Abstract
Plants are the main source of folate in human diets, but many fruits, tubers, and seeds are poor in this vitamin, and folate deficiency is a worldwide problem. Plants synthesize folate from pteridine, p-aminobenzoate (PABA), and glutamate moieties. Pteridine synthesis capacity is known to drop in ripening tomato fruit; therefore, we countered this decline by fruit-specific overexpression of GTP cyclohydrolase I, the first enzyme of pteridine synthesis. We used a synthetic gene based on mammalian GTP cyclohydrolase I, because this enzyme is predicted to escape feedback control in planta. This engineering maneuver raised fruit pteridine content by 3- to 140-fold and fruit folate content by an average of 2-fold among 12 independent transformants, relative to vector-alone controls. Most of the folate increase was contributed by 5-methyltetrahydrofolate polyglutamates and 5,10-methenyltetrahydrofolate polyglutamates, which were also major forms of folate in control fruit. The accumulated pteridines included neopterin, monapterin, and hydroxymethylpterin; their reduced forms, which are folate biosynthesis intermediates; and pteridine glycosides not previously found in plants. Engineered fruit with intermediate levels of pteridine overproduction attained the highest folate levels. PABA pools were severely depleted in engineered fruit that were high in folate, and supplying such fruit with PABA by means of the fruit stalk increased their folate content by up to 10-fold. These results demonstrate that engineering a moderate increase in pteridine production can significantly enhance the folate content in food plants and that boosting the PABA supply can produce further gains.
Tetrahydrofolate (THF) and its derivatives (collectively termed folates) are essential cofactors for the one-carbon transfer reactions needed to form methionine, serine, purines, and thymidylate (1, 2). Whereas plants and microorganisms can synthesize folates, humans and other animals lack a complete folate synthesis pathway and so require a dietary supply. For humans, this supply comes mostly from plant sources (2, 3). Green leafy vegetables and legume seeds are folate-rich, but staple foods such as cereals, tubers, and many fruits are relatively poor in folate; related to this, folate intakes are seriously inadequate in poor countries and suboptimal even in rich ones (3–5). The consequences of low folate intake include birth defects, anemia, and increased risk of vascular disease and some cancers (5–7).
Dietary folate deficiency can be rectified by adding chemically synthesized folic acid to staple foods (fortification), or by taking folic acid pills (supplementation), but these solutions have significant recurrent costs and are hard to implement in developing countries. A promising alternative strategy (biofortification) is to enhance the folate content of food organisms by metabolic engineering (2, 8, 9). The validity of this approach has been demonstrated in food-grade lactic acid bacteria, in which engineering tripled folate production (10). Because the plant folate synthesis pathway is now known (2, 11–15), folate engineering can be attempted with plants, and several groups have begun doing this (16–19). We have chosen to engineer the folate pathway in tomatoes, in view of their worldwide importance as a food crop, their well developed molecular genetics, and their relatively low folate content (3, 13).
Folates are tripartite molecules that consist of pteridine, p-aminobenzoate (PABA), and one or more glutamate moieties (Fig. 1A). In plants, the pteridine moiety is produced in the cytosol, PABA is formed in plastids, and the two are coupled together and glutamylated in mitochondria (Fig. 1B) (11–15). In tomato fruit, the first enzyme of pteridine synthesis, GTP cyclohydrolase I (GCHI), disappears at the onset of ripening (13), cutting off the pteridine supply. As ripening tomatoes contain large pools of glutamate (20) and moderate pools of PABA (14, 21), a rational gambit to enhance folate synthesis is thus to engineer sustained GCHI activity (13). An attractive way to achieve sustained GCHI activity is to use a synthetic gene based on mammalian GCHI because this enzyme is unlikely to be subject to retroinhibition in planta. The reason is 2-fold. First, retroinhibition of mammalian GCHI is mediated by a feedback regulatory protein (22), which plants appear not to have (13). Second, the inhibitory end product of mammalian GCHI is tetrahydrobiopterin (22), which plants apparently lack (23). We report here the successful engineering of the tomato folate pathway by using a synthetic GCHI gene.
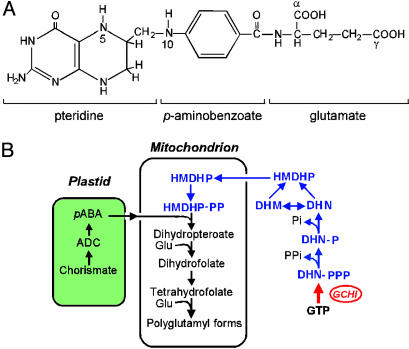
Fig. 1.
Structure and biosynthesis of folates. (A) Chemical structure of THF, monoglutamyl form. Most plant folates have γ-linked polyglutamyl tails of up to approximately six residues attached to the first glutamate. One-carbon units at various levels of oxidation (from formate to methyl) are attached to N-5 and/or N-10. The pteridine ring of folates and free pteridines can exist in tetrahydro, dihydro, and fully oxidized forms. (B) Outline of the plant folate synthesis pathway. Pteridines are in blue; note that one enzyme, dihydroneopterin (DHN) aldolase, interconverts dihydroneopterin and dihydromonapterin (DHM) and cleaves both to 6-hydroxymethyldihydropterin (HMDHP) (15). The red arrow is the engineered GCHI reaction. ADC, aminodeoxychorismate; -P, monophosphate; -PP, pyrophosphate; -PPP, triphosphate.
Materials and Methods
Expression Vector Construction. The pMON10086 vector (24) containing the tomato E8 promoter (25) and pea Rubisco small subunit terminator, and the npt II kanamycin resistance gene was modified by ablating the NotI site and ligating a polylinker (5′-GGATCCGCGGCCGCGAATTCAGGCCTGGTACCGGCGCGCCAGATCT-3′) into the unique BamHI site between the E8 promoter and the terminator. A mammalian GCHI cDNA (GenBank accession no. BE136861) was modified by using PCR primers to change the sequence context of the start codon to the plant consensus TAAACAATG (26) and to replace 17 rare codons (Fig. 2A). This synthetic cDNA was inserted between the NotI and AscI sites of the modified vector and introduced into Agrobacterium tumefaciens strain ABI by electroporation.
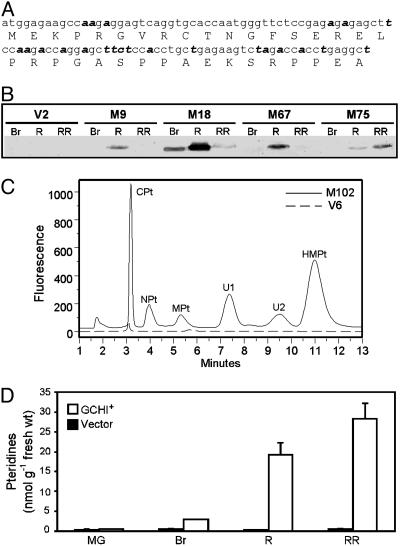
Fig. 2.
GCHI overexpression increases pteridine content in tomato fruit. (A) The 5′ region of the synthetic GCHI cDNA ORF, with the recoded nucleotides in bold italics. (B) Western analysis of recombinant GCHI protein in the fruit of four representative GCHI+ transformants and a vector-alone control (V2) at breaker (Br), red (R), and red-ripe (RR) stages. Tracks contained 80 μg of protein. (C) Fluorometric HPLC analysis (Ultremex C18 IP column) of oxidized pteridines in red-ripe fruit from a representative GCHI+ transformant (M102) and a vector control (V6). CPt, 6-carboxypterin; NPt, neopterin; MPt, monapterin, U1, unknown 1; U2, unknown 2; HMPt, 6-hydroxymethylpterin. (D) Changes in total pteridine content of control (V2) and GCHI+ (M9) fruit during ripening. Data are means of three replicates and SE. MG, mature green stage.
Transgenic Plants. The sequence-verified GCHI construct and the modified vector were used to transform tomato (Lycopersicon esculentum Mill., cv. MicroTom) essentially as described in ref. (27). Transformants were selected and regenerated on media containing 100 μg·ml–1 kanamycin. Kanamycin-resistant plantlets were screened for the GCHI construct by PCR with a forward primer located in the E8 promoter (5′-CTTTCTTGTTCCCATTTCTC-3′) and a reverse primer from the GCHI coding region (5′-ATGCACATGTGTGTCGCTTC-3′); vector-alone transformants were verified by using primers for the npt II gene. Plants positive for the GCHI construct were transplanted to soil and grown to maturity in a growth chamber (16-h day; 23°C; photosynthetic flux density, 200 μmol of photons per m2 per s per 8-h night; 20°C) and irrigated with nutrient solution. Fruit was harvested at the stages indicated in the text. For uniformity, the red and red-ripe stages were respectively defined as 3 and 7 d after breaker stage. In PABA-feeding experiments, breaker-stage fruit were cut at the base of the stalk, supplied through the stalk with 2 μmol of PABA in 100 μl of water or water only for 1 d, then destalked and allowed to ripen for a further 6 d at 23°C.
Antibodies and Western Analysis. To produce recombinant antigen, the synthetic GCHI cDNA was cloned into the EcoRI and XhoI sites of pET28b (Novagen), which adds hexahistidine tags to both termini. The construct was electroporated into Escherichia coli BL21 (DE3) CodonPlus-RIL cells (Stratagene), and the recombinant protein was isolated from isopropyl β-d-thiogalactoside-induced cells by Ni2+ affinity chromatography under denaturing conditions followed by preparative SDS/PAGE. Rabbit antibodies were prepared by Cocalico Biologicals (Reamstown, PA). To screen for GCHI expression, proteins were extracted from pericarp tissue by grinding in 0.1 M Tris·HCl (pH 8.0) containing 15 mM ascorbate, 2 mM DTT, and 3% (wt/vol) polyvinylpolypyrrolidone. After being cleared by centrifugation, samples (80 μg of protein) were separated on SDS-polyacrylamide gels, blotted to nitrocellulose, and probed as described in ref. 28, with antiserum diluted to 1:2,000. No crossreaction with tomato GCHI was detectable.
Pteridine and PABA Analysis. Representative fruit segments (0.70 g) were pulverized in liquid N2 and homogenized with 7 ml of methanol. For pteridine analysis, a 600-μl portion of the homogenate was mixed with 250 μl of CHCl3 and 50 μl of water and shaken for 40 min before adding another 225 μl of CHCl3 and 340 μl of water. After shaking for 20 min and centrifuging to break the emulsion, the aqueous phase was removed, dried in vacuo, and redissolved in 200 μl of water. Samples were oxidized by adding 0.1 vol of a solution of 1% I2 and 2% KI (wt/vol) in 1 M HCl and incubating in darkness for 1 h; excess I2 was then removed by adding 10 μlof5%(wt/vol) Na-ascorbate (29). The oxidized samples were separated by HPLC with a 5-μm, 250 × 4.6-mm Ultremex C18 IP column (Phenomenex, Belmont, CA) or a4-μm, 250 × 4.6-mm Synergi Fusion-RP 80 column (Phenomenex) eluted isocratically with 10 mM Na-phosphate (pH 6.0) at 1.5 ml·min–1. Peaks were detected by a Waters 2475 fluorescence detector (350-nm excitation and 450-nm emission) and identified by reference to standards and by spectral properties. To investigate the pteridine oxidation state, 10 mM 2-mercaptoethanol and 2% (wt/vol) Na-ascorbate were added to the methanol extraction medium, and the oxidation step was omitted. Pteridine conjugate peaks were recovered from the mobile phase by an ion exchange procedure (30) and treated with HCl (1 M, 100°C, 1 h) or with yeast α-glucosidase or almond β-glucosidase (2 units in 40 μl of 10 mM Na-phosphate, pH 6.0, 37°C, 5–120 min). The 6-carboxypterin, neopterin, monapterin, and 6-hydroxymethylpterin peaks were quantified relative to standard pteridines; conjugate peaks were quantified relative to the corresponding free pteridine because hydrolysis did not change fluorescence yield. Pteridine standards were from Schircks Laboratories (Jona, Switzerland). A 5-ml portion of the methanol extract above was used for analysis of total PABA (i.e., free PABA plus PABA glucose ester) by acid hydrolysis, cation exchange chromatography, ethyl acetate partitioning, and fluorometric HPLC as described in ref. 21.
Folate Analysis. Folates were extracted from representative fruit segments (0.5–1.0 g) by polytron homogenization in 10 ml of 50 mM Na-Hepes/50 mM 2-(N-cyclohexylamino)ethanesulfonic acid adjusted to pH 7.9 with HCl containing 1 mM CaCl2, 2% (wt/vol) Na-ascorbate, and 10 mM 2-mercaptoethanol, followed by boiling for 10 min and centrifugation (13,000 × g) for 10 min. The pellet was reextracted the same way. The combined extracts were treated with 1 ml of dialyzed rat plasma at 37°C for 2 h to deglutamylate folates. Samples were then boiled for 15 min, centrifuged as above, filtered through glass wool, and applied to folate affinity columns prepared as described in ref. 31. After being washed with 5 ml of 25 mM K-phosphate (pH 7.0)/1% Na-ascorbate (buffer 1) containing 1 M NaCl, then with 5 ml of buffer 1 alone, the columns were eluted with 5 ml of HPLC mobile phase A (see below) containing 1% ascorbic acid. Samples of the eluate (400 μl) were taken for HPLC analysis with electrochemical detection (32), using a 5-μm, 150 × 3.2-mm Prodigy ODS2 column (Phenomenex) and a four-channel detector (CoulArray Model 5600A, ESA, Chelmsford, MA) with potentials set at 0, 300, 500, and 600 mV. The mobile phase was a binary mixture of 28 mM K2HPO4 and 0.59 mM H3PO4 (pH 2.5) (buffer A) and a mixture of 75% (vol/vol) buffer A and 25% CH3CN (buffer B) with a 55-min nonlinear elution program from 90% buffer A to 100% buffer B at 1 ml·min–1. Detector response was calibrated by using THF, 5-methyl-THF, 5,10-methenyl-THF, 5-formyl-THF, and folic acid standards from Schircks. For analysis of polyglutamyl tail length (32), the rat plasma treatment was omitted. Folate extracts from human erythrocytes were used to identify the retention times of 5-methyl-THF polyglutamates (33).
Results
Overexpressing GCHI in Ripening Fruit Increases Pteridine Levels. A synthetic GCHI gene was constructed by partially recoding a mammalian GCHI cDNA (Fig. 2 A) and by adjusting the sequence context around the start codon to fit the plant consensus (26). This synthetic gene was placed behind the ripening-specific E8 promoter (25) in an Agrobacterium binary vector and introduced into tomato plants. Screening fruit by Western analysis identified transformants that expressed a protein of the correct size (27 kDa) in a ripening-specific fashion (Fig. 2B). Twelve such GCHI+ transformants, spanning a range of protein expression levels, were chosen for further analysis along with 10 empty vector controls. GCHI+ plants and their fruit were not visibly different from control plants.
Pteridines were analyzed by fluorometric HPLC after conversion to their fully oxidized, fluorescent forms. The resulting pteridine profiles of GCHI+ fruits differed greatly from those of vector controls (Fig. 2C). Measured at the red-ripe stage, the total pteridine content of the GCHI+ fruit was 3- to 140-fold higher than the average for controls. Monitoring pteridines during ripening showed that GCHI+ fruit began to diverge from controls after the mature green stage and that their pteridine contents were highest at the red-ripe stage (Fig. 2D). This pattern of pteridine accumulation fits with the expected activity of the E8 promoter that was used to drive GCHI expression (25).
Pteridine Speciation. The major pteridines detected in GCHI+ fruit included neopterin, monapterin, 6-hydroxymethylpterin, which are oxidized forms of folate synthesis intermediates, and 6-carboxypterin, a catabolite of pteridines and folates (Fig. 2C). The identity of 6-hydroxymethylpterin was confirmed by using HPLC MS to check its mass ([M + H]+ = m/z 194) and fragmentation pattern relative to an authentic standard. GCHI+ fruit also showed peaks at ≈7.5 and ≈9.5 min that did not match any of our standards (Fig. 2C). Acid hydrolysis converted these unknowns to neopterin and monapterin, respectively, as did treatment with almond β-glucosidase but not yeast α-glucosidase (Fig. 3A). This result indicates that both unknown peaks are β-d-glycosides. The relative proportions of the various pteridines were similar in all GCHI+ fruit, regardless of absolute levels (Fig. 3B). Control fruit had small peaks with the same retention times as most of the pteridines in GCHI+ fruit (data not shown) but contained too little material to identify spectrally.
Fig. 3.
Pteridine species accumulated by GCHI+ fruit. (A) HPLC fluorometric analysis of unknown peaks 1 and 2 before treatment (control) and after treatment with HCl, α-glucosidase, or β-glucosidase. Glucosidase treatments were for 5 min for peak 1 and 2 h for peak 2. (B) Quantitation of the major pteridines in red-ripe fruit of three representative GCHI+ transformants. NPt-G, neopterin glycoside; MPt-G, monapterin glycoside; other abbreviations are as in Fig. 2C.
The in vivo oxidation states of the pteridines in representative GCHI+ fruit were studied by comparing samples extracted in the presence of 2-mercaptoethanol and ascorbate (to maintain native oxidation state) and given no oxidation treatment, with sister samples that were extracted and oxidized as usual. Because reduced pteridines do not fluoresce, the difference in peak size between oxidized and nonoxidized treatments measures reduced pteridines. This method indicated that pteridine pools were typically ≈50–90% reduced at breaker and red stages, but ≤20% reduced at the red-ripe stage. The reduced pteridines appeared to be dihydro, not tetrahydro, forms because alkaline I2 treatment did not yield any pterin, a diagnostic cleavage product of tetrahydro pteridines in these conditions (29).
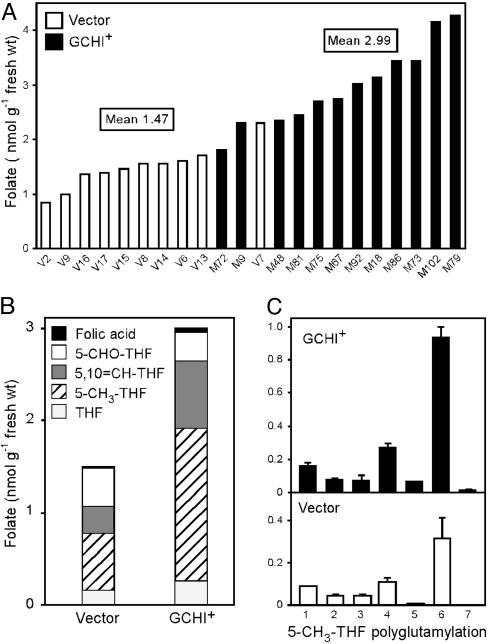
Pteridine Accumulation Enhances Folate Content. The total folate contents of control fruit fell in the range of 0.8–2.3 nmol per g of fresh weight (FW), whereas almost all of the GCHI+ fruit exceeded this range (Fig. 4A) and the mean for the GCHI+ population was double that for the controls (2.99 versus 1.47 nmol per g of FW; difference significant at P < 0.001 by t test). The increase in folate was contributed mainly by 5-methyl-THF and 5,10-methenyl-THF, both of which were major forms in vector-alone control fruit (Fig. 4B). (The acidic mobile phase used in our HPLC analysis causes quantitative conversion of 10-formyl-THF to 5,10-methenyl-THF so that the 5,10-methenyl-THF peak included both 10-formyl-THF and any preexisting 5,10-methenyl-THF.) Analysis of the polyglutamyl forms of 5-methyl-THF showed a similar distribution in GCHI+ and control fruit, with hexaglutamate dominant in both (Fig. 4C). Other folates also existed mainly as polyglutamates in both GCHI+ and control fruit (data not shown). Taken together, these data show that the extra folate in GCHI+ fruit was all in normal forms.
Fig. 4.
Analysis of folates in GCHI+ and vector-control fruits by HPLC with electrochemical detection. (A) Total folate contents of red-ripe fruit of 12 independent GCHI+ and 10 independent control transformants. Values for each transformant are averages of two fruit. (B) Analysis of the one-carbon substituents of folates from GCHI+ and control fruits. Data are mean values for all 12 GCHI+ and all 10 control transformants. Note that the analytical procedures used converted 10-formyl-THF to 5,10-methenyl-THF. (C) Polyglutamyl tail length of 5-methyl-THF from control and GCHI+ fruit. Values are means for three GCHI+ fruit (M73, M86, and M102) and three controls (V6, V13, and V15).
A scatter plot of total pteridine level versus folate content (Fig. 5A) shows that maximal folate was attained with a total pteridine level of ≈25 nmol per g of FW and that further increase in pteridine was of no benefit. In fact, regression analysis indicated a significant negative correlation between folate content and pteridine level for fruit with >25 nmol of pteridine per g of FW (linear correlation coefficient r = 0.66, significant at P = 0.015) (Fig. 5A). Similar negative correlations were also found between folate and each of the individual pteridines except hydroxylmethylpterin. Such negative trends raise the possibility that excess pteridines, beyond being unnecessary for high folate production, may actually antagonize it.
Fig. 5.
Relationships between the levels of folate, pteridines, and PABA. (A) Scatter plot of folate content versus total pteridine content. Data are for two different fruit from each of the 10 control and 12 GCHI+ transgenics. Note that above a threshold of 25 nmol of pteridines per g of FW (white area of frame), folate content in GCHI+ fruit is negatively correlated with pteridine level (dashed line; r = 0.66, P = 0.015). (B) Scatter plot of PABA content versus folate content for the same fruit as in A. Note that PABA content is negatively correlated with folate content (dashed line; r = 0.45, P = 0.003). (C) Effect of exogenously supplied PABA on folate levels in the fruit of seven GCHI+ transgenics. Two matched, breaker-stage fruits from each transgenic were supplied through the stalk with either 2 μmol of PABA in 100 μl of water or water alone for 1 d. The stalks were then removed, and the fruit were left to ripen for another 6 d. The half of the fruit nearest the stalk was taken for analysis because a pilot experiment with [14C]PABA showed that 96% of the PABA taken up by the fruit was located in this half. The mean folate contents of the water-treated and PABA-treated fruit tissue (1.20 and 5.65 nmol per g of FW, respectively) are significantly different at P < 0.0001 by t test.
Folate Accumulation Is Linked to PABA Depletion. Because the total PABA pool in nonengineered tomatoes is about the same size (1–2 nmol per g of FW) (14, 21) as the folate increase in GCHI+ fruit, we examined the relationship between the levels of folate and PABA. A significant negative correlation was evident, and the GCHI+ fruit with the highest folate contents had very little PABA (Fig. 5B). Furthermore, it can be calculated from the data of Fig. 5B that in the five fruits richest in folate, 90–97% of the PABA moieties present in the fruit had been incorporated into folate.
Exogenous PABA Increases Folate Accumulation. To test whether PABA depletion limits folate accumulation, GCHI+ fruit were harvested at the breaker stage, supplied by means of the cut stalk with PABA (or water as a control), and allowed to ripen. Fruit tissue was then analyzed for PABA and folates. Among fruit from seven different transformants, those fed PABA had much higher PABA levels than controls (57 ± 26 versus 1.8 ± 0.5 nmol per g of FW) and were 2.5- to 10-fold richer in folate (Fig. 5C). Feeding PABA to fruit harboring the vector alone did not significantly raise folate level (data not shown). The folate levels in the water-control GCHI+ fruit (Fig. 5C, open bars), which ripened off the plant, were below those in similar fruit that ripened on the plant (Fig. 4A). We revisit this effect below.
Discussion
Installing a feedback-insensitive enzyme at the head of a pathway is a standard engineering strategy to increase metabolic flux (34), and we show here that it works for the pteridine branch of folate synthesis in tomato fruit. The mammalian-type GCHI that we introduced was predicted to be free of feedback control in planta (13, 22) and was highly effective in increasing flux to pteridines, which reached levels 140 times the average value in control fruit. The same basic strategy, but using bacterial GCHI, was similarly successful in enhancing pteridine production in Arabidopsis leaves (16). The folate synthesis intermediates dihydroneopterin, dihydromonapterin, and hydroxymethyldihydropterin were among the pteridines that accumulated in GCHI+ tomatoes, showing that the three enzymes directly downstream of GCHI all remain active in ripening fruit and can accommodate increased flux (these enzymes are the two that dephosphorylate dihydroneopterin triphosphate and dihydroneopterin aldolase; see Fig. 1B). Dihydroneopterin phosphates or other phosphorylated pteridines would not have been detected in our HPLC analyses because they elute close to the void volume peak. It therefore remains to be determined whether these also accumulate in GCHI+ fruit.
Acid-labile conjugates of neopterin and monapterin were also among the pteridines found in GCHI+ fruit and, like other pteridines, appeared to be largely in the dihydro form in vivo. Based on their susceptibility to hydrolysis by almond β-glucosidase, they were identified as β-d-glycosides. Although glycosides of various pteridines, including neopterin and monapterin, are known from cyanobacteria and other prokaryotes (35, 36), there appears to be no previous reports of their occurrence in plants. Because vector-control fruit showed small peaks with the same retention times as the glycosides, these compounds may serve as natural storage pools for the pteridine precursors of folates. If so, a parallel can be drawn with the precursor PABA, which exists in vivo mainly as a glucose conjugate (21).
Compared with the massive overproduction of pteridines in GCHI+ fruit, the increase in folates was modest (2-fold for the GCHI+ population as a whole, and ≈3-fold in the highest cases), so there was clearly a constraint on flux in the folate pathway in tomato fruit at some point downstream from hydroxymethyldihydropterin (Fig. 1B). The same constraint applies to the folate pathway in Arabidopsis leaves, because an ≈1,000-fold increase in pteridines gave only a 3-fold increase in folates (16). A strong candidate for this constraint is the supply of PABA (Fig. 1B). The negative correlation between PABA and folate levels, the severe depletion of PABA in the GCHI+ fruits with high folate contents (Fig. 5B), and the increase in folate when PABA was supplied (Fig. 5C) all indicate that PABA becomes limiting as flux to folates increases in ripening fruit. This interpretation is further supported by the finding that expression of the first gene of PABA synthesis, aminodeoxychorismate synthase, ceases abruptly after the breaker stage (14). Our next engineering objective is accordingly to enhance the biosynthesis of PABA in tomato fruit already engineered to overproduce pteridines. Interestingly, the PABA supply also limits folate synthesis in lactic acid bacteria (37).
The hyperaccumulation of intermediates that occurs when feedback inhibition is abolished by engineering can sometimes be counterproductive (34). There is some evidence for this in our study, because in the most pteridine-rich fruit, folate levels were negatively correlated with the total pteridine level and with most individual pteridine levels. It is conceivable that the accumulated pteridines act as negative effectors in the folate pathway, for example by competitively inhibiting enzymes or transporters. Whatever the case, an excessive buildup of pteridines is clearly not needed to enhance folate levels. Furthermore, pteridine levels outside the natural range for plants are undesirable from a nutritional standpoint because the effects of high pteridine intake are unknown. Compared with the natural range of pteridine levels reported in food plants (23), the total pteridine levels in our GCHI+ fruit were up to 10-fold higher, and those in engineered Arabidopsis leaves were as much as 1,000-fold higher (16).
Our GCHI+ transgenic fruit are a significant first step toward a viable biofortified product, because the highest folate level we achieved so far (≈4 nmol per g of FW) is equivalent to 180 μg per standard 100-g serving. This level would provide the entire recommended dietary allowance for a young child and almost half of that for an adult. However, these elevated folate levels were seen in GCHI+ fruit that ripened on the plant, and equivalent fruit that ripened after detaching at breaker stage contained far less folate. This negative effect of detachment can be ascribed to greater net folate breakdown in the detached fruit (13) and may explain why the folate contents of store-bought tomatoes (3, 18) are so low, because these are ripened after harvesting. In any case, the folate loss after harvest shows that engineering attention should be paid to folate breakdown as well as synthesis.
Acknowledgments
We thank H. J. Klee for advice, Monsanto (St. Louis) for pMON10086, and M. J. Ziemak and J. Williamson for technical assistance. This work was supported by the Florida Agricultural Experiment Station, National Science Foundation Grant MCB-0129944, and an endowment from the C. V. Griffin, Sr., Foundation. This paper was approved for publication as Florida Agricultural Experiment Station Journal Series R-10382.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FW, fresh weight; PABA, p-aminobenzoate; GCHI, GTP cyclohydrolase I; THF, tetrahydrofolate.
References
- 1.Cossins, E. A. & Chen, L. (1997) Phytochemistry 45, 437–452. [DOI] [PubMed] [Google Scholar]
- 2.Scott, J., Rebeille, F. & Fletcher, J. (2000) J. Sci. Food Agric. 80, 795–824. [Google Scholar]
- 3.Konings, E. J., Roomans, H. H., Dorant, E., Goldbohm, R. A., Saris, W. H. & van den Brandt, P. A. (2001) Am. J. Clin. Nutr. 73, 765–776. [DOI] [PubMed] [Google Scholar]
- 4.de Bree, A., van Dusseldorp, M., Brouwer, I. A., van het Hof, K. H. & Steegers-Theunissen, R. P. (1997) Eur. J. Clin. Nutr. 51, 643–660. [DOI] [PubMed] [Google Scholar]
- 5.Krishnaswamy, K. & Madhavan Nair, K. (2001) Br. J. Nutr. 85, Suppl., S115–S124. [DOI] [PubMed] [Google Scholar]
- 6.Lucock, M. (2000) Mol. Genet. Metab. 71, 121–138. [DOI] [PubMed] [Google Scholar]
- 7.Molloy, A. M. & Scott, J. M. (2001) Public Health Nutr. 4, 601–609. [DOI] [PubMed] [Google Scholar]
- 8.Bouis, H. E. (2002) J. Nutr. 132, Suppl., 491S–494S. [DOI] [PubMed] [Google Scholar]
- 9.DellaPenna, D. (1999) Science 285, 375–379. [DOI] [PubMed] [Google Scholar]
- 10.Sybesma, W., Starrenburg, M., Kleerebezem, M., Mierau, I., de Vos, W. M. & Hugenholtz, J. (2003) Appl. Environ. Microbiol. 69, 3069–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson, A. D. & Gregory, J. F., III (2002) Curr. Opin. Plant Biol. 5, 244–249. [DOI] [PubMed] [Google Scholar]
- 12.Ravanel, S., Cherest, H., Jabrin, S., Grunwald, D., Surdin-Kerjan, Y., Douce, R. & Rébeillé, F. (2001) Proc. Natl. Acad. Sci. USA 98, 15360–15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basset, G., Quinlivan, E. P., Ziemak, M. J., Díaz de la Garza, R., Fischer, M., Schiffmann, S., Bacher, A., Gregory, J. F., III, & Hanson, A. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12489–12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basset, G. J., Quinlivan, E. P., Ravanel, S., Rébeillé, F., Nichols, B. P., Shinozaki, K., Seki, M., Adams-Phillips, L. C., Giovannoni, J. J., Gregory, J. F., III, et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyer, A., Illarionova, V., Roje, S., Fischer, M., Bacher, A. & Hanson, A. D. (2004) Plant Physiol. 135, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hossain, T., Rosenberg, I., Selhub, J., Kishore, G., Beachy, R. & Schubert, K. (2004) Proc. Natl. Acad. Sci. USA 101, 5158–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rafalski, J. A., Weng, Z. & Harvell, L. T. (2003) U.S. Patent 6,642,435.
- 18.Zhang, G. F., Maudens, K. E., Storozhenko, S., Mortier, K. A., Van Der Straeten, D. & Lambert, W. E. (2003) J. Agric. Food Chem. 51, 7872–7878. [DOI] [PubMed] [Google Scholar]
- 19.van der Meer, I. M., Bovy, A. G. & Bosch, D. (2001) Curr. Opin. Biotechnol. 12, 488–492. [DOI] [PubMed] [Google Scholar]
- 20.Boggio, S. B., Palatnik, J. F., Heldt, H. W. & Valle, E. M. (2000) Plant Sci. 159, 125–133. [DOI] [PubMed] [Google Scholar]
- 21.Quinlivan, E. P., Roje, S., Basset, G., Shachar-Hill, Y., Gregory, J. F., III, & Hanson, A. D. (2003) J. Biol. Chem. 278, 20731–20737. [DOI] [PubMed] [Google Scholar]
- 22.Yoneyama, T. & Hatakeyama, K. (1998) J. Biol. Chem. 273, 20102–20108. [DOI] [PubMed] [Google Scholar]
- 23.Kohashi, M., Tomita, K. & Iwai, K. (1980) Agric. Biol. Chem. 44, 2089–2094. [Google Scholar]
- 24.Klee, H. J. & Kishore, G. M. (1996) U.S. Patent 5,512,466.
- 25.Deikman, J., Kline, R. & Fischer, R. L. (1992) Plant Physiol. 100, 2013–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koziel, M. G., Carozzi, N. B. & Desai, N. (1996) Plant Mol. Biol. 32, 393–405. [DOI] [PubMed] [Google Scholar]
- 27.Tieman, D. M., Ciardi, J. A., Taylor, M. G. & Klee, H. J. (2001) Plant J. 26, 47–58. [DOI] [PubMed] [Google Scholar]
- 28.Nuccio, M. L. & Thomas, T. L. (1999) Plant Mol. Biol. 39, 1153–1163. [DOI] [PubMed] [Google Scholar]
- 29.Fukushima, T. & Nixon, J. C. (1980) Anal. Biochem. 102, 176–188. [DOI] [PubMed] [Google Scholar]
- 30.Stea, B., Halpern, R. M., Halpern, B. C. & Smith, R. A. (1980) J. Chromatogr. 188, 363–375. [DOI] [PubMed] [Google Scholar]
- 31.Gregory, J. F., III, & Toth, J. P. (1988) Anal. Biochem. 170, 94–104. [DOI] [PubMed] [Google Scholar]
- 32.Bagley, P. J. & Selhub, J. (2000) Clin. Chem. 46, 404–111. [PubMed] [Google Scholar]
- 33.Pfeiffer, C. M. & Gregory, J. F., III (1996) Clin. Chem. 42, 1847–1854. [PubMed] [Google Scholar]
- 34.Fell, D. A. (1998) Biotechnol. Bioeng. 58, 121–124. [DOI] [PubMed] [Google Scholar]
- 35.Forrest, H. S. & Van Baalen, C. (1970) Annu. Rev. Microbiol. 24, 91–108. [DOI] [PubMed] [Google Scholar]
- 36.Lin, X. L. & White, R. H. (1988) J. Bacteriol. 170, 1396–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sybesma, W., Starrenburg, M., Tijsseling, L., Hoefnagel, M. H. & Hugenholtz, J. (2003) Appl. Environ. Microbiol. 69, 4542–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]