Abstract
Coxsackievirus A21 (CAV21) is classified within the species Human enterovirus C (HEV-C) of the Enterovirus genus of picornaviruses. HEV-C share striking homology with the polioviruses (PV), their closest kin among the enteroviruses. Despite a high level of sequence identity, CAV21 and PV cause distinct clinical disease typically attributed to their differential use of host receptors. PV cause poliomyelitis, whereas CAV21 shares a receptor and a propensity to cause upper respiratory tract infections with the major group rhinoviruses. As a model for CAV21 infection, we have developed transgenic mice that express human intercellular adhesion molecule 1, the cell-surface receptor for CAV21. Surprisingly, CAV21 administered to these mice via the intramuscular route causes a paralytic condition consistent with poliomyelitis. The virus appears to invade the CNS by retrograde axonal transport, as has been demonstrated to occur in analogous PV infections. We detected human intercellular adhesion molecule 1 expression on both transgenic mouse and human spinal cord anterior horn motor neurons, indicating that members of HEV-C may share PV's potential to elicit poliomyelitis in humans.
The Enterovirus genus of Picornaviridae is a pathogenically diverse group of small nonenveloped positive-sense RNA viruses that primarily infect humans (1). Because most serotypes were identified in the mid-20th century, the enteroviruses (EV) were originally classified according to their pathogenicity for primates and suckling mice, resulting in four traditional taxonomic groups: polioviruses (PV), Coxsackie A viruses, Coxsackie B viruses, and echoviruses (2).
The advent of modern molecular techniques prompted considerable reassessment of conventional EV taxonomy, and since the 1970s, newly identified serotypes have simply been labeled “enterovirus” and numbered sequentially, starting with EV68. The current classification scheme recognizes five distinct Enterovirus species (3): Poliovirus and Human enterovirus A, B, C, and D (HEV-A to -D). The three PV serotypes constitute the species Poliovirus, whereas Coxsackie A virus serotypes 1, 11, 13, 15, 17, 18–22, and 24 make up Human enterovirus C (HEV-C) (3). Despite distinct pathogenic profiles (the main clinical manifestation of PV infection is poliomyelitis, whereas most HEV-C are associated with upper respiratory infections), these viruses show surprising homogeneity at the sequence level. Sequence comparisons consistently yielded mixed clusters in which individual PV serotypes align more closely with certain HEV-C than with other PV types (4–6) and have provided evidence of intimate genetic intermingling between PV and HEV-C to the extent that it is impossible to distinguish whether sequences of PV nonstructural proteins originated from HEV-C or vice versa (6). The striking molecular similarity between PV and HEV-C has led to the suggestion that they be merged into a single EV species designation (6).
Although virus classification according to pathogenic profile has given way to analyses of genetic diversity, sequence variability remains of most immediate concern as a correlate of the ability to cause disease. The capsid sequences comprise the only genome region to clearly delineate PV and HEV-C, corresponding to the groups' differential receptor preferences: PVs use CD155 (7), whereas many HEV-C recognize human intercellular adhesion molecule 1 (HsICAM-1) (4, 8, 9). It is generally believed that receptor specificity underlies the apparent pathogenic disparity between PV and HEV-C. This assumption is supported by the fact that, although evolutionarily distant, the major group human rhinoviruses are most similar to HEV-C among the Picornaviridae in pathogenic profile, route of transmission, and shared utilization of HsICAM-1 for viral entry (10, 11).
Excellent transgenic (tg) mouse models have been developed to aid the study of the pathogenesis of poliomyelitis and its genetic determinants (12, 13). The fact that PV-infected CD155 tg mice develop poliomyelitis bearing the hallmarks of the primate disease (12–14) underscores the cardinal role of receptor preference in EV disease specificity.
Coxsackievirus A21 (CAV21) is a prototypical HEV-C serotype that uses HsICAM-1 as a cellular receptor (15). Mouse cells are refractory to CAV21 infection, but ectopic expression of HsICAM-1 renders them susceptible (15). This suggested that HsICAM-1 tg mice could serve as permissive hosts to study CAV21 pathogenesis, similar to established mouse models for PV infection. In the present study, we have derived CAV21-susceptible mice expressing HsICAM-1. Surprisingly, CAV21 infection of these mice by the i.m. route produced classic paralytic poliomyelitis. Because only i.m. inoculation led to paralysis, and prior ipsilateral nerve dissection prevented CNS involvement, retrograde axonal transport may be instrumental in CAV21-caused poliomyelitis. Our findings reveal unexpected pathogenic properties for CAV21 mediated by its receptor, HsICAM-1, and induced retrograde axonal transport, a known provocation factor for PV-caused poliomyelitis (16). These observations further blur the boundaries separating HEV-C and PV and reveal a previously unrecognized overlap in pathogenic potential.
Materials and Methods
Derivation of tg Mice. Escherichia coli harboring bacterial artificial chromosome (BAC) CTD-2369P2 (California Institute of Technology Human BAC Library D; GenBank accession no. AC011511) were propagated, and BAC DNA was purified as described elsewhere (17). CTD-2369P2 DNA was digested with NruI, and the fragment containing HsICAM-1 was separated by field inversion gel electrophoresis and prepared for pronuclear injection (18). tg mice were identified with a PCR-based HsICAM-1 genotyping protocol by using the following primers: (i) 5′-cccccggcccgcgctgcccgc-3′ and (ii) 5′-cctgggcaacaaaacgaggcccg-3′. The HsICAM-1 transgene was maintained in a hemizygous state by breeding founders with BALB/cAnNTac mice (Taconic Farms). Embryo fibroblast cultures were established from day 13 postcoitum embryos after dispersion in DMEM containing trypsin/EDTA (Invitrogen) and triple aspiration through an 18-gauge needle, then infected with CAV21 at a multiplicity of infection of 10.
Antigen Capture Immunoblot. Human and murine tissues were homogenized in lysis buffer [100 mM KCl/25 mM EDTA/5mM MgCl2/10 mM Hepes, pH 7.0/0.5% Nonidet P-40/protease inhibitors (Roche Applied Science, Indianapolis)] by using Potter–Elvehjem tissue grinders. Polystyrene 96-well plates were coated with 500 ng per well of α-HsICAM-1 monoclonal antibody (RR1/1; BioSource International, Camarillo, CA) and blocked with 3% nonfat dried milk in TBST (tris-buffered saline, Tween-20 0.5%). Homogenate protein (150 μg per well) was applied, and plates were incubated at room temperature for 4 h, then rinsed. Bound proteins were released with 95°C NuPAGE SDS sample buffer (Invitrogen), resolved on a 7% Tris-acetate gel (Invitrogen), and transferred to nitrocellulose (Schleicher & Schuell). Membranes were probed by using a biotinylated α-HsICAM-1 monoclonal antibody (15.2; Calbiochem), followed by streptavidin–peroxidase conjugate (Roche Applied Science) and chemiluminescent detection (Amersham Pharmacia).
Immunohistochemistry. Human and mouse spinal cord cryosections (10 μm) were stained by using a Biotin Tyramide Signal Amplification kit (PerkinElmer). Briefly, sections were fixed in cold acetone and blocked with TNB buffer. Endogenous biotin and peroxidase were blocked by using an avidin/biotin kit (Vector Laboratories) and 1% H2O2 in PBS, respectively. To detect HsICAM-1, biotinylated antibody RR1/1 was applied for 1 h at room temperature, followed by streptavidin–peroxidase, biotinyl tyramide, and ABC Elite (Vector Laboratories), according to the manufacturer's instructions. Finally, sections were stained with metal-enhanced diaminobenzidine substrate (Pierce) and hematoxylin. Gastrocnemius muscle cryosections (10 μm) were fixed in cold acetone, blocked by using an M.O.M. kit (Vector Laboratories), then incubated for 1.5 h at room temperature with FITC-conjugated antibody 15.2 and 20 min with Alexa Fluor 594-conjugated α-bungarotoxin (Molecular Probes).
Virus Infections. Plasmid pCAV21 (strain Kuykendall) was cut with MluI and transcribed in vitro with T7 RNA polymerase (Stratagene) to derive virus, as described (14). Anesthetized mice (10–28 wk old) were injected with CAV21 [2.8 × 108 plaque-forming units (pfu) in 30–100 μl of PBS] into the gastrocnemius muscle with a 27-gauge needle. Infected animals were monitored daily for signs of disease. Virus levels in gastrocnemius muscle (average mass, 0.135 g) and spinal cord (average mass, 0.047 g) were determined from two animals at each designated time point, as described (14). Sciatic nerve transection (16) and intracerebral stereotaxic injections (19) were carried out as described. All animal procedures were performed according to Institutional Animal Care and Use Committee-approved protocols.
Results
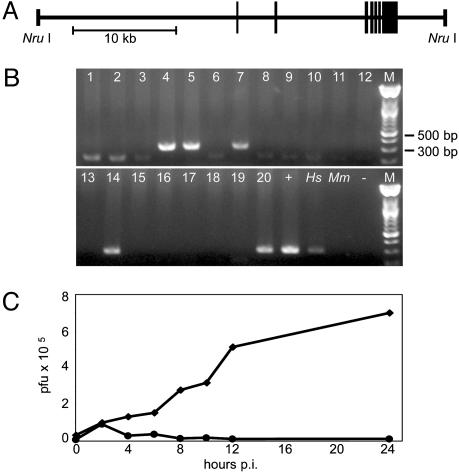
Derivation of HsICAM-1 tg Mice. A BAC carrying genomic sequence from human chromosome 19 was digested with NruI to isolate a ≈39-kb segment containing the HsICAM-1 locus (Fig. 1A). This large fragment was chosen to minimize position effects and encourage faithful expression patterns by preserving native regulatory elements within extensive 5′ and 3′ flanking regions of ≈18.9 and ≈4.6 kb, respectively. The transgene was purified and introduced into fertilized mouse oocytes via pronuclear injection. Of the resulting 20 pups, we identified 5 tg founders by PCR from genomic DNA (Fig. 1B). Fibroblasts derived from tg embryos supported CAV21 replication (Fig. 1C), indicating that the integrated transgene is functional and able to confer virus susceptibility to mouse cells.
Fig. 1.
Generation of tg mice. (A) Schematic 5′–3′ representation of the 39-kb DNA fragment containing the HsICAM-1 gene used for microinjection to derive tg mice (boxes signify HsICAM-1 exons). (B) PCR from founder genomic DNA (lanes 1–20) yields a specific 312-bp product for tg animals. +, BAC DNA; Hs, human genomic DNA; Mm, wild-type mouse genomic DNA; –, no template. (C) CAV21 replication in embryo fibroblast cultures established from HsICAM-1 tg embryos (♦) and non-tg littermates (•).
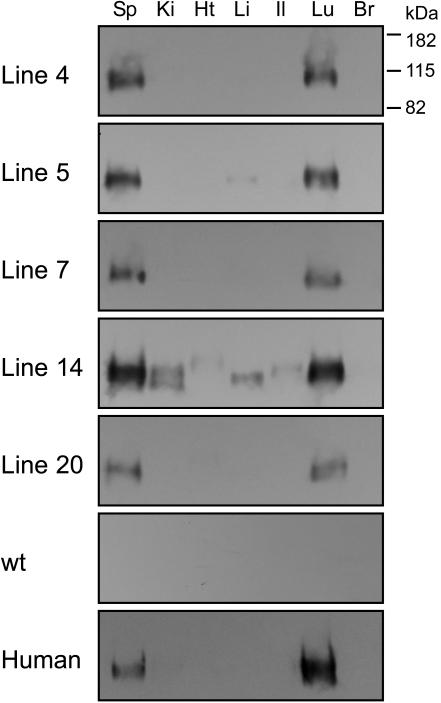
From the founders, we established five separate tg lines (Fig. 2). Using an antigen capture–immunoblot assay, we observed HsICAM-1 expression in a tissue-specific pattern that was consistent from line to line (Fig. 2). Transgene expression was highest in lung and spleen; lower to absent in liver, ileum, kidney, and heart; and undetectable in brain. The magnitude of expression was similar between lines, except line 14, which consistently exhibited the strongest signals in each positive tissue (Fig. 2). Importantly, the expression pattern was similar between tg and corresponding human tissues (Fig. 2) and in agreement with published data on the native distribution of HsICAM-1 (20). This result corroborates previous reports of strong conservation between human and mouse ICAM-1 regulatory elements (21). We selected line 4 for further experiments because of the parallel HsICAM-1 expression intensity and tissue distribution between these animals and humans (Fig. 2).
Fig. 2.
Antigen capture/immunoblot detection of HsICAM-1 in tissue homogenates from HsICAM-1 tg mouse lines 4, 5, 7, 14, and 20, a wild-type mouse, and corresponding human tissues. Sp, spleen; Ki, kidney; Ht, heart; Li, liver; Il, ileum; Lu, lung; Br, brain.
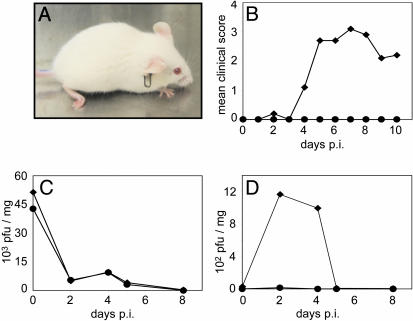
Characterization of CAV21-Induced Poliomyelitis. To assess susceptibility of HsICAM-1 tg mice to CAV21, we administered virus to groups of animals by several routes: i.v. (1.2 × 109 pfu), intranasal (1.6 × 108 pfu), and i.m. (2.8 × 108 pfu). No clinically evident disease resulted from i.v. or intranasal infection (not shown). However, i.m.-infected tg animals unexpectedly developed flaccid paralysis affecting the injected limb (Fig. 3A). Neurological dysfunction appeared 3–4 days after virus administration and involved dropfoot, lower hindlimb paresis, and abnormal gait. To substantiate this observation, 10 tg and 8 age-matched control mice were given identical i.m. doses of CAV21 and scored daily for symptoms of paralytic disease. All tg animals developed paralytic disease, whereas the control mice remained free of symptoms (Fig. 3B). Clinical symptoms peaked on day 7 postinfection, after which some animals partially regained motor function of the affected limb (Fig. 3B). In all cases, involvement was restricted to the inoculated limb, and affected animals appeared otherwise healthy.
Fig. 3.
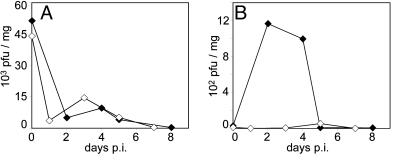
CAV21 pathogenicity in HsICAM-1 tg mice. (A) tg mouse 7 days after i.m. CAV21 injection. Note the characteristic drop-foot posture of the right hind limb. (B) Average clinical progression of CAV21-induced poliomyelitis in 10 tg mice (♦) and 8 wild-type controls (•). Severity of paresis was scored on an arbitrary scale (0, no symptoms; 1, minor lower paresis; 2, slight gait abnormality; 3, severe lower paresis/gait abnormality/drop foot; 4, ipsilateral paralysis). (C and D) CAV21 titers in gastrocnemius muscle (C) and spinal cord (D) of tg (♦) and wild-type mice (•) infected i.m. with CAV21 and killed at the indicated intervals.
We assessed viral titers in gastrocnemius muscle (the injection site; Fig. 3C) and spinal cord tissues (Fig. 3D) to assess the kinetics of viral replication. Viral titers in muscle tissue were indistinguishable between tg and control mice. Muscle titers were highest immediately after injection [day 0 postinfection (p.i.)], then sharply decreased (Fig. 3C). In contrast, spinal cord titers demonstrated a clear distinction between tg and control groups. CAV21 reached significantly elevated titer in spinal cords of tg mice between days 2 and 4 p.i., whereas virus remained negligible in spinal cord samples from non-tg mice (Fig. 3D).
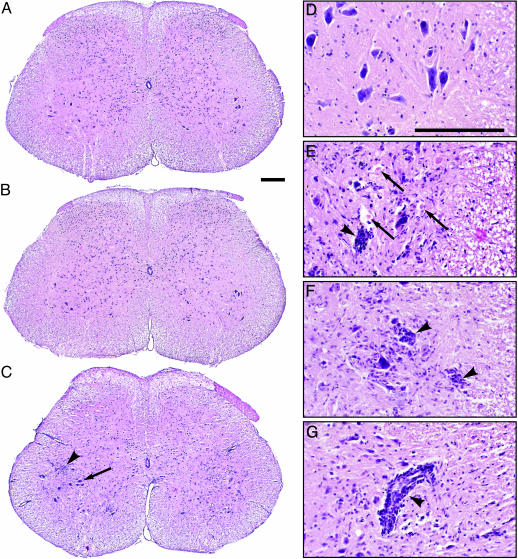
Spinal cord histopathology correlated with clinical observations and virus replication data. All postinfection samples processed from tg mice (six total) revealed the histopathological signature of classic poliomyelitis (Fig. 4C; ref. 22). There was loss of motor neurons with concurrent microglial infiltration in the anterior horn (Fig. 4C). Neuronal destruction selectively affected lumbar spinal anterior horn motor neurons and did not extend to the adjacent dorsal horn (Fig. 4C), the thoracic and cervical spinal cord, brainstem, or the brain (not shown). In accordance with clinically apparent ipsilateral pareses of the lower extremity, signs of spinal cord damage were concentrated in the anterior horn gray matter ipsilateral to the injection site (Fig. 4C).
Fig. 4.
Histopathology of CAV21-mediated poliomyelitis in the lumbar spinal cord of HsICAM-1 tg mice. (A and B) The spinal cords of mock-infected (A) and CAV21-infected non-tg (B) mice were histologically normal. (C) In the spinal cord of CAV21-infected HsICAM-1 tg mice, motor neuron destruction occurred ipsilaterally. Although contralateral motor neurons mostly remained unscathed (arrow), there was evidence of cellular infiltrates in their vicinity (arrowhead). (D–G) Histological detail of the anterior horn of CAV21-infected wild-type (D) and HsICAM-1 tg (E–G) mice. The latter had prominent evidence of neuronophagia (E and F, arrowheads) and eosinophilic bodies suggesting destroyed motor neurons (E, arrows). Mononuclear infiltrates occurred both diffusely in the anterior horn (C), surrounding affected neurons (E and F), and as perivascular cuffs (G, arrowhead). (Bars = 200 μm.)
Affected areas contained motor neurons in various stages of destruction (Fig. 4 E and F) and prominent signs of inflammation (Fig. 4 C and E–G), previously described with PV-caused poliomyelitis in patients (22). We detected mononuclear cell infiltrates of predominantly microglial/macrophage origin diffusely distributed in the anterior horn gray matter (Fig. 4 C and E–G), clustered around pyknotic neurons (Fig. 4 E and F), and in perivascular cuffs (Fig. 4G).
Marginal CAV21 replication in gastrocnemius muscle after i.m. injection produced minor signs of inflammation and tissue damage, which may partly derive from the injection procedure (not shown). There were no signs of rhabdomyolysis that has been described with CAV21 infection of wild-type suckling mice (23), reflecting the adult age of the animals in our study.
CNS Invasion via Retrograde Axonal Transport. Exposure of skeletal muscle to virus by i.m. inoculation (24, 25) or through viremia concomitant with skeletal muscle injury (16) is a known factor facilitating PV CNS invasion. Retrograde axonal transport of virus mediated by peripheral entry at the neuromuscular junction (NMJ) is suspected to account for this effect (16), because paralysis typically originates in the injected limb (the “localization” phenomenon; ref. 26).
In HsICAM-1 tg mice, CAV21 neuroinvasion is contingent upon i.m. virus administration and produces the localization phenomenon, indications that CAV21 might enter the CNS via the neural route. To assess this possibility, we performed sciatic nerve transection on a group of five tg mice, thereby disrupting the neural conduit between the spinal cord and the injected gastrocnemius muscle. Animals with transected left sciatic nerves received CAV21 injections into the left gastrocnemius muscle. We were unable to evaluate these mice clinically, because the transection itself results in mild symptoms of motor dysfunction similar to the neurological condition induced by i.m. CAV21 infection of HsICAM-1 tg mice (Fig. 3A).
Sciatic nerve transection had no discernible impact on virus levels in the gastrocnemius muscle (Fig. 5A). However, the procedure did prevent CAV21 from reaching elevated titers in the spinal cord (Fig. 5B), and spinal cord tissue from these animals appeared histopathologically normal with no detectable inflammation or motor neuron damage (not shown). These results indicate that discontinuity between the spinal cord and the site of i.m. virus administration blocks viral CNS access and subsequent incidence of poliomyelitis. This is consistent with a neural route of CNS invasion operative in HsICAM-1 tg mice infected i.m. with CAV21.
Fig. 5.
CAV21 titers in gastrocnemius muscle (A) and spinal cord (B) of i.m.-infected tg mice with intact (♦) and severed (⋄) sciatic nerve.
To assess whether direct CNS administration of CAV21 can circumvent the retrograde axonal route and produce poliomyelitis in HsICAM-1 tg mice, we performed intracerebral inoculations. Groups of four mice, consisting of two tg animals and two non-tg littermates, each received stereotaxic inoculations of 1.3 × 105, 1.3 × 106, or 1.3 × 107 pfu. None of the animals developed any neurological symptoms. Histopathologic evaluation of the brain and spinal cord revealed inflammatory reactions in the vicinity of the inoculation site. These consisted of microglial proliferation and lymphocytic infiltration in a predominantly perivascular pattern (not shown). There were no histopathological abnormalities in the spinal cord in either HsICAM-1 tg mice or controls.
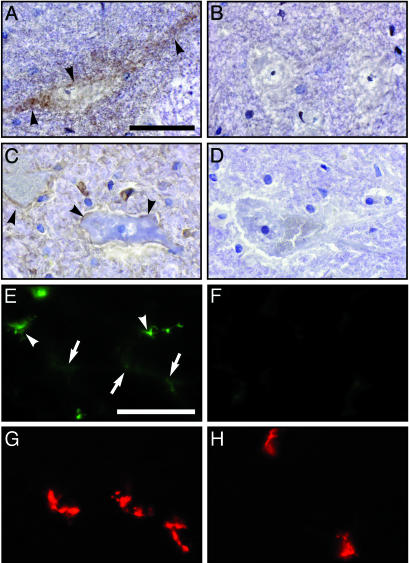
HsICAM-1 Expression on Motor Neurons. Susceptibility to CAV21-induced poliomyelitis clearly required the HsICAM-1 transgene (Fig. 3). To ascertain the role of HsICAM-1 in directing CAV21 toward spinal cord motor neurons, we conducted immunohistochemical analyses of HsICAM-1 expression in spinal cord and skeletal muscle tissue (Fig. 6). These assays revealed specific surface staining on ventral horn motor neurons in HsICAM-1 tg mice (Fig. 6A) as well as in the human spinal cord (Fig. 6C). We speculate that the expression level of HsICAM-1 in the spinal cord of tg mice and humans is low, because the immunohistochemical signal was weak and we were unable to detect signal in spinal cord homogenates subjected to our antigen capture-immunoblot assay (not shown). Immunohistochemical analyses of gastrocnemius muscle of HsICAM-1 tg mice (Fig. 6E) revealed HsICAM-1 in known expression sites (vascular endothelium) as well as structures specifically labeled by the NMJ marker α-bungarotoxin (Fig. 6G). Compared with capillary endothelium, the HsICAM-1 signal at the NMJ was faint, suggesting evenly low levels of HsICAM-1 expression on extended motor neurons.
Fig. 6.
Immunohistochemical detection of HsICAM-1. HsICAM-1 staining in the ventral horn of HsICAM-1 tg mouse (A) and human (C) lumbar spinal cord. Specificity was controlled by staining parallel sections from wild-type mice (B) and by incubating sections of human spinal cord with isotype-matched nonspecific primary antibody (D). HsICAM-1 signal was visualized with diaminobenzidine substrate (brown; arrowheads), and sections were counterstained with hematoxylin (blue). Fluorescent detection of HsICAM-1 (E and F) and NMJ (G and H) in gastrocnemius muscle of tg (E and G) and wild-type (F and H) mice. Two patterns of HsICAM-1 signal appeared in muscle (E): bright staining corresponding to capillary endothelium (arrowheads) and a more delicate signal that colocalized with NMJ (arrows; compare E and G). (Bars = 50 μm.)
Discussion
EVs have been linked to a bewildering array of clinical conditions affecting many organ systems within the human body. With the exception of fulminant generalized infections in neonates, all EVs target specific cell populations. Cell type-specific EV susceptibility may reflect cell-internal restrictions to virus propagation as well as the distribution of cellular receptors that mediate particle attachment, cell membrane passage, and capsid disassembly.
Resolution of the near-atomic level structures of receptors complexed to CAV21, PV, and human rhinoviruses (HRV) (27–30) demonstrated an intricate association of the receptor moiety with the “canyon”, a narrow depression on the capsid surface. The precise fit of receptors into the capsid architecture suggests that receptor specificity would be conserved among related picornaviruses. However, genetic kinship poorly correlates with receptor usage; CAV21 shares ≈70% and ≈50% amino acid sequence of the capsid-coding region with PV1 and HRV14, respectively; yet it shares receptor specificity with the latter.
Here we report that the genetic kinship of HEV-C and PV is reflected in common pathogenic properties, despite a preference for discrete receptor entities. Poliomyelitis in patients has been observed with other nonpolio EVs (31–33); however, it has never been observed with any HEV-C serotypes. It is unknown which cellular attachment protein(s) is responsible for paralytic disease due to CAV7, EV70, or EV71, and a role for the PV receptor CD155 has not been ruled out. Our report indicates that poliomyelitis can be mediated specifically by a receptor entity other then CD155. Interestingly, this precedent involves the closest relatives of PV, the HEV-C, and HsICAM-1, a viral attachment protein never implicated in the pathogenesis of viral CNS infection.
Although the pathognomonic features of CAV21- and PV-induced poliomyelitis in HsICAM-1 tg and CD155 tg mice, respectively, are identical, the symptoms and course of progression are far more benign in the former. i.m. inoculation with CAV21 produced the hallmarks of “provocation poliomyelitis” that were observed in PV vaccinees treated with multiple i.m. injections (34), namely enhanced susceptibility to neurological complications and localization of initial paresis to the injected limb. The pathogenic mechanism responsible for this effect, retrograde axonal transport, facilitates, but is not essential for, CNS invasion by PV. In contrast, CAV21 seems to require peripheral nerve entry to cross the threshold into the CNS. Once established in PV-infected CD155 tg mice, poliomyelitis regularly spreads throughout the entire spinal cord, leading to respiratory failure and death (12, 13). CAV21-infected HsICAM-1 tg mice developed regionally restricted spinal cord lesions and mild motor dysfunction that never progressed beyond the initially affected limb. These differences may relate to the distribution of the respective viral receptors or may reflect PV's superior ability to persevere and spread in the CNS. Indeed, even intracerebral inoculation of CAV21 failed to induce poliomyelitis in HsICAM-1 tg mice, suggesting an inability to penetrate CNS tissue to reach its target in the spinal cord anterior horn.
Despite CAV21's lesser neuroinvasive properties, our results demonstrate that it shares PV's inherent potential to cause the distinctive pattern of motor neuron damage clinically manifest as paralytic poliomyelitis. It is tempting to speculate that the capacity for propagation in motor neurons is determined by noncoding regulatory elements or sequences coding for nonstructural gene products, considering the genetic homogeneity of PV and CAV21 in these regions. This assumption is supported by the isolation of PV variants from acute cases of flaccid paralysis harboring sequences of suspected HEV-C origin mapping to these regions (35).
Because it exhibits the most genetic divergence between HEV-C and PV, the capsid may be responsible for deficient neuroinvasion by CAV21. Apart from recognition of diverse receptor entities, capsid structure may influence pathogenicity by affecting particle stability, tissue penetration, and immune recognition. Therefore, it is possible that if CAV21 were to acquire a more “PV-like” capsid, an enhanced capacity for neuroinvasion could result. The extreme genetic plasticity of EVs, due to their characteristically high mutation rates and propensity for genetic recombination, makes this scenario conceivable.
Disease was limited to the CNS after i.m. injection and not readily apparent elsewhere and with other routes of administration. However, the characteristically mild course of CAV21 respiratory infections in humans, combined with the similarity, particularly in lung, between human and tg mouse HsICAM-1 expression, suggests that HsICAM-1 tg mice may be susceptible to respiratory infection. Thus, these mice may prove useful for in vivo studies of respiratory infection by CAV21 or other picornaviruses that initiate infection via HsICAM-1, including recently described mouse-adapted human rhinovirus strains (36).
CAV21-mediated poliomyelitis in HsICAM-1 tg mice depends on HsICAM-1 expression, presumably on motor neurons within the spinal cord and/or on their axonal extensions. Although the detection of HsICAM-1 on human spinal cord motor neurons suggests CAV21 susceptibility, our observations indicate that factors other than receptor expression, such as peripheral nerve entry, are required for CAV21 to produce poliomyelitis. The extreme adaptability of EV genomes has led to the suggestion that HEV-C could represent a genetic pool from which novel PV-like variants could emerge to fill the evolutionary void generated by the planned worldwide eradication of PV (37). This hypothesis, for obvious reasons, is difficult to verify, but our finding of common pathogenic properties for these viruses further reinforces this possibility.
Acknowledgments
We are grateful to E. Wimmer (Stony Brook University, Stony Brook, NY) for supplying valuable materials. We obtained human CNS specimens from the New York Brain Bank (Columbia University, New York), which is supported by Public Health Service Grants P50 AG08702 and NS16367. We thank R. McLendon (Duke University) for neuropathological analysis. This work was supported by Public Health Services Grants AI054216 (to M.G.) and P30 AI051445 (to Bart Haynes, Duke University). A.T.D. was the recipient of a Grants-in-Aid of Research Award from Sigma Xi. M.G. is a recipient of a Burroughs Wellcome Fund Career Award.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAV21, Coxsackievirus A21; HEV-C, Human enterovirus C; PV, poliovirus; HsICAM-1, human intercellular adhesion molecule 1; EV, enterovirus; tg, transgenic; CNS, central; BAC, bacterial artificial chromosome; NMJ, neuromuscular junction; pfu, plaque-forming units.
References
- 1.Grist, N. R., Bell, E. J. & Assaad, A. (1978) Prog. Med. Virol. 24, 114–157. [PubMed] [Google Scholar]
- 2.Dalldorf, G. & Melnick, J. L. (1965) in Viral and Rickettsial Infections of Man, eds. Horsfall, F. L & Tamm, I. (Lippincott, Philadelphia), 4th Ed., pp. 474–512.
- 3.King, A. M. Q., Brown, F., Christian, P., Hovi, T., Hyypia, T., Knowles, N. J., Lemon, S. M., Minor, P. D., Palmenberg, A. C., Skern, T., et al. (2000) in Virus Taxonomy, Seventh Report of the International Committee on Taxonomy of Viruses, eds. Van Regenmortel, M. H. V., Fanquet, C. M., Bishop, D. H. L., Carstens, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R., et al. (Academic, San Diego), pp. 657–678.
- 4.Pulli, T., Koskimies, P. & Hyypia, T. (1995) Virology 212, 30–38. [DOI] [PubMed] [Google Scholar]
- 5.Pöyry, T., Kinnunen, L., Hyypiä, T., Brown, B., Horsnell, C., Hovi, T. & Stanway, G. (1996) J. Gen. Virol. 77, 1699–1717. [DOI] [PubMed] [Google Scholar]
- 6.Brown, B., Oberste, S., Maher, K. & Pallansch, M. A. (2003) J. Virol. 77, 8973–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendelsohn, C. L., Wimmer, E. & Racaniello, V. R. (1989) Cell 56, 855–865. [DOI] [PubMed] [Google Scholar]
- 8.Colonno, R. (1987) BioEssays 5, 270–274. [DOI] [PubMed] [Google Scholar]
- 9.Newcombe, N. G., Andersson, P., Johansson, E. S., Au, G. G., Lindberg, A. M., Barry, R. D. & Shafren, D. R. (2003) J. Gen. Virol. 84, 3041–3050. [DOI] [PubMed] [Google Scholar]
- 10.Staunton, D. E., Merluzzi, V. J., Rothlein, R., Barton, R., Marlin, S. D. & Springer, T. A. (1989) Cell 56, 849–853. [DOI] [PubMed] [Google Scholar]
- 11.Greve, J. M., Davis, G., Meyer, A. M., Forte, C. P., Yost, S. C., Marlor, C. W., Kamarck, M. E. & McClelland, A. (1989) Cell 56, 839–847. [DOI] [PubMed] [Google Scholar]
- 12.Ren, R., Costantini, F., Gorgacz, E. J., Lee, J. J. & Racaniello, V. R. (1990) Cell 63, 353–362. [DOI] [PubMed] [Google Scholar]
- 13.Koike, S., Taya, C., Kurata, T., Abe, S., Ise, I., Yonekawa, H. & Nomoto, A. (1991) Proc. Natl. Acad. Sci. USA 88, 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gromeier, M., Alexander, L. & Wimmer, E. (1996) Proc. Natl. Acad. Sci. USA 93, 2370–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafren, D. R., Dorahy, D. J., Greive, S. J., Burns, G. F. & Barry, R. D. (1997) J. Virol. 71, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gromeier, M. & Wimmer, E. (1998) J. Virol. 72, 5056–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrast, R., Scott, H. S. & Antonarakis, S. E. (1999) Transgenic Res. 8, 147–150. [DOI] [PubMed] [Google Scholar]
- 18.Hammes, A. & Schedl, A. (2000) in Mouse Genetics and Transgenics: A Practical Approach, eds. Jackson, I. J. & Abbott, C. M. (Oxford Univ. Press, New York), pp. 217–245.
- 19.Gromeier, M., Lachmann, S., Rosenfeld, M. R., Gutin, P. H. & Wimmer, E. (2000) Proc. Natl. Acad. Sci. USA 97, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Stolpe, A. & van der Saag, P. T. (1996) J. Mol. Med. 74, 13–33. [DOI] [PubMed] [Google Scholar]
- 21.Ballantyne, C. M., Sligh, J. E., Dai, X. Y. & Beaudet, A. L. (1992) Genomics 14, 1076–1080. [DOI] [PubMed] [Google Scholar]
- 22.Bodian, D. (1949) Am. J. Med. 6, 563–578. [DOI] [PubMed] [Google Scholar]
- 23.Mufson, M., Kawana, R., Bloom, H. H., Gorstein, F. & Chanock, R. M. (1968) Proc. Soc. Exp. Biol. Med. 128, 237–246. [DOI] [PubMed] [Google Scholar]
- 24.Ren, R. & Racaniello, V. R. (1992) J. Infect. Dis. 166, 747–752. [DOI] [PubMed] [Google Scholar]
- 25.Ohka, S., Yang, W., Terada, E., Iwasaki, K. & Nomoto, A. (1998) Virology 250, 67–75. [DOI] [PubMed] [Google Scholar]
- 26.Nathanson, N. & Bodian, D. (1961) Bull. Johns Hopkins Hosp. 108, 308–333. [PubMed] [Google Scholar]
- 27.Xiao, C., Bator, C. M., Bowman, V. D., Rieder, E., He, Y., Hebert, B., Bella, J., Baker, T. S., Wimmer, E., Kuhn, R. J., et al. (2001) J. Virol. 75, 2444–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, Y., Bowman, V. D., Mueller, S., Bator, C. M., Bella, J., Peng, X. Z., Baker, T. S., Wimmer, E., Kuhn, R. J. & Rossman, M. G. (2000) Proc. Natl. Acad. Sci. USA 97, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belnap, D. M., McDermott, B. M., Filman, D. J., Cheng, N., Trus, B. L., Zuccola, H. J., Racaniallo, V. R., Hogle, J. M. & Steven, A. C. (2000) Proc. Natl. Acad. Sci. USA 97, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolatkar, P. R., Bella, J., Olson, N. H., Bator, C. M., Baker, T. S. & Rossmann, M. G. (1999) EMBO J. 18, 6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voroshilova, M. K. & Chumakov M. P. (1959) Prog. Med. Virol. 2, 106–170. [Google Scholar]
- 32.Shindarov, L. M., Chumakov, M. P., Voroshilova, M. K., Bojinov, S., Vasilenko, S. M., Iordanov, I., Kirov, I. D., Kamenov, E., Leshchinskaya, E. V., Mitov, G., et al. (1979) J. Hyg. Epid. Microb. Im. 23, 284–295. [PubMed] [Google Scholar]
- 33.Wadia, N. H., Katrak, S. M., Misra, V. P., Wadia, P. N., Miyamura, K., Hashimoto, K., Ogino, T., Hikiji, T. & Kono, R. (1983) J. Infect. Dis. 147, 660–668. [DOI] [PubMed] [Google Scholar]
- 34.Strebel, P. M., Ion-Nedelcu, N., Baughman, A. L., Sutter, R. W. & Cochi, S. L. (1995) N. Engl. J. Med. 332, 500–506. [DOI] [PubMed] [Google Scholar]
- 35.Kew, O., Morris-Glasgow, V., Landaverde, M., Burns, C., Shaw, J., Garib, Z., Andre, J., Blackman, E., Freeman, C. J., Jorba, J., et al. (2002) Science 296, 356–359. [DOI] [PubMed] [Google Scholar]
- 36.Harris, J. R. & Racaniello, V. R. (2003) J. Virol. 77, 4773–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieder, E., Gorbalenya, A. E., Xiao, C., Baker, T. S., Kuhn, R. J., Rossmann, M. G. & Wimmer, E. (2001) Dev. Biol. 105, 111–122. [PubMed] [Google Scholar]