Abstract
It has long been believed that the retina of mature mammals is incapable of regeneration. In this study, using the N-methyl-d-aspartate neurotoxicity model of adult rat retina, we observed that some Müller glial cells were stimulated to proliferate in response to a toxic injury and produce bipolar cells and rod photoreceptors. Although these newly produced neurons were limited in number, retinoic acid treatment promoted the number of regenerated bipolar cells. Moreover, misexpression of basic helix–loop–helix and homeobox genes promoted the induction of amacrine, horizontal, and rod photoreceptor specific phenotypes. These findings demonstrated that retinal neurons regenerated even in adult mammalian retina after toxic injury. Furthermore, we could partially control the fate of the regenerated neurons with extrinsic factors or intrinsic genes. The Müller glial cells constitute a potential source for the regeneration of adult mammalian retina and can be a target for drug delivery and gene therapy in retinal degenerative diseases.
The retina is a component of the central nervous system (CNS), and retinal neuronal death causes a severe loss of visual function. Although significant progress has been made in identifying factors that regulate retinal neurogenesis, no treatment is presently available to treat retinal degenerative diseases such as retinitis pigmentosa. Embryonic or adult retinal cells have been transplanted into patients with retinitis pigmentosa or age-related macular degeneration (1), but the outcomes of these procedures are not sufficient and the cell supplies are limited. Stem cell transplantation to retina has been under investigation; however, few donor cells expressed retinal neuronal markers in the host retina after transplantation (2–5). Therefore, a new strategy for retinal cell replacement is needed.
It has long been thought that the CNS is incapable of regeneration. However, recent efforts revealed that stem cells remain in the adult CNS and have the ability to produce new neurons and glia (6, 7). Furthermore, various injuries stimulate the proliferation of endogenous progenitors in locations where neurogenesis normally does not occur (8). Several studies have focused on neural regeneration after acute damage (9, 10). It is reasonable to hypothesize that, if inactive endogenous progenitors could be stimulated by some manipulation, they may produce new neurons and ultimately repair damaged compartments of the CNS.
When considering the adult mammalian retina, it has been believed that the retina of the mature mammal has no regenerative capacity. However, adult retinal stem cells have been identified in the nonpigmented ciliary margin of rodents (11, 12), implying that adult mammalian retina have some degree of regenerative potential. Furthermore, recent work has demonstrated that Müller glia play a role for regenerative responses of the fish and postnatal chicken retina (13–15).
Here we show that Müller glia in the central retina of adult mammals proliferate in response to neurotoxin-induced damage and generate new retinal neurons, which indicates the regenerative potential of the adult mammalian retina.
Materials and Methods
Animals. The use of animals in this study was in accordance with the Guidelines for Animal Experiments of Kyoto University. All animal experiments in this study were conducted with the approval of the Animal Research Committee, Graduate School of Medicine, Kyoto University. Six-week-old male Sprague–Dawley rats were obtained from Shimizu Laboratory Supplies (Kyoto, Japan) and housed at room temperature on a cycle of 12 h light/12 h dark.
Intraocular Injections. Rats were anesthetized by inhalation of diethyl ether and i.p. injection of pentobarbital. The eyes were injected with 5 μl of 40 mM N-methyl-d-aspartate (NMDA) (total 200 nmol), 30 nmol BrdUrd, basic fibroblast growth factor (100 ng per injection), epidermal growth factor (100 ng per injection), retinoic acid (RA) (100 ng per injection), Activin-A (1–100 ng per injection), and bovine insulin (100 ng to 2 μg per injection). All drugs were obtained from Sigma.
Fixation, Sectioning, and Immunohistochemistry. Tissue sectioning and immunohistochemistry were performed as described (5, 16). The primary antibodies and working dilutions in this study were as follows: mouse anti-BrdUrd (1:50, Roche Applied Science), rat anti-BrdUrd (1:10, Oxford Biotechnology, Kidlington, U.K.), mouse anti-glutamine synthetase (GS) (1:1,000, Chemicon), mouse anti-ED1 (1:100, Serotec), rabbit anti-GS (1:500, Sigma), rabbit anti-PKC (1:1,000, Sigma), mouse anti-PKC (1:500, Sigma), rabbit anti-neuron-specific enolase (NSE) (1:2, Immunotech, Luminy, France), rabbit anti-calbindin (1:200, Chemicon), mouse anti-calbindin (1:200, Sigma), mouse anti-HPC1 (1:2,000, Sigma), rabbit anti-Thy1.2 (Pharmingen), mouse anti-RET-P1 (1:2,000, Sigma), rabbit anti-rhodopsin (1:1,000, LSL, Tokyo), rabbit anti-recoverin, mouse anti Ki67(1:200, Pharmingen), mouse anti-nestin (1:1,000, Pharmingen), rabbit anti-GFP (1:500, Molecular Probes), and mouse anti-Myc (Invitrogen). The secondary antibodies and working dilutions were as follows: labeled goat anti-mouse IgG antibodies (Alexa Fluor 488, Alexa Fluor 594, 1:500, Molecular Probes), labeled goat anti-rabbit IgG antibodies (Alexa Fluor 488, Alexa Fluor 594, 1:500, Molecular Probes), and labeled goat anti-rat IgG antibodies (Alexa Fluor 488, Alexa Fluor 594, 1:500, Molecular Probes). Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay was performed with a detection kit (Roche Diagnostics). The specimens were observed and photographed with a laser-scanning confocal microscope (Leica).
Retinal Explant Culture. The retinal explant culture was conducted as described (17). Briefly, the retina without pigmented epithelium was placed on a Millicell chamber filter (Millipore, 0.4-μm culture plate insert and 30-mm diameter) with the ganglion cell layer (GCL) upwards. The Millicell-CM chamber was moved to a six-well culture plate. Each well contained 1 ml of culture medium [50% minimum essential medium with Hepes (Invitrogen)/25% Hanks' solution (Invitrogen)/25% heat-inactivated horse serum/200 μM l-glutamine/5.75 mg/ml glucose]. Explants were cultured at 34°C in 5% CO2, and the media were changed every other day.
Preparation and Infection of Recombinant Retrovirus. The method for construction of recombinant retrovirus CLIG, CLIG-Pax6, CLIG-Crx, CLIG-Math3, CLIG-NeuroD, CLIG-Pax6-Math3, CLIG-Pax6-NeuroD, and CLIG-Crx-NeuroD genes was described (17). For construction of CLIG-Math3, CLIG-NeuroD, CLIG-Pax6, and CLIG-Crx, cDNAs were cloned into the EcoRI site of pCLIG, which directs expression of the cloned genes together with enhanced GFP from the upstream LTR promoter. For construction of CLIG-Pax6-Math3, CLIG-Pax6-NeuroD, and CLIG-Crx-NeuroD, the basic helix–loop–helix (bHLH) genes were cloned into the BsrGI and ClaI sites of pCLIG-Pax6 and pCLIG-Crx, which are located in the 3′ region of GFP gene, so that GFP gene is fused in frame with each bHLH gene. The retroviral DNAs were transfected with LopofectAMINE (GIBCO/BRL) into the ectopic packaging cells. Two days later, the media were collected and concentrated with Centricon Plus-20 (Amicon). The explants were infected with 40 μl of the virus containing 20 mg/ml polybrene for 3 h at 34°C. After viral infection, the media were replaced.
Results
Müller Glia Could Be Endogenous Progenitors. To induce neurotoxic injury, we injected 200 nmol NMDA into the vitreous chamber of adult Sprague–Dawley rat (postnatal 6–7 weeks) eyes. In the adult rat, this dose of NMDA is known to cause loss of the cells in the GCL and a reduction in the thickness of the inner plexiform layer (IPL) without affecting other retinal layers (18). To observe dividing cells, we injected BrdUrd into the vitreous chamber (30 nmol) and i.p. space (150 mg/kg body weight) 5 h before death at 1, 1.5, 2, 2.5, 3, and 4 days after NMDA treatment (DAN) (Fig. 1A). As a control, we injected PBS into the vitreous chamber instead of NMDA, and found no BrdUrd-labeled cells (Fig. 1B).
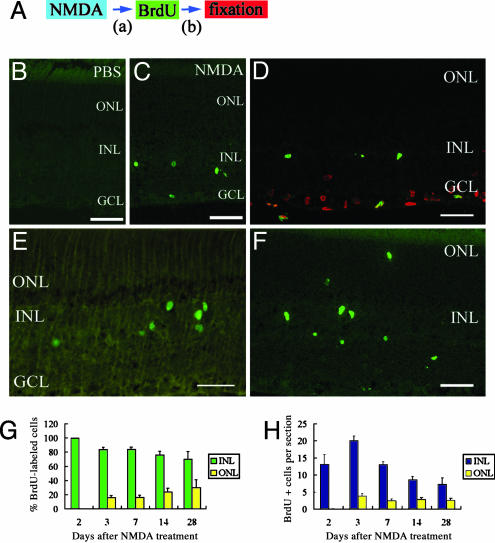
Fig. 1.
Some cells undergo proliferative response to NMDA-induced neurotoxic damage. (A) Animal protocols. (B) Section of retina treated with PBS and BrdUrd. (C–E) Sections of retina treated with NMDA and BrdUrd (2 days after NMDA treatment; DAN2) and fixed (DAN2) (i.e., a = 2 days and b = 5hin A). (C) BrdUrd-labeling (green) was detected in the INL and GCL. (D) BrdUrd-labeled cells (green) in the GCL were positive for TUNEL (red) whereas those in the INL were negative. (E) Ki-67-labeled cells (green) were detected in the INL, but not in the GCL. (F) Sections of retina treated with NMDA and BrdUrd (DAN2) and fixed (DAN3) (i.e., a = 2 days and b = 1 day in A). Some BrdUrd-labeled cells (green) were detected in the ONL. (G) Percentage of BrdUrd-labeled cells in the INL and ONL at different times after NMDA treatment. (H) Numbers of BrdUrd-labeled cells in the INL and ONL per section at different times after NMDA treatment. All data are mean ± SD obtained from at least four animals. (Scale bar, 50 μm.)
When we injected BrdUrd at DAN1, -1.5, -3, and -4, there was no difference between NMDA- and control-treated rats (data not shown). However, when we injected BrdUrd at DAN2, increased numbers of BrdUrd-positive cells were detected in the central retina (Fig. 1C). Most of the BrdUrd-positive cells were found in the inner nuclear layer (INL). Some of the cells in the GCL were labeled with BrdUrd, but TUNEL assay indicated that these cells were labeled in the process of apoptosis (Fig. 1D). BrdUrd-labeled cells in the INL were negative for TUNEL. Moreover, cells immunoreactive for Ki-67, a nuclear antigen expressed by proliferating cells, were also found in the INL, but not in the GCL (Fig. 1E). These data suggest that at DAN2 some cells in the INL entered the mitotic cell cycle.
When we injected BrdUrd at DAN2.5, we found some BrdUrd-labeled cells in the INL, but the numbers of cells were smaller than that seen at DAN2. In all following experiments, we injected BrdUrd at DAN2, and followed the fate of BrdUrd-positive cells by killing the rats at different intervals (3–28 days) after NMDA treatment. At DAN3, some BrdUrd-labeled cells were detected in the outer nuclear layer (ONL) (Fig. 1F). The percentage of BrdUrd-labeled cells in the ONL increased by degrees (Fig. 1G), suggesting that proliferating cells in the INL migrated to the ONL. At DAN2, none of the BrdUrd-labeled cells were detected in the ONL (524 cells, 40 sections examined, n = 15 animals), but at DAN14, 24 ± 5% (324 cells, 29 sections examined, n = 5 animals) of the BrdUrd-labeled cells were in the ONL. The total number of BrdUrd-labeled cells increased at DAN3, and from that day decreased gradually (Fig. 1H), which might be due to the cell death. It is likely that most BrdUrd-labeled cells in the INL divided only once. BrdUrd-labeled cells remained for at least 9 months after NMDA treatment (data not shown).
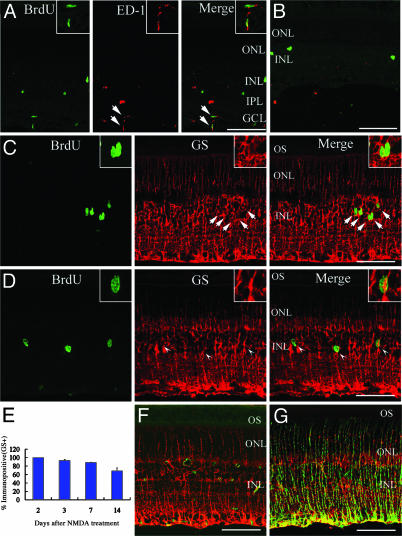
To identify the cell type that was labeled with BrdUrd, we next performed immunohistochemistry double staining for BrdUrd and cell-specific markers. At DAN2, some BrdUrd-labeled cells in the GCL and IPL expressed ED-1 (marker for activated phagocytes; Fig. 2A), consistent with previous reports that microglia are activated after neurotoxic damage (19). However, none of the BrdUrd-labeled cells in the INL expressed ED-1. At DAN14, the number of ED-1-labeled activated microglia significantly decreased, and few BrdUrd-labeled cells expressed ED-1 (Fig. 2B). Thus, microglia were activated in the early period after neurotoxic injury, but most of them did not exist longer than 2 weeks. We next used anti-GS (Müller glial marker). At DAN2, all of the BrdUrd-labeled cells in the INL expressed GS (Fig. 2C, 126 cells examined, n = 6 animals). Thus, Müller glia reentered the cell cycle at DAN2 in the adult rat retina, which is identical to results obtained in the chick (15). Furthermore, we found that some BrdUrd-labeled cells did not express GS at DAN3 (Fig. 2D). We followed the percentage of BrdUrd-labeled cells in the INL positive for GS (Fig. 2E). At DAN3, 93 ± 2% (137 cells examined, n = 6 animals) of BrdUrd-positive cells expressed GS, whereas only 69 ± 7% (125 cells examined, n = 4 animals) of BrdUrd-positive cells were GS positive at DAN14, suggesting that the proliferating Müller glial cells converted to some other cell type. We next used the anti-nestin (marker for progenitors) antibody for immunofluorescent analysis. In control retinas (treated with PBS), only a small number of cells expressed nestin (Fig. 2F), but numerous nestin-positive cells were detected in the retinal sections after NMDA treatment (Fig. 2G). Many nestin-labeled cells coexpressed GS, suggesting that Müller glial cells became active.
Fig. 2.
Müller glia proliferate, become active, and convert to other cell types. (A) At DAN2, some BrdUrd-labeled cells (green) in the GCL and IPL expressed ED-1 (red). (B) At DAN14, most BrdUrd-labeled cells (green) did not express ED-1 (red). (C) At DAN2, all BrdUrd-labeled cells (green) in the INL expressed GS (red). Note that GS labeling is detected from GCL to ONL, but not in OS (outer segments of photoreceptors). (D) At DAN3, some BrdUrd-labeled cells (green) did not express GS (red). Arrowhead, BrdUrd-positive/GS-negative cell. (Inset) Magnified view. (E) Percentage of BrdUrd-labeled cells in the INL that expressed GS at different times after NMDA treatment. Data are mean ± SD obtained from at least four animals. (F) Sections of retina treated with PBS and labeled with nestin (green) and GS (red). (G) Sections of retina obtained at DAN2 and labeled with nestin (green) and GS (red). (Insets in A and C) Magnified view of double labeling cells (arrows). (Scale bar, 100 μm.)
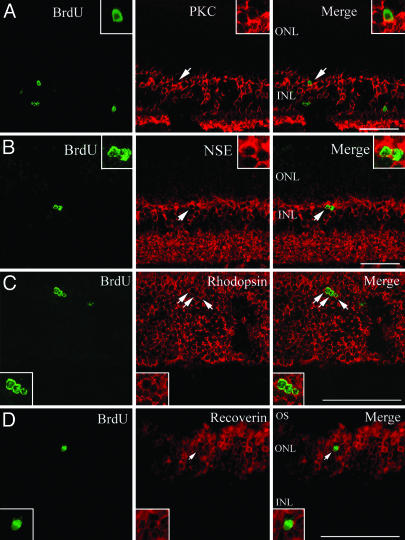
To further identify the fate of BrdUrd-labeled cells that did not express GS, we examined whether BrdUrd-positive cells expressed neuronal markers. We used anti-protein kinase C (PKC; marker for bipolar cells), anti-NSE (in the INL, specifically expressed by bipolar cells), anti-rhodopsin (marker for rod photoreceptors), and anti-recoverin (marker for rod photoreceptors, cone photoreceptors, and cone bipolar cells). At DAN14 or -28, we found the expression of PKC, NSE, rhodopsin, and recoverin in some BrdUrd-labeled cells (Fig. 3), suggesting that Müller glia-derived cells differentiated to bipolar cells and rod photoreceptors. Interestingly, the nuclei of the rhodopsin and recoverin-labeled cells were round and small, which is characteristic of rod photoreceptors. We also labeled with anti-calbindin (marker for horizontal cells), anti-HPC-1 (marker for amacrine cells), and Thy1.2 (marker for ganglion cells), but did not find BrdUrd labeling in these cell types (data not shown). Assuming that factors required to generate all cell types are absent in the adult retina, we next examined whether intrinsic cues and extrinsic signals control the differentiation of Müller glia-derived cells.
Fig. 3.
Some BrdUrd-labeled cells express neuronal markers. Shown are sections of retina obtained at DAN14. Double labeling with BrdUrd (green) and PKC (red in A), NSE (red in B), rhodopsin (red in C), and recoverin (red in D). (Inset) Magnified view of double labeling cells (arrows). (Scale bar, 100 μm.)
RA Promotes Bipolar Cell Genesis. To induce the differentiation of neural cells more effectively, we tested whether exogenous growth factors stimulate neural regeneration. We injected BrdUrd along with growth factors into the vitreous chamber of adult Sprague–Dawley rat (postnatal 6–7 weeks) eyes at DAN2. From that day, we performed three injections of growth factors every other day, and killed rats at DAN14 (Fig. 4A). We selected fibroblast growth factor 2 (FGF-2), epidermal growth factor (EGF), RA, Activin-A, and bovine insulin for analysis. We also tested the combinations of FGF-2/EGF and FGF-2/bovine insulin. We could not find any increase of BrdUrd-labeled cells either in the central or peripheral region of the retina after treatment with each growth factor (data not shown). There was no significant difference in the percentage of BrdUrd-labeled cells located in INL and ONL among the retina after treatment with each growth factor (data not shown).
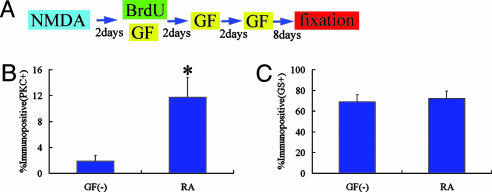
Fig. 4.
RA treatment promotes bipolar cell differentiation. (A) Animal protocols. (B and C) Percentage of BrdUrd-labeled cells in the INL that expressed PKC (B) and GS (C). Cell counts were made from retinas treated with RA and control retinas (without growth factor treatment). *, P < 0.01 compared to the control retina. All data are mean ± SD obtained from four animals. GF, growth factor.
However, 1 week after the last injection of RA (DAN14), we found significantly increased numbers of BrdUrd/PKC positive cells in the INL of the central retina (Fig. 4B), whereas the overall BrdUrd-labeled cell number remained constant. In the control retinas (without growth factor treatment), BrdUrd/PKC-labeled cells comprised only 2 ± 1% (312 cells examined, n = 4 animals) of the population. We found that 12 ± 3% (204 cells examined, n = 4 animals) of the BrdUrd-labeled cells in the INL were immunoreactive for PKC in the RA-treated retina (P < 0.01). RA treatment did not change the BrdUrd/GS-positive cell population (Fig. 4C), and we did not find any increase in the number of BrdUrd-labeled cells positive for other neuronal markers. BrdUrd-labeled cells in the INL were TUNEL-negative in the RA-treated retina (data not shown). These data suggest that RA predominantly promoted bipolar cell differentiation. Some of the uncommitted cells that would have remained immature may have been induced to differentiate into bipolar cells. We also examined another possibility that RA caused some fraction of the Müller glia to express PKC. However, we could not find double labeled cells for GS and PKC either in the RA-treated retina or the control retina (Fig. 8, which is published as supporting information on the PNAS web site).
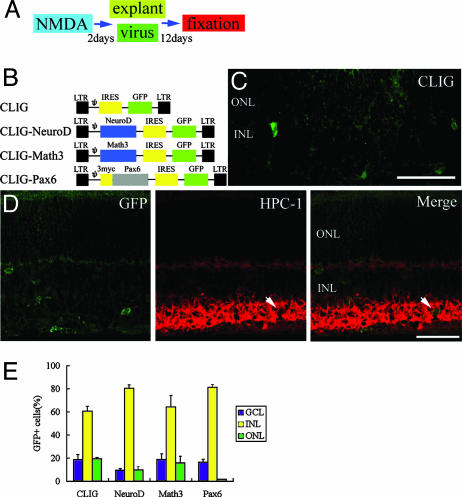
Math3 and NeuroD Promote Amacrine Cell Genesis. Recent studies have demonstrated that the bHLH gene Math3/NeuroD and the homeobox gene Pax6 regulate amacrine cell fate specification (20, 21). To define the role of intrinsic cues in the differentiation of Müller glia-derived cells, we misexpressed these genes in NMDA-treated retinas by using a retroviral expression system. We applied virus to retinal explants, which were isolated from adult Sprague–Dawley rat (postnatal 6–7 weeks) eyes at DAN2 (Fig. 5A). Retroviruses are infectious only to mitotic cells, so the cells that were infected on the second day after NMDA treatment and survived for 2 weeks should be derived from activated Müller glia. We used a replication-incompetent retrovirus, CLIG, which directs expression of GFP. Pax6, Math3,or NeuroD cDNA were inserted upstream of the internal ribosomal entry site (IRES) (Fig. 5B). After 14 days, we examined the fates of the infected cells by monitoring GFP-positive cells. When we applied the control virus CLIG, 69 ± 4% (117 cells examined, n = 6 animals) of the infected cells expressed GS, and only a small number of them expressed retinal neuronal markers (Fig. 5C). These data are similar to the results observed in the in vivo studies described above (Figs. 2E and 3).
Fig. 5.
NeuroD and Math3 induce differentiation to amacrine cells. (A) Animal protocols. (B) Retroviral vectors. LTR, long terminal repeat. (C and D) Sections of retinal explants isolated from eyes and infected with CLIG (C) and CLIG-NeuroD (D) at DAN2. After 2 weeks of culture, the explants were subjected to immunohistochemistry with anti-GFP (green) and anti-HPC-1 (red) antibodies. GFP is expressed in the cytoplasm of the infected cell. Arrows, double labeling cells. (Scale bar, 100 μm.) (E) Percentage of infected cells that were located in the GCL, INL, and ONL. All data are mean ± SD obtained from at least three animals.
When we applied CLIG-NeuroD and CLIG-Math3, we found the expression of HPC-1 in some GFP-labeled cells, suggesting that misexpression of NeuroD or Math3 induced Müller glia-derived cells to differentiate to amacrine cells (Figs. 5D and 6D). When we applied CLIG-Pax6, we did not find HPC-1/GFP labeled cells. However, misexpression of Pax6 promoted generation of INL cells (Fig. 5E). It is likely that Pax6 alone prevents migration to the ONL and induces the generation of the INL cells, whereas most of these cells did not become mature interneurons.
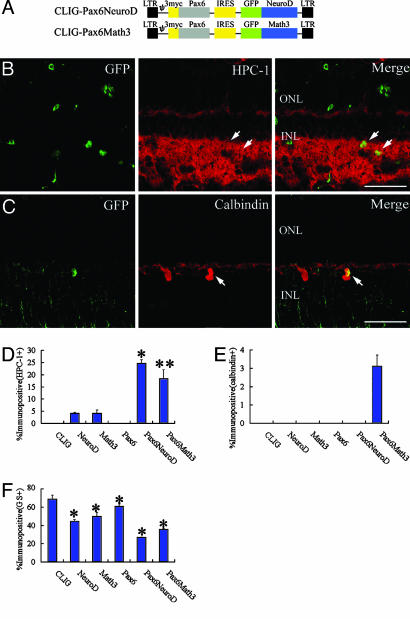
Fig. 6.
NeuroD/Math3 together with Pax6 promote differentiation to amacrine cells. (A) Structures of the recombinant retrovirus. The bHLH genes were fused with GFP, and Pax6 has three repeats of Myc tag. (B and C) Sections of retinal explants isolated (DAN2) and infected with CLIG-Pax6-NeuroD (B) and CLIG-Pax6-Math3 (C) (DAN2). After 2 weeks of culture, the explants were subjected to immunohistochemistry with anti-GFP (green), anti-HPC-1 (B; red), and anti-calbindin (C; red) antibodies. GFP gene is fused with each bHLH gene, so GFP is expressed in the nucleus of the infected cell. Arrows, double labeling cells. (Scale bar = 100 μm.) (D–F) Percentage of cell types induced by misexpression of bHLH genes and Pax6. (D) HPC-1+ cells; *, P < 0.001 compared to NeuroD; **, P < 0.01 compared to Math3. (E) Calbindin+ cells. (F) GS+ cells; *, P < 0.05 compared to CLIG. All data are mean ± SD obtained from at least three animals.
Because the combination of homeobox and bHLH genes has been shown to be important for retinal cell type specification (21, 22), Pax6 was coexpressed with Math3 or NeuroD in the NMDA-treated retinal explant cultures. For coexpression of the two factors, the bHLH factors were fused with GFP and each was coexpressed with Pax6 (Fig. 6A). More than 99% of the cells infected with these retroviruses coexpressed both bHLH and homeobox genes (data not shown). The majority of the infected cells were TUNEL-negative after 1 week of culture (data not shown). Coexpression of NeuroD and Pax6 predominantly promoted the differentiation to amacrine cells (HPC-1+; Fig. 6 B and D). A total of 25 ± 2% of the infected cells expressed HPC-1 (331 cells examined, n = 4 animals). Coexpression of Math3 and Pax6 promoted not only amacrine cell genesis (19 ± 4%, 329 cells examined, n = 4 animals; Fig. 6D) but also the horizontal cell genesis (3 ± 0.6%, 192 cells examined, n = 4 animals; Fig. 6 C and E). In addition, these genes significantly decreased the population of Müller glia (Fig. 6F), suggesting that Math3/NeuroD and Pax6 regulate the neuronal versus glial fate choice.
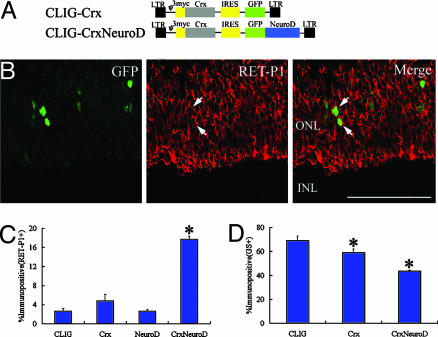
Coexpression of Crx and NeuroD Promotes Rhodopsin Expression. It has been shown that the homeobox gene Crx and the bHLH gene NeuroD code for photoreceptor specification (20, 23–25). We misexpressed each gene in the NMDA-treated retina. Crx cDNA was inserted upstream of the IRES in the CLIG retroviral vector (Fig. 7A). We applied CLIG-Crx and CLIG-NeuroD to NMDA-treated retinal explants. When we applied CLIG, 3 ± 0.5% (182 cells examined, n = 6 animals) of the infected cells expressed RET-P1 (rhodopsin; rod photoreceptors; Fig. 6C), which is identical with the results of the in vivo studies described above (Fig. 3 C and D). When we applied CLIG-Crx or CLIG-NeuroD, we found a decreased population of GS-positive Müller glial cells (Fig. 7D). However, Crx or NeuroD alone did not promote rhodopsin expression significantly (Fig. 7C).
Fig. 7.
Coexpression of Crx and NeuroD promotes rhodopsin expression. (A) Retroviral vectors. (B) Sections of retinal explants isolated (DAN2) and infected with CLIG-Crx-NeuroD (DAN2). After 2 weeks of culture, the explants were subjected to immunohistochemistry with anti-GFP (green) and anti-RET-P1 (red) antibodies. GFP gene is fused with NeuroD gene, so GFP is expressed in the nucleus of the infected cell. Arrows, double labeling cells. (Scale bar = 100 μm.) (C and D) Percentage of cell types induced by expression of Crx, NeuroD, and Crx-NeuroD. (C) RET-P1+ cells. (D) GS+ cells. All data are mean ± SD obtained from at least three animals. *, P < 0.01 and < 0.05 compared to CLIG in C and D, respectively.
We next examined the effects of combined expression of Crx and NeuroD on Müller glia differentiation. For coexpression of the two genes, NeuroD was fused with GFP and coexpressed with Crx (Fig. 7A). Coexpression of Crx and NeuroD significantly increased the expression of rhodopsin (Fig. 6 B and C). A total of 18 ± 0.6% of the infected cells expressed rhodopsin (P < 0.001, 174 cells examined, n = 4 animals). The majority of the infected cells were TUNEL-negative after 1 week of culture (data not shown). Thus, the increased rhodopsin expression induced by Crx and NeuroD was not the result of apotosis of other cell types. These data suggest that the combination of Crx and NeuroD is important for the expression of photoreceptor-specific phenotype from Müller glia-derived cells.
Discussion
We demonstrated that Müller glia of adult mammalian retina proliferated in response to acute damage with NMDA, and some of the proliferated cells expressed bipolar-specific or rod photoreceptor-specific markers. RA treatment increased bipolar cell genesis. Misexpression of homeobox and bHLH genes promoted the regeneration of various retinal neural cells.
There have been some reports that indicate the participation of glial cells in neural regeneration. Radial glial cells of rat embryo are considered as neuronal progenitors, and after neurogenesis, they shift toward the exclusive generation of astrocytes (26). They also act as migrational guides for their neuronal progeny (27). The subventricular zone astrocytes of the adult mammalian brain act as neural stem cells in both the normal and regenerating brain (28).
With regard to the retina, it has been proposed that Müller glial cells might behave as stem cells after retinal injury in chicken and fish (13–15). In this study, we found Müller glia of adult mammalian retina also have a potential to dedifferentiate and produce retinal neurons. Müller glia recognize a variety of neuronal signals and actively control levels of K+, H+, and neurotransmitters in the extracellular space. Beyond that, in response to retinal damage, Müller glia could play a role as retinal progenitors and regenerate some retinal neurons. In this study, the number of nestin-positive Müller glial cells were higher than that of BrdUrd-positive ones. It is likely that many Müller glial cells became active glia, but only a part of them underwent proliferation. It also should be thought that we could not detect all proliferative cells by a single BrdUrd injection.
It has been shown that interactions between the progenitor cells and their microenvironment play an essential role in the determination of cell phenotype (29–34). RA can influence the development of rod photoreceptor cells in retinal cell cultures (35). Several studies have shown that RA may have a role in the control of phenotypic choice in the developing retina in vivo (36, 37). In this study, RA predominantly promoted the differentiation of bipolar cells from stimulated Müller glia in vivo. We could not find an increase in rod photoreceptor differentiation. This discrepancy could be due to different experimental conditions: in vitro embryonic retinal cell cultures (35) and in vivo adult retina after acute injury (this study).
Recent studies have demonstrated that the differentiation of retinal neurons is controlled by intrinsic cues, such as bHLH and homeobox-type transcription factors (20–25, 38, 39). This study indicated that Math3 and NeuroD induce amacrine cell genesis from Müller glia, the combination of Math3 or NeuroD with Pax6 promoted amacrine genesis, and the combination of Math3 and Pax6 induced horizontal cells. In addition, the combination of Crx and NeuroD promoted the expression of photoreceptor-specific phenotype. These data suggest that the bHLH genes and homeobox genes control the differentiation of Müller glia-derived progenitors. Homeobox and bHLH genes might give the different information on cells. Misexpression of Pax6 promoted generation of INL cells, but most of them were immature interneurons or Müller glia. Thus, Pax6 is not sufficient to generate mature amacrine or horizontal cells. Pax6 may provide the INL-specific identity to Müller glia-derived progenitors. Although Math3 and NeuroD alone can induce amacrine cell genesis, their effect is still small. However, they can more effectively specify the neuronal subtypes when the homeobox gene Pax6 is coexpressed. NeuroD and Math3 may give the neuronal lineage information, whereas Pax6 gives the position-specific identity.
In conclusion, the adult mammalian retina has the potential for regeneration. Müller glia could proliferate after neurotoxic damage, and produce new neurons in the adult mammalian retina. Intrinsic and extrinsic cues promoted and partially controlled the differentiation of these cells into retinal neurons. Our results may provide an alternative strategy for cell replacement therapies that make use of the regenerative capacity of endogenous progenitors. Müller glia may become a target for drug delivery and gene therapies for retinal degenerative diseases. However, we did not confirm whether the produced neurons integrated into the retinal neuronal circuit. Further investigations to efficiently activate Müller glia to produce more neurons and make them integrate in the retinal circuit would expand the possibility of retinal regeneration therapies.
Supplementary Material
Acknowledgments
We thank Masato Nakafuku for technical advice, Peter Gruss (Max Planck Institute of Biophysical Chemistry, Göttingen, Germany) for Pax6 cDNA, and J. F. McGinnis (University of Oklahoma Health Sciences Center, Norman) and R. J. Elias (University of Manitoba, Winnipeg, Canada) for anti-recoverin antibody. This study was performed through the Advanced and Innovational Research program in Life Sciences from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NMDA, N-methyl-d-aspartate; DAN, days after NMDA treatment; RA, retinoic acid; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; bHLH, basic helix–loop–helix; GS, glutamine synthetase; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; ONL, outer nuclear layer; NSE, neuron-specific enolase; IRES, internal ribosome entry site.
References
- 1.Lund, R. D., Kwan, A. S., Keegan, D. J., Sauve, Y., Coffey, P. J. & Lawrence, J. M. (2001) Prog. Retin. Eye Res. 20, 415–449. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi, M., Palmer, T. D., Takahashi, J. & Gage, F. H. (1998) Mol. Cell. Neurosci. 12, 340–348. [DOI] [PubMed] [Google Scholar]
- 3.Young, M. J., Ray, J., Whiteley, S. J., Klassen, H. & Gage, F. H. (2000) Mol. Cell. Neurosci. 16, 197–205. [DOI] [PubMed] [Google Scholar]
- 4.Nishida, A., Takahashi, M., Tanihara, H., Nakano, I., Takahashi, J. B., Mizoguchi, A., Ide, C. & Honda, Y. (2000) Invest. Ophthalmol. Vis. Sci. 41, 4268–4274. [PubMed] [Google Scholar]
- 5.Akita, J., Takahashi, M., Hojo, M., Nishida, A., Haruta, M. & Honda, Y. (2002) Brain Res. 954, 286–293. [DOI] [PubMed] [Google Scholar]
- 6.Temple, S. (2001) Nat. Rev. Neurosci. 2, 513–520. [DOI] [PubMed] [Google Scholar]
- 7.Gould, E. & Gross, C. G. (2002) J. Neurosci. 22, 619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson, C. B., Momma, S., Clarke, D. L., Risling, M., Lendahl, U. & Frisen, J. (1999) Cell 96, 25–34. [DOI] [PubMed] [Google Scholar]
- 9.Magavi, S. S., Leavitt, B. R. & Macklis, J. D. (2000) Nature 405, 951–955. [DOI] [PubMed] [Google Scholar]
- 10.Nakatomi, H., Kuriu, T., Okabe, S., Yamamoto, S., Hatano, O., Kawahara, N., Tamura, A., Kirino, T. & Nakafuku, M. (2002) Cell 110, 429–441. [DOI] [PubMed] [Google Scholar]
- 11.Tropepe, V., Coles, B. L., Chiasson, B. J., Horsford, D. J., Elia, A. J., McInnes, R. R. & van der Kooy, D. (2000) Science 287, 2032–2036. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad, I., Tang, L. & Pham, H. (2000) Biochem. Biophys. Res. Commun. 270, 517–521. [DOI] [PubMed] [Google Scholar]
- 13.Braisted, J. E., Essman, T. F. & Raymond, P. A. (1994) Development (Cambridge, U.K.) 120, 2409–2419. [DOI] [PubMed] [Google Scholar]
- 14.Raymond, P. A. & Hitchcock, P. F. (1997) Adv. Neurol. 72, 171–184. [PubMed] [Google Scholar]
- 15.Fischer, A. J. & Reh, T. A. (2001) Nat. Neurosci. 4, 247–252. [DOI] [PubMed] [Google Scholar]
- 16.Ooto, S., Haruta, M., Honda, Y., Kawasaki, H., Sasai, Y. & Takahashi, M. (2003) Invest. Ophthalmol. Vis. Sci. 44, 2689–2693. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama, J. & Kageyama, R. (2002) Methods 28, 387–395. [DOI] [PubMed] [Google Scholar]
- 18.Siliprandi, R., Canella, R., Carmignoto, G., Schiavo, N., Zanellato, A., Zanoni, R. & Vantini, G. (1992) Vis. Neurosci. 8, 567–573. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, A. J., Seltner, R. L., Poon, J. & Stell, W. K. (1998) J. Comp. Neurol. 393, 1–15. [PubMed] [Google Scholar]
- 20.Morrow, E. M., Furukawa, T., Lee, J. E. & Cepko, C. L. (1999) Development (Cambridge, U.K.) 126, 23–36. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, T., Hojo, M., Bessho, Y., Tano, Y., Lee, J. E. & Kageyama, R. (2002) Development (Cambridge, U.K.) 129, 831–842. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama, J., Tomita, K., Inoue, T. & Kageyama, R. (2001) Development (Cambridge, U.K.) 128, 1313–1322. [DOI] [PubMed] [Google Scholar]
- 23.Furukawa, T., Morrow, E. M. & Cepko, C. L. (1997) Cell 91, 531–541. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa, T., Morrow, E. M., Li, T., Davis, F. C. & Cepko, C. L. (1999) Nat. Genet. 23, 466–470. [DOI] [PubMed] [Google Scholar]
- 25.Haruta, M., Kosaka, M., Kanegae, Y., Saito, I., Inoue, T., Kageyama, R., Nishida, A., Honda, Y. & Takahashi, M. (2001) Nat. Neurosci. 4, 1163–1164. [DOI] [PubMed] [Google Scholar]
- 26.Malatesta, P., Hartfuss, E. & Gotz, M. (2000) Development (Cambridge, U.K.) 127, 5253–5263. [DOI] [PubMed] [Google Scholar]
- 27.Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. (2001) Nature 409, 714–720. [DOI] [PubMed] [Google Scholar]
- 28.Doetsch, F., Caille, I., Lim, D. A., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. (1999) Cell 97, 703–716. [DOI] [PubMed] [Google Scholar]
- 29.Reh, T. A. & Levine, E. M. (1998) J. Neurobiol. 36, 206–220. [PubMed] [Google Scholar]
- 30.Anchan, R. M. & Reh, T. A. (1995) J. Neurobiol. 28, 133–145. [DOI] [PubMed] [Google Scholar]
- 31.Fuhrmann, S., Kirsch, M. & Hofmann, H. D. (1995) Development (Cambridge, U.K.) 121, 2695–2706. [DOI] [PubMed] [Google Scholar]
- 32.Altshuler, D., Lo Turco, J. J., Rush, J. & Cepko, C. (1993) Development (Cambridge, U.K.) 119, 1317–1328. [DOI] [PubMed] [Google Scholar]
- 33.Jensen, A. M. & Wallace, V. A. (1997) Development (Cambridge, U.K.) 124, 363–371. [DOI] [PubMed] [Google Scholar]
- 34.Davis, A. A., Matzuk, M. M. & Reh, T. A. (2000) Mol. Cell. Neurosci. 15, 11–21. [DOI] [PubMed] [Google Scholar]
- 35.Kelley, M. W., Turner, J. K. & Reh, T. A. (1994) Development (Cambridge, U.K.) 120, 2091–2102. [DOI] [PubMed] [Google Scholar]
- 36.Ruberte, E., Dolle, P., Chambon, P. & Morriss-Kay, G. (1991) Development (Cambridge, U.K.) 111, 45–60. [DOI] [PubMed] [Google Scholar]
- 37.Hyatt, G. A., Schmitt, E. A., Marsh-Armstrong, N. R. & Dowling, J. E. (1992) Proc. Natl. Acad. Sci. USA 89, 8293–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kageyama, R. & Nakanishi, S. (1997) Curr. Opin. Genet. Dev. 7, 659–665. [DOI] [PubMed] [Google Scholar]
- 39.Cepko, C. L. (1999) Curr. Opin. Neurobiol. 9, 37–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.