Abstract
Many insects are highly resistant to plant toxins, such as the cardiac glycoside ouabain. How can the epithelia that must handle such toxins, also be refractory to them? In Drosophila, the Malpighian (renal) tubule contains large amounts of Na+,K+ ATPase that is known biochemically to be exquisitely sensitive to ouabain, yet the intact tissue is almost unaffected by even extraordinary concentrations. The explanation is that the tubules are protected by an active ouabain transport system, colocated with the Na+,K+ ATPase, thus preventing ouabain from reaching inhibitory concentrations within the basolateral infoldings of principal cells. These data show that the Na+,K+ ATPase, previously thought to be unimportant, may be as vital in insect tissues as in vertebrates, but can be cryptic to conventional pharmacology.
Keywords: Na+,K+ ATPase; organic anion transporting polypeptide; oatp; Drosophila melanogaster; Malpighian tubule
Many insects have diets that expose them to highly toxic plant metabolites, such as glycosides, and even those that do not may be refractory to their actions. Historically, this has been explained by several mechanisms, such as detoxification (1, 2), direct excretion (3), or storage excretion (4), sometimes coupled by active resorption of the toxin (3). In some cases, critical residues in target proteins have been found to have mutated to confer resistance (5). These plant and insect strategies can be seen as an “arms race” between herbivores and their host plants (6).
Ouabain, a known, potent inhibitor of the Na+,K+ ATPase (7), fails to inhibit fluid secretion by the Malpighian (renal) tubules of many insects, including Drosophila melanogaster (8, 9), Formica polyctena (10), the New Zealand Alpine weta (11) and Glossina morsitans (12). Indeed, in the bloodsucker Rhodnius, it can mildly increase fluid secretion rates (13). Accordingly (and in contrast to the vertebrate literature), the universal basolateral Na+,K+ ATPase is not seen as important in most insect epithelia, which are almost invariably energized by an apical plasma membrane proton-motive V ATPase (14–17). This pump provides a convenient explanation for the apparent absence of ouabain-sensitive ATPase; the basolateral membrane is seen as being dominated by potassium channels, rather than pumps.
However, there are problems with this model. In Drosophila the Na+,K+ ATPase α-subunit is encoded by a single copy gene, contains high-affinity ouabain binding sites, and is exquisitely sensitive to ouabain in vitro (18), with IC50 ≈ 10–7 M. Significantly, immunocytochemistry has also shown that Na+,K+ ATPase is abundantly expressed in Malpighian tubule (18). However (as in other insect tubules), ouabain has either no (8) or little (9) effect on fluid secretion, even at concentrations of 10–3 M. This is the “ouabain paradox.”
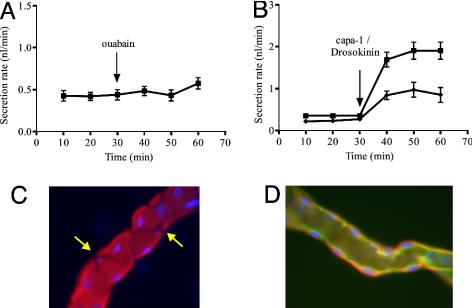
Using Malpighian tubules from wild-type (Oregon R) flies, secretion assays confirm that ouabain, even at 10–3 M, has no effect on the rate of fluid secretion (8) (Fig. 1A). However, immunocytochemistry (Fig. 1 C and D) shows that, within the main, fluid-secreting segment of tubules, the Na+,K+ ATPase is abundant, but only in principal cells [as seen by the excluded stellate cells, identified by their smaller nuclei (19) Fig. 1C], and that it is localized to the basolateral membrane. The basolateral localization was confirmed by comparison with an apically localized protein, a GFP-tagged V ATPase subunit (Fig. 1D). These results confirm earlier reports of Na+,K+ ATPase expression in tubule (18), but additionally identify the cell type and the subcellular localization.
Fig. 1.
The ouabain paradox. (A) Even at 10–3 M, ouabain fails to inhibit secretion by adult Drosophila Malpighian tubules. [Fluid secretion assays were performed as described elsewhere (8). Data as mean ± SEM, n = 10 tubules.] (B) Na+,K+ ATPase is nonetheless important for fluid secretion. Tubules were maximally stimulated at 30 min with 10–7 M of each of the diuretic neuropeptides CAPA1 (41) and Drosokinin (42). In P{ATPα}/+ heterozygotes (diamonds), stimulated secretion rates are significantly depressed, compared with wild-type flies (squares). (Data as mean ± SEM, n = 7 tubules.) (C and D) Localization of the Na+,K+ ATPase in the Drosophila Malpighian tubule by immunocytochemistry with anti-Na+,K+ ATPase α-subunit antibody. (C) Na+,K+ ATPase is confined to principal cells and appears basolateral. Note exclusion from stellate cells (arrows), which can be distinguished both on the basis of shape and smaller nuclear size (19, 27). (D) Confocal section, showing basolateral location of Na+,K+ ATPase (red). The apical domain is marked by a transgenic vhaSFD-GFP construct [kind gift of X. Morin: (28)], marking the apical microvilli (green). To aid discrimination of apical and basal domains, nuclei are labeled blue with 4′,6-diamidino-2-phenylindole (DAPI). In all micrographs, the tubule diameter can be taken as 35 μm.
If the Na+,K+ ATPase in principal cells is playing a functional role, then flies mutant for the Na+,K+ α-subunit might show defective urinary function. Accordingly, fluid secretion assays were performed on flies heterozygous for a lethal P-element insertion (20); these thus carry only one functional copy of the α-subunit. Basal secretion rates were similar to wild-type, but after neuropeptide stimulation with capa-1/Drosokinin (10–7 M), the P{ATPα}/TM3 tubules show a significantly reduced fluid secretion response compared with wild-type tubules (Fig. 1B). The Na+,K+ ATPase is thus abundant in the main ion-transporting cell type and functionally important, but anomalously refractory to ouabain.
To resolve this paradox, we hypothesized that the basolateral Na+,K+ ATPase might be protected by a colocated active ouabain excretory mechanism. The basolateral membrane of the tubule is thrown into deep infoldings and underlain by a basement membrane (21), so it is possible to imagine a standing gradient of ouabain such that most of the Na+,K+ ATPase is not exposed to inhibitory concentrations of ouabain, even when very high concentrations are added to the bathing solution. This postulated transport is plausible, because Malpighian tubules excrete organic solutes, such as dyes and acylamides, by an active transport process (22). In addition, the Malpighian tubules of Oncopeltus fasciatus were reported to excrete ouabain, although only passively (3). However, here, we show that active transport exists, characterize it, localize it to the same cell type and membrane as the Na+,K+ ATPase, and identify by reverse genetics at least one gene responsible.
Experimental Procedures
Drosophila. Oregon R flies were kept on standard medium in tubes at 25°C, 12:12 h photoperiod, and 55% relative humidity. For dissection, flies were anaesthetized by chilling on ice and decapitated before dissecting out tubules in Schneider's medium. Fluid secretion assays were performed as described (8). All chemicals and drugs were obtained from Sigma.
For RNA interference (RNAi) constructs, a hairpin loop (23, 24), directed against 283 bp of the coding region of 58Db, and with a 501-bp spacer, were cloned into pP{UAST} (25), and germ-line transformed into w1118 embryos, according to standard protocols. Expression in principal cells was driven with line c42 (26, 27).
Tubule Ouabain Transport Assays. Ouabain was labeled 3H ouabain (Amersham Pharmacia) and added to tubule secretion assays as described above. Secreted fluid was collected over the next 2 h, the volume measured, and counted in Optiflow SAFE scintillant. Transport ratios were calculated as the ratio of specific activities of secreted:bathing fluid. Values >1 thus imply concentration of ouabain by the tubule.
TLC. After ouabain transport assays as described above, 1-μl aliquots of secreted fluid and of authentic 3H ouabain were dried onto a 20 × 20 cm Polygram Sil G/UV254 plate (Machery-Nagel GmbH, Düren, Germany), and run out by using PBS (pH 7) as eluent. This system produces Rf values around 0.5 for ouabain, and so should be capable of resolving small differences in mobility. Plates were visualized with a Fuji PhosphorImager with tritium plates, and Rf values measured with nih image 6.1.
Imaging. For Na+,K+ ATPase, monoclonal antibody α5, developed by D. M. Fambrough (18), was obtained from the Developmental Studies Hybridoma Bank (University of Iowa), and visualized with Texas red anti-mouse. To distinguish Na+,K+ ATPase localization from apical microvilli, tubules were taken from a GFP-fusion gene-trap of the V ATPase vhaSFD subunit (28). For visualization of ouabain transport, tubules were incubated in 2 × 10–5 M 9-anthroyl ouabain (Molecular Probes) (29) for 10–15 min, and tubules were examined immediately by confocal microscopy (Zeiss 510 Meta).
Oatp anti-peptide antibodies were raised in rabbit (by Genosphere, Paris) and affinity purified according to standard protocols. The epitopes used were: 30B, YTNPSFEQESDQPPD; 33Ea, DQPITPLLAKKSEQE; 33Eb, QPESPRPQSPETDF; 58Db, DEKTVQAKQSDDIE; 58Dc, LKIFDEDVKEVEMK; and 74D, PASNGRTLEVSESK. An N-terminal cysteine was added to permit conjugation to BSA. Tubule immunocytochemistry was as described (30), by using either a Zeiss Ortholux or Zeiss 510 Meta confocal microscope.
Expression in NIH 3T3 Cells. NIH 3T3 cells were grown in DMEM at 37°C in 5% CO2. ORFs were amplified, cloned into pcDNA3.1/V5-His-TOPO (Invitrogen), and verified by sequencing. Cells were transiently transfected by the calcium phosphate method, and grown on for 24 h before use. Cells were harvested for Western analysis, by standard protocols, to verify the specificity of the antibodies. For transport assays, cells were seeded into multiwell dishes, and 3H-labeled ouabain (Amersham Pharmacia) was added to 10–6 M for 30 min. Cells were ruptured with ice-cold 5% trichloroacetic acid and counted by using Optiflow SAFE scintillant. Uptake was expressed as a percentage increase in counts, compared with matched mock-transfected controls.
Results and Discussion
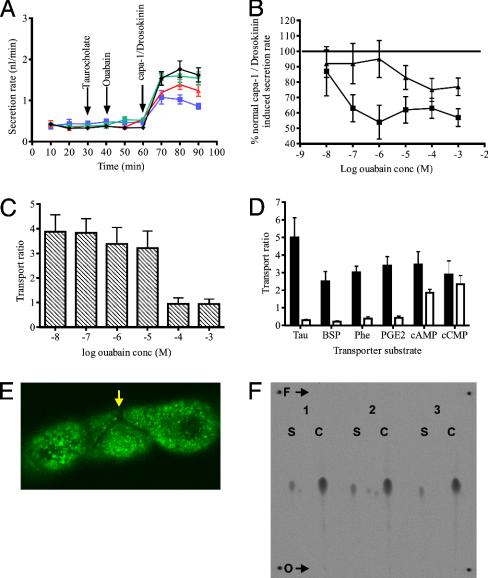
Tubule Sensitivity to Ouabain Can Be Unmasked by Organic Anion-Transporting Polypeptide (oatp) Substrates. In vertebrates, the oatp family transport a wide range of organic solutes, and some members are known to transport ouabain. Accordingly a cardinal oatp substrate, taurocholate, was used in competition assays to determine whether it could unmask the tubules' sensitivity to ouabain. Fig. 2A shows that this is the case: taurocholate at 6 × 10–4 M [chosen as the highest concentration that does not affect secretion: (31)] unmasks Malpighian tubule sensitivity to ouabain. [It should be noted that, in these and other results, ouabain reduced fluid secretion by 50–60%, but not by 100%; the ouabain-insensitive component is believed to reflect K+ entry through basolateral potassium channels and Na+/K+/2Cl– cotransporters (9, 32, 33).]
Fig. 2.
Tubule sensitivity to ouabain can be unmasked by competitive oatp substrates. (A) Taurocholate unmasks tubule sensitivity to ouabain. Fluid secretion was measured every 10 min. At 30 min, taurocholate (6 × 10–4 M) was added; and at 40 min, ouabain (10–3 M). Tubules were then maximally stimulated at 60 min by the joint application of the neurohormones CAPA and DLK (each at 10–7 M). Ouabain alone (green) is indistinguishable from control (black). Taurocholate (red) slightly depresses maximal stimulation, but ouabain in the presence of taurocholate (blue) cuts stimulated secretion by over 50%. (Data as mean ± SEM, n = 10 tubules.) (B) Taurocholate reveals potent ouabain sensitivity of fluid secretion. Data from experiments such as A were used to prepare dose-response curves to ouabain, both in the presence (squares) and absence (triangles) of taurocholate. Addition of taurocholate reduced the apparent IC50 for ouabain from around 10–5 to 10–7 M. This result compares with the IC50 for ouabain inhibition of the Drosophila Na+,K+ATPase in vitro, of 2.5 × 10–7 M. (Data as mean ± SEM, n = 10 tubules.) (C) [3H]ouabain is actively transported. Tubules were incubated in radioactively labeled ouabain of various concentrations, and the concentrations of ouabain in the secreted drop compared with the bathing solution, to calculate the “transport ratio” at each concentration. Transport ratios >1 are indicative of active transport. The apparent k1/2 for ouabain transport is ≈10–5 M. (Data as mean ± SEM, n = 10 tubules.) (D) Alternative oatp substrates abolish active transport of [3H]ouabain. Transport ratios were measured as described for ouabain transport both with (open bars) and without (filled bars) a range of classical oatp substrates, at maximal concentrations shown not to affect tubules directly (data not shown). Tau, taurocholate (6 × 10–4 M); BSP, sulfobromopthalein (2 × 10–6 M); Phe, phenol red (6 × 10–5 M); PGE2, prostaglandin E2 (1 × 10–4 M) each block ouabain transport, whereas cAMP (1 × 10–7 M) and cytidine 2′,3′-cyclic monophosphate (cCMP) (4 × 10–5 M) (although actively transported themselves) do not compete with ouabain. (Data as mean ± SEM, n = 10 tubules.) (E) Cell specificity of ouabain transport. 9-anthroyl ouabain, a fluorescent derivative of ouabain (29), is transported only into principal cells; note the absence of staining in the stellate cell (arrow). Glancing confocal section. (F) Ouabain is transported unchanged through the tubule. Secreted fluid from 20 tubules, incubated in [3H]ouabain, was pooled over 1–2 h, until a volume of 1 μl had been accumulated. Secreted fluid (S) and authentic 3H-labeled ouabain controls (C) were compared by TLC. O, origin; F, eluent front. Rf values: 0.46 ± 0.009 secreted compared with 0.47 ± 0.003 (n = 3 replicates), not significant by Student's t test for paired values, two-tailed.
The experiment was repeated by using various concentrations of ouabain (10–8 M to 10–3 M) in the presence or absence of taurocholate at 6 × 10–4 M to generate ouabain dose-response curves (Fig. 2B). In the presence of taurocholate, the IC50 for ouabain is between 10–7 M and 10–8 M. This result is in close agreement with the published IC50 for Drosophila Na+,K+ ATPase to ouabain in vitro (2.5 × 10–7 M).
Ouabain Is Actively Excreted by the Tubule. Passive ouabain transport, as has been reported (3), would not suffice to bring ouabain to low levels in the basolateral space; an active transport mechanism would be needed. To test this possibility, the excretion of radiolabeled ouabain in urine was followed at concentrations ranging from 10–8 M to 10–3 M. Transport ratios (34) for each concentration are shown in Fig. 2C. At low bathing concentrations, ouabain is concentrated in the secreted fluid: the transport ratio for each of these concentrations is between 3 and 4. At very high concentrations, saturation of the active transport is reflected by the transport ratio tending to 1, implying that diffusional processes dominate.
The oatps are multispecific transporters, with broad substrate ranges. Several such substrates were selected for competitition assays with ouabain. Taurocholate, sulfobromopthalein (BSP), prostaglandin E2 (PGE2) and phenol red completely abolish active transport of ouabain (Fig. 2D), whereas the cyclic nucleotides cAMP and cytidine 2′,3′-cyclic monophosphate (cCMP) do not. Additionally, the fluorescent ouabain analogue, 9-anthroyl ouabain (29), is taken up by principal cells only (Fig. 2E), locating the transport to the same cell type that contains high levels of Na+,K+ ATPase. Although ouabain could be removed from the haemolymph by intracellular “storage excretion,” or modified before being excreted, the counts in the secreted fluid have precisely the same mobility by TLC in aqueous solvent as labeled ouabain (Fig. 2F), and fewer than 1% of counts remain in the tubule at the end of the experiment (data not shown). We thus believe that virtually all transported ouabain passes into the lumen unmodified.
Strictly, active transport is demonstrated by net flux against an electrochemical gradient. Ouabain is a rhamnose glucoside, and thus uncharged at neutral pH (35, 36); thus, its transport should be unaffected by transepithelial potential. Consistent with this finding, transport of ouabain was similar both at rest and when the tubule was stimulated by the diuretic neuropeptide leucokinin, which collapses the transepithelial potential to near zero (37) (data not shown). So concentration of ouabain in secreted fluid, and a demonstration that it passes unchanged through the tubule, are sufficient to assert that ouabain moves by (primary or secondary) active transport.
Identification and Functional Characterization of the Drosophila oatp Family. From Fig. 2D, it is clear that ouabain is actively transported with a pharmacology characteristic of the oatp family: in vertebrates, oatps can transport ouabain, taurocholate, sulfobromopthalein, and prostaglandin E2, but not cyclic nucleotides.
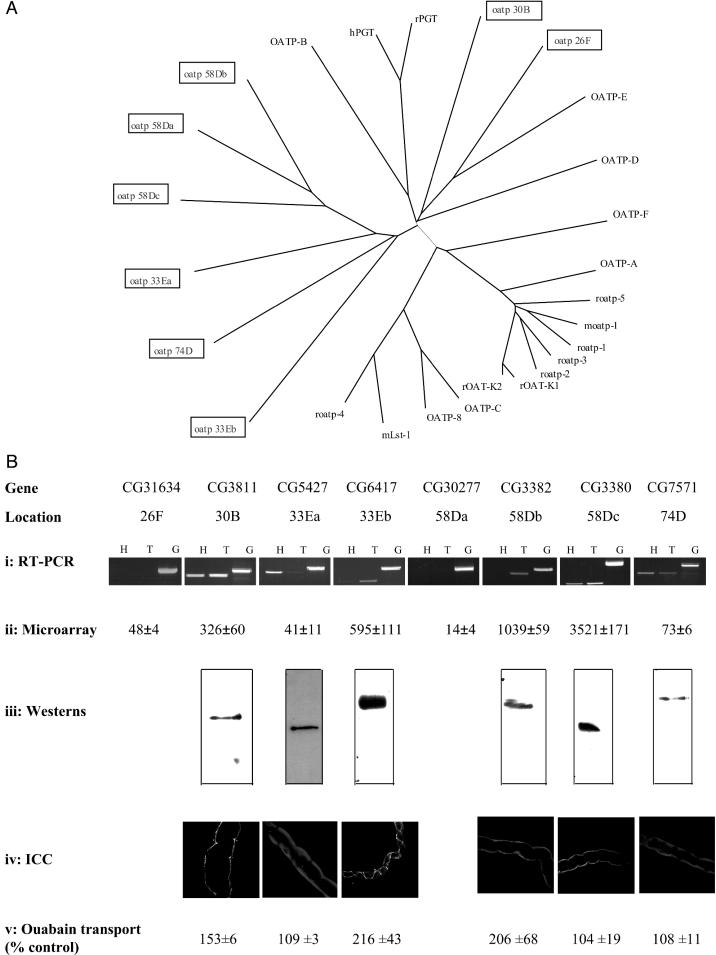
Eight putative Drosophila oatp genes were identified by sequence similarity to vertebrate members. Each of these putative Drosophila members was named according to the chromosome region to which it mapped (Fig. 3A). Two of these genes are located together at chromosomal location 33E, with another three genes clustered together at chromosomal location 58D. The remaining oatp genes are dispersed through the genome. A similar number of oatp genes are conspicuous in the Anopheles genome (data not shown), confirming that the oatps are not unique to Drosophila among insects.
Fig. 3.
Summary of the oatp gene family and its role in Malpighian tubules. (A) Rooted phylogenetic tree representation of relatedness between Drosophila and human oatp families. Tree was prepared by using clustal x (43), and plotted by using treeview (44), using default parameters. (B) Characterization of Drosophila oatps. For each gene (arranged as columns), expression data are shown as follows. (i) RT-PCR, gene-specific primers were designed to bracket introns, and amplified from head (H) or tubule (T) cDNA, or genomic (G) DNA. (ii) Affymetrix microarray signal intensities, from a study on adult Drosophila tubule (39). (iii) For Western blots, affinity purified antibodies against nonconserved regions of each protein were used on NIH 3T3 cell extracts. In each case, the single band is of the predicted size (30B, 126; 33Ea, 79; 33Eb, 75; 58Db, 81; 58Dc, 88; and 74D, 85 kDa), also confirming specificity. (iv) Immunocytochemistry (ICC): the same antibodies were used to identify protein and its location in tubules. (v) Ouabain transport: each reading frame was expressed heterologously in NIH 3T3 cells, and radioactive ouabain uptake was measured and expressed as a percentage relative to mock-transfected controls (mean ± SEM, n = 6).
The similarity between these Drosophila genes and vertebrate oatp members is shown in the gene tree in Fig. 3A. Interestingly, although six of the eight Drosophila genes contain the oatp family consensus (38), and all are clearly oatps, the insect sequences are relatively divergent from human. This finding might be expected if there is a selective pressure on insects to adapt to new plant defenses. Oatps 33Ea and 33Eb share a relatively high level of sequence similarity, although they are also closely related to oatp 74D. This is also the case with oatps 58Da, 58Db and 58Dc.
RT-PCR, using primers designed specifically for each Drosophila oatp gene, showed that six of the eight oatp genes (namely oatps 30B, 33Ea, 33Eb, 58Db, 58Dc, and 74D) were expressed in the Malpighian tubules (Fig. 3Bi). Subsequently, it proved possible to validate these results with a microarray dataset for tubule (39); the agreement is excellent (Fig. 3Bii). Antibodies were generated against the tubule-expressed proteins, and Western blot analysis confirmed expression of these oatps at the protein level (Fig. 3Biii). Immunocytochemical analysis (Fig. 3Biv) placed oatps 30B, 58Db, and 58Dc at the basolateral membrane, oatps 33Ea and 74D to intracellular organelles, and oatp 33Eb to the boundaries between stellate and principal cells and also between principal cells.
Each of the tubule-expressed oatps was expressed in NIH 3T3 cells, and ouabain uptake was measured. Three of them (30B, 33Eb, and 58Db) were capable of significant ouabain transport (Fig. 3Bv).
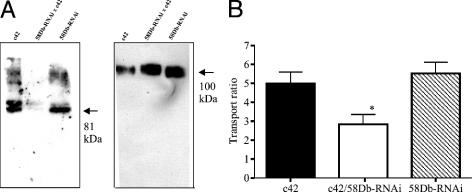
RNAi Confirms a Role for oatp58Db in Malpighian Tubule Active Ouabain Transport. The observation that three of the oatps were capable of taking up ouabain made them candidates for the basolateral entry step in the ouabain transport pathway. The gene oatp58Db was selected for further study, as it was abundant, localized to the basolateral membrane, and showed high rates of active transport in vitro (Fig. 3). Existing mutants of this gene were unavailable, so flies were generated (UAS-oatp58Db-RNAi) that were transgenic for a hairpin loop construct that would be transcribed to double-stranded RNA, under control of the UAS promoter (24). Expression of the RNAi construct was achieved by crossing to the c42 GAL4 enhancer trap line, which drives expression in principal cells only (26, 27). Western blot analysis confirms strong down-regulation of oatp58Db protein levels in the c42/UAS-oatp58Db-RNAi Malpighian tubules, with normal expression of the protein level apparent in the c42 and UAS-oatp58Db-RNAi parental lines (Fig. 4A). The anti-Na+,K+ ATPase antibody was used as a loading control and showed that there was no nonspecific knockdown of transcription. Phenotypic analysis of the c42/UAS-oatp58Db-RNAi Malpighian tubules reveal that they transport significantly less ouabain than either the c42 or UAS-oatp58Db-RNAi parental lines (Fig. 4B). The residual transport activity may reflect residual oatp58Db protein or additional contributions from other transporters; nonetheless, the data show the importance of oatp58Db in ouabain excretion.
Fig. 4.
Knockdown of oatp58Db by RNAi reduces active ouabain transport. (A) Western blot analysis of oatp 58Db expression in RNAi tubules. Tubules were dissected from lines containing a principal cell-specific GAL4 driver (c42), the RNAi construct under UAS control (58Db-RNAi), or both (58Db-RNAi/c42). Only in this last case, RNAi should be activated in the principal cells. (Left) Blot probed with α-oatp58Db. (Right) Blot stripped and reprobed with α-Na, K ATPase, as a loading control, and to verify the specificity of RNAi action. Arrows denote the predicted sizes of each protein. (B) RNAi to oatp58Db reduces active uptake of ouabain by around 50%. Tubules were dissected from parental lines as described in A, and assayed for ouabain transport as described. (data as mean ± SEM, n = 7.)
Conclusions
Taken together, these results both delineate and resolve the ouabain paradox in a genetically tractable model organism. It will be interesting to test the generality of this model in related insect species, for example the Dipteran vectors of human and animal disease. The oatps may be significant targets for insecticides, not just in their own right, but because their broad specificities may permit excretion of otherwise useful insecticides.
More generally, these results illustrate the general principle that basic problems in physiology can be addressed powerfully and fairly quickly by a combination of forward and reverse genetics, bioinformatics, biochemistry, and physiology in an appropriate genetic model organism, an approach we have termed “integrative physiology” (40).
Acknowledgments
We thank Dr. Chris Brett (University of Glasgow) for loan of thin-layer chromatography facilities, and Dr. Marshall Stark (University of Glasgow) for use of the PhosphorImager. This work was supported by grants from the British Biotechnology and Biological Sciences Research Council (BBSRC), and by a Cooperative Award in Science and Engineering studentship (sponsored by Syngenta) (to L.S.T.). J.C.R. was supported by a Wellcome Trust studentship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAi, RNA interference; oatp, organic anion transporting polypeptide.
References
- 1.Berenbaum, M. R. (2002) J. Chem. Ecol. 28, 873–896. [DOI] [PubMed] [Google Scholar]
- 2.Ratzka, A., Vogel, H., Kliebenstein, D. J., Mitchell-Olds, T. & Kroymann, J. (2002) Proc. Natl. Acad. Sci. USA 99, 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meredith, J., Moore, L. & Scudder, G. G. (1984) Am. J. Physiol. 246, R705–R715. [DOI] [PubMed] [Google Scholar]
- 4.Dussourd, D. E., Harvis, C. A., Meinwald, J. & Eisner, T. (1989) Experientia 45, 896–898. [DOI] [PubMed] [Google Scholar]
- 5.Holzinger, F., Frick, C. & Wink, M. (1992) FEBS Lett. 314, 477–480. [DOI] [PubMed] [Google Scholar]
- 6.Berenbaum, M. R. & Zangerl, A. R. (1998) Proc. Natl. Acad. Sci. USA 95, 13743–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glynn, I. M. (1957) J. Physiol. 136, 148–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dow, J. A., Maddrell, S. H., Gortz, A., Skaer, N. J., Brogan, S. & Kaiser, K. (1994) J. Exp. Biol. 197, 421–428. [DOI] [PubMed] [Google Scholar]
- 9.Linton, S. M. & O'Donnell, M. J. (1999) J. Exp. Biol. 202, 1561–1570. [DOI] [PubMed] [Google Scholar]
- 10.Leyssens, A., Dijkstra, S., Van Kerkhove, E. & Steels, P. (1994) J. Exp. Biol. 195, 123–145. [DOI] [PubMed] [Google Scholar]
- 11.Leader, J. P. & Neufeld, D. S. (1997) J. Insect Physiol. 44, 39–48. [DOI] [PubMed] [Google Scholar]
- 12.Gee, J. D. (1976) J. Exp. Biol. 65, 323–332. [DOI] [PubMed] [Google Scholar]
- 13.Maddrell, S. H. P. & Overton, J. A. (1988) J. Exp. Biol. 137, 265–276. [DOI] [PubMed] [Google Scholar]
- 14.Wieczorek, H. (1992) J. Exp. Biol. 172, 335–343. [DOI] [PubMed] [Google Scholar]
- 15.Wieczorek, H., Brown, D., Grinstein, S., Ehrenfeld, J. & Harvey, W. R. (1999) Bioessays 21, 637–648. [DOI] [PubMed] [Google Scholar]
- 16.Harvey, W. R. & Wieczorek, H. (1997) J. Exp. Biol. 200, 203–216. [DOI] [PubMed] [Google Scholar]
- 17.Dow, J. A. T., Davies, S. A., Guo, Y., Graham, S., Finbow, M. E. & Kaiser, K. (1997) J. Exp. Biol. 202, 237–245. [DOI] [PubMed] [Google Scholar]
- 18.Lebovitz, R. M., Takeyasu, K. & Fambrough, D. M. (1989) EMBO J. 8, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dow, J. A. T., Maddrell, S. H. P., Görtz, A., Skaer, N. V., Brogan, S. & Kaiser, K. (1994) J. Exp. Biol. 197, 421–428. [DOI] [PubMed] [Google Scholar]
- 20.Sözen, M. A., Armstrong, J. D., Yang, M. Y., Kaiser, K. & Dow, J. A. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubiger, M., Feng, Y., Fambrough, D. M. & Palka, J. (1994) Neuron 12, 373–381. [DOI] [PubMed] [Google Scholar]
- 22.Wessing, A. & Eichelberg, D. (1978) in The Genetics and Biology of Drosophila, eds. Ashburner, A. & Wright, T. R. F. (Academic, London), Vol. 2c, pp. 1–42. [Google Scholar]
- 23.Maddrell, S. H., Gardiner, B. O., Pilcher, D. E. & Reynolds, S. E. (1974) J. Exp. Biol. 61, 357–377. [DOI] [PubMed] [Google Scholar]
- 24.Kennerdell, J. R. & Carthew, R. W. (2000) Nat. Biotechnol. 18, 896–898. [DOI] [PubMed] [Google Scholar]
- 25.Piccin, A., Salameh, A., Benna, C., Sandrelli, F., Mazzotta, G., Zordan, M., Rosato, E., Kyriacou, C. P. & Costa, R. (2001) Nucleic Acids Res. 29, E55–E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 27.Rosay, P., Davies, S. A., Yu, Y., Sozen, M. A., Kaiser, K. & Dow, J. A. T. (1997) J. Cell Sci. 110, 1683–1692. [DOI] [PubMed] [Google Scholar]
- 28.Broderick, K. E., Kean, L., Dow, J. A., Pyne, N. J. & Davies, S. A. (2004) J. Biol. Chem. 279, 8159–8168. [DOI] [PubMed] [Google Scholar]
- 29.Morin, X., Daneman, R., Zavortink, M. & Chia, W. (2001) Proc. Natl. Acad. Sci. USA 98, 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortes, P. A. G. (1977) Biochemistry 16, 531–540. [DOI] [PubMed] [Google Scholar]
- 31.Radford, J. C., Davies, S. A. & Dow, J. A. (2002) J. Biol. Chem. 277, 38810–38817. [DOI] [PubMed] [Google Scholar]
- 32.Torrie, L. S. (2004) in Division of Molecular Genetics (University of Glasgow, Glasgow).
- 33.Wiehart, U. I., Klein, G., Steels, P., Nicolson, S. W. & Van Kerkhove, E. (2003) J. Exp. Biol. 206, 959–965. [DOI] [PubMed] [Google Scholar]
- 34.Doring, F., Wischmeyer, E., Kuhnlein, R. P., Jackle, H. & Karschin, A. (2002) J. Biol. Chem. 277, 25554–25561. [DOI] [PubMed] [Google Scholar]
- 35.Maddrell, S. H. P. (1976) J. Exp. Biol. 64, 267–281. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs, W. A. & Bigelow, N. M. (1932) J. Biol. Chem. 96, 647–658. [Google Scholar]
- 37.Stecher, P. G. (1960) The Merck Index of Chemicals and Drugs (Merck & Co., Inc., Rahway, NJ), 7th Ed.
- 38.O'Donnell, M. J., Dow, J. A. T., Huesmann, G. R., Tublitz, N. J. & Maddrell, S. H. P. (1996) J. Exp. Biol. 199, 1163–1175. [DOI] [PubMed] [Google Scholar]
- 39.Tamai, I., Nezu, J., Uchino, H., Sai, Y., Oku, A., Shimane, M. & Tsuji, A. (2000) Biochem. Biophys. Res. Commun. 273, 251–260. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J., Kean, L., Yang, J., Allan, A. K., Davies, S. A., Herzyk, P. & Dow, J. A. T. (2004) Genome Biol. 5, R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dow, J. A. T. & Davies, S. A. (2003) Physiol. Rev. 83, 687–729. [DOI] [PubMed] [Google Scholar]
- 42.Kean, L., Pollock, V. P., Broderick, K. E., Davies, S. A., Veenstra, J. & Dow, J. A. T. (2002) Am. J. Physiol. 282, R1297–R1307. [DOI] [PubMed] [Google Scholar]
- 43.Terhzaz, S., Oconnell, F. C., Pollock, V. P., Kean, L., Davies, S. A., Veenstra, J. A. & Dow, J. A. T. (1999) J. Exp. Biol. 202, 3667–3676. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Page, R. D. M. (1996) Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]