Future Oncol. 2010;6:117–125. doi: 10.2217/fon.09.135. [DOI] [PubMed] [Google Scholar]
- 153.Banks WA, Farr SA, Butt W, Kumar VB, Franko MW, Morley JE. Delivery across the blood-brain barrier of antisense directed against amyloid beta: reversal of learning and memory deficits in mice overexpressing amyloid precursor protein. J Pharmacol Exp Ther. 2001;297:1113–1121. [PubMed] [Google Scholar]
- 154.Jaeger LB, Dohgu S, Hwang MC, Farr SA, Murphy MP, Fleegal-DeMotta MA, Lynch JL, Robinson SM, Niehoff ML, Johnson SN, Kumar VB, Banks WA. Testing the neurovascular hypothesis of Alzheimer’s disease: LRP-1 antisense reduces blood-brain barrier clearance, increases brain levels of amyloid-beta protein, and impairs cognition. J Alzheimers Dis. 2009;17:553–570. doi: 10.3233/JAD-2009-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 156.Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: Fit to deliver small RNA. Commun Integr Biol. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kalani A, Tyagi A, Tyagi N. Exosomes: mediators of neurodegeneration, neuroprotection and therapeutics. Mol Neurobiol. 2014;49:590–600. doi: 10.1007/s12035-013-8544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 160.Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 161.Jadhav VM, Scaria V, Maiti S. Antagomirzymes: oligonucleotide enzymes that specifically silence microRNA function. Angew Chem Int Ed Engl. 2009;48:2557–2560. doi: 10.1002/anie.200805521. [DOI] [PubMed] [Google Scholar]
- 162.Karnati HK, Yalagala RS, Undi R, Pasupuleti SR, Gutti RK. Therapeutic potential of siRNA and DNAzymes in cancer. Tumour Biol. 2014;35:9505–9521. doi: 10.1007/s13277-014-2477-9. [DOI] [PubMed] [Google Scholar]
- 163.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 164.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 166.Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 170.Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2012;133:142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chan AW, Kocerha J. The Path to microRNA Therapeutics in Psychiatric and Neurodegenerative Disorders. Front Genet. 2012;3:82. doi: 10.3389/fgene.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Patel N, Hoang D, Miller N, Ansaloni S, Huang Q, Rogers JT, Lee JC, Saunders AJ. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008:3. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhu HC, Wang LM, Wang M, Song B, Tan S, Teng JF, Duan DX. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res Bull. 2012;88:596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 175.Cheng PH, Li CL, Chang YF, Tsai SJ, Lai YY, Chan AW, Chen CM, Yang SH. miR-196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am J Hum Genet. 2013;93:306–312. doi: 10.1016/j.ajhg.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 177.Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Long JM, Ray B, Lahiri DK. MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012;287:31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fang M, Wang J, Zhang X, Geng Y, Hu Z, Rudd JA, Ling S, Chen W, Han S. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 182.Lau P, Bossers K, Janky R, Salta E, Frigerio CS, Barbash S, Rothman R, Sierksma AS, Thathiah A, Greenberg D, Papadopoulou AS, Achsel T, Ayoubi T, Soreq H, Verhaagen J, Swaab DF, Aerts S, De Strooper B. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grimm MO, Grosgen S, Rothhaar TL, Burg VK, Hundsdorfer B, Haupenthal VJ, Friess P, Muller U, Fassbender K, Riemenschneider M, Grimm HS, Hartmann T. Intracellular APP Domain Regulates Serine-Palmitoyl-CoA Transferase Expression and Is Affected in Alzheimer’s Disease. Int J Alzheimers Dis. 2011;2011:695413. doi: 10.4061/2011/695413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 186.Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT. MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One. 2010;5:e15546. doi: 10.1371/journal.pone.0015546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Citron BA, Dennis JS, Zeitlin RS, Echeverria V. Transcription factor Sp1 dysregulation in Alzheimer’s disease. J Neurosci Res. 2008;86:2499–2504. doi: 10.1002/jnr.21695. [DOI] [PubMed] [Google Scholar]
- 189.Villa C, Ridolfi E, Fenoglio C, Ghezzi L, Vimercati R, Clerici F, Marcone A, Gallone S, Serpente M, Cantoni C, Bonsi R, Cioffi S, Cappa S, Franceschi M, Rainero I, Mariani C, Scarpini E, Galimberti D. Expression of the transcription factor Sp1 and its regulatory hsa-miR-29b in peripheral blood mononuclear cells from patients with Alzheimer’s disease. J Alzheimers Dis. 2013;35:487–494. doi: 10.3233/JAD-122263. [DOI] [PubMed] [Google Scholar]
- 190.Gaudreault SB, Dea D, Poirier J. Increased caveolin-1 expression in Alzheimer’s disease brain. Neurobiol Aging. 2004;25:753–759. doi: 10.1016/j.neurobiolaging.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 191.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol. 2011;22:1087–1098. doi: 10.1681/ASN.2010090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Perry G, Roder H, Nunomura A, Takeda A, Friedlich AL, Zhu X, Raina AK, Holbrook N, Siedlak SL, Harris PL, Smith MA. Activation of neuronal extracellular receptor kinase (ERK) in Alzheimer disease links oxidative stress to abnormal phosphorylation. Neuroreport. 1999;10:2411–2415. doi: 10.1097/00001756-199908020-00035. [DOI] [PubMed] [Google Scholar]
- 193.Hyman BT, Elvhage TE, Reiter J. Extracellular signal regulated kinases. Localization of protein and mRNA in the human hippocampal formation in Alzheimer’s disease. Am J Pathol. 1994;144:565–572. [PMC free article] [PubMed] [Google Scholar]
- 194.Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 195.Connor B, Beilharz EJ, Williams C, Synek B, Gluckman PD, Faull RL, Dragunow M. Insulin-like growth factor-I (IGF-I) immunoreactivity in the Alzheimer’s disease temporal cortex and hippocampus. Brain Res Mol Brain Res. 1997;49:283–290. doi: 10.1016/s0169-328x(97)00192-7. [DOI] [PubMed] [Google Scholar]
- 196.Hu YK, Wang X, Li L, Du YH, Ye HT, Li CY. MicroRNA-98 induces an Alzheimer’s disease-like disturbance by targeting insulin-like growth factor 1. Neurosci Bull. 2013;29:745–751. doi: 10.1007/s12264-013-1348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem. 2013;127:739–749. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cardo LF, Coto E, Ribacoba R, Menendez M, Moris G, Suarez E, Alvarez V. MiRNA profile in the substantia nigra of Parkinson’s disease and healthy subjects. J Mol Neurosci. 2014;54:830–836. doi: 10.1007/s12031-014-0428-y. [DOI] [PubMed] [Google Scholar]
- 200.Hoglinger GU, Breunig JJ, Depboylu C, Rouaux C, Michel PP, Alvarez-Fischer D, Boutillier AL, Degregori J, Oertel WH, Rakic P, Hirsch EC, Hunot S. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sinha M, Ghose J, Bhattarcharyya NP. Micro RNA -214,-150,-146a and-125b target Huntingtin gene. RNA Biol. 2011;8:1005–1021. doi: 10.4161/rna.8.6.16035. [DOI] [PubMed] [Google Scholar]
- 204.Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Parain K, Murer MG, Yan Q, Faucheux B, Agid Y, Hirsch E, Raisman-Vozari R. Reduced expression of brain-derived neurotrophic factor protein in Parkinson’s disease substantia nigra. Neuroreport. 1999;10:557–561. doi: 10.1097/00001756-199902250-00021. [DOI] [PubMed] [Google Scholar]
- 206.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jin J, Cheng Y, Zhang Y, Wood W, Peng Q, Hutchison E, Mattson MP, Becker KG, Duan W. Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J Neurochem. 2012;123:477–490. doi: 10.1111/j.1471-4159.2012.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 210.Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, O’Duibhir E, de Jager W, Van Rijen PC, Gosselaar PH, de Graan PN, Pasterkamp RJ. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci. 2012;69:3127–3145. doi: 10.1007/s00018-012-0992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Liu YW, Curtis MA, Gibbons HM, Mee EW, Bergin PS, Teoh HH, Connor B, Dragunow M, Faull RL. Doublecortin expression in the normal and epileptic adult human brain. Eur J Neurosci. 2008;28:2254–2265. doi: 10.1111/j.1460-9568.2008.06518.x. [DOI] [PubMed] [Google Scholar]
- 212.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 213.Jimenez-Mateos EM, Bray I, Sanz-Rodriguez A, Engel T, McKiernan RC, Mouri G, Tanaka K, Sano T, Saugstad JA, Simon RP, Stallings RL, Henshall DC. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am J Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huang H, Wang H, Yuan J, Wu X, Huang Y, Zhou X, Chen Y. Protein expression of phospho-lim kinase-1 in patients and an experimental rat model with intractable temporal lobe epilepsy. Int J Clin Exp Med. 2015;8:625–633. [PMC free article] [PubMed] [Google Scholar]
- 215.Engel T, Murphy BM, Schindler CK, Henshall DC. Elevated p53 and lower MDM2 expression in hippocampus from patients with intractable temporal lobe epilepsy. Epilepsy Res. 2007;77:151–156. doi: 10.1016/j.eplepsyres.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao F, Hu Y, Zhang Y, Zhu Q, Zhang X, Luo J, Xu Y, Wang X. Abnormal expression of stathmin 1 in brain tissue of patients with intractable temporal lobe epilepsy and a rat model. Synapse. 2012;66:781–791. doi: 10.1002/syn.21567. [DOI] [PubMed] [Google Scholar]