Abstract
Oxidative DNA damage causes blocks and errors in transcription and replication, leading to cell death and genomic instability. Although repair mechanisms of the damage have been extensively analyzed in vitro, the actual in vivo repair processes remain largely unknown. Here, by irradiation with an UVA laser through a microscope lens, we have conditionally produced single-strand breaks and oxidative base damage at restricted nuclear regions of mammalian cells. We showed, in real time after irradiation by using antibodies and GFP-tagged proteins, rapid and ordered DNA repair processes of oxidative DNA damage in human cells. Furthermore, we characterized repair pathways by using repair-defective mammalian cells and found that DNA polymerase β accumulated at single-strand breaks and oxidative base damage by means of its 31- and 8-kDa domains, respectively, and that XRCC1 is essential for both polymerase β-dependent and proliferating cell nuclear antigen-dependent repair pathways of single-strand breaks. Thus, the repair of oxidative DNA damage is based on temporal and functional interactions among various proteins operating at the site of DNA damage in living cells.
Oxidative base damage and single-strand breaks (SSBs) are the most frequent types of DNA damage caused by reactive oxygen species, and such DNA damage can cause transcription and replication block, leading to cell death and genomic instability (1, 2). In cells without high-dose exposure of ionizing radiation, accumulated oxidative base damage and SSBs may be major causes for the production of double-strand breaks. The importance of the repair of oxidative base damage and SSBs is further implied by the observation that mice deficient in the genes involved in the repair DNA polymerase β (POL β) and the SSB-repair protein XRCC1 are embryonic lethal (3, 4) and that cells deficient in these genes are hypersensitive to exposures producing base damage and/or SSBs (5, 6). DNA repair mechanisms of oxidative base damage in mammalian cells have been analyzed extensively in vitro by using model DNA substrates and purified proteins or cell extracts, and several alternative pathways of the repair processes have been proposed (2, 5, 7). Base damage is removed by various DNA glycosylases and processed by POL β-dependent short-patch and/or proliferating cell nuclear antigen (PCNA)/polymerase δ/ε-dependent long-patch repair pathways, which are termed base excision repair (BER) (8). For repair of SSBs, SSB-induced activation of poly(ADP-ribose) polymerases (PARPs) and poly(ADP-ribosyl)ation of proteins surrounding SSBs triggers accumulation of XRCC1, which seems to play the role of a matchmaker for recruitment of other proteins involved in SSB repair (9, 10). However, the processes that actually operate in response to oxidative base damage and SSBs within cells remain largely unknown. Fundamental questions remain about the repair process in living cells, such as the following: What is the time scale for the repair of base damage and SSBs? How do repair proteins come to be localized to damage sites? How are SSBs processed after accumulation of XRCC1? How do the repair pathways for base damage and SSBs differ from each other? And, finally, how much are the in vitro data obtained up until now reflective of the in vivo situation? These important questions can be answered only by in vivo analysis of the repair processes. Here, we present an experimental system for real-time analysis of repair processes and show how cells respond to base damage and SSBs in living cells.
Methods
Microscopy and Laser-Light Irradiation. Fluorescence images were obtained and processed by using an FV-500 confocal scanning laser microscopy system (Olympus, Tokyo). A laser interface system (365 nm; Photonic Instruments, St. Charles, IL) was coupled to the epifluorescence path of the microscope. A 365-nm pulse laser was focused through a ×40 objective lens to yield a spot size of ≈1 μm. The power of the laser can be adjusted with a filter before the mirror, and filter transparencies (F) 20, 25, and 30 were used. Cells were incubated with Opti-MEM (GIBCO) in glass-bottom dishes that were covered with a chamber to prevent evaporation on a 37°C heating plate. The energy of fluorescent light was measured with a laser power/energy monitor (ORION, Ophir Optronics, Jerusalem). The mean intensity of each focus was obtained after subtraction of the background intensity in the irradiated cell. Each experiment was done at least three times, and data presented here are mean values obtained in a given experiment.
Immunocytochemistry and Chemicals. HeLa cells were stained by anti-poly(ADP-ribose) (1:200; Trevigen, Gaithersburg, MD), anti-γH2AX (1:200; Upstate Biotechnology, Lake Placid, NY), anti-8-hydroxy-2′-deoxyguanosine (8-OHdG) (1:20; Japan Institute for the Control of Aging, Shizuoka, Japan), anti-XRCC1 (1:50, Neomarker, Fremont, CA), anti-PCNA (1:100, Merck), anti-chromatin assembly factor 1 p150 subunit (CAF1-p150) (1:50, Merck), and anti-ligase IIIα (LIGIIIα) (1:100, GeneTex, San Antonio, TX). Cells were fixed within 5 min after irradiation. The anti-8-OHdG recognizes both modified base and deoxyribose structure of 8-OHdG in DNA (11). Immunofluorescence studies were performed as described in ref. 10. RO-19-8022, kindly provided by Pierre Weber and Elmer Gocke (Roche), was dissolved in ethanol, added into the medium, and incubated at 37°C for 5 min at a final concentration of 250 nM. 1,5-Dihydroxyisoquinoline (DIQ) (Sigma) was added with the final concentration of 500 μM for 1 h before irradiation.
Plasmid Construction for GFP-Fused Genes. Human genes (cDNA) amplified from HeLa cDNA with PCR based on the National Center for Biotechnology Information database were cloned into pEGFP-C1 or -N1 vectors (Clontech). GFP was fused at the C terminus of NEIL glycosylases to ensure that the enzymatic activity present in the N terminus would remain intact (12). In the cases of other proteins, there was no difference between N- and C-terminal fusions in their accumulation and dissociation properties, and data obtained by GFP fused at the N terminus of repair proteins are shown in this paper.
Cell Lines and Transfections. The following cell lines were used: HeLa, EM9 [Xrcc1-deficient Chinese hamster ovary (CHO)], AA8 (parental CHO cell line of EM9), EFX (EM9 cells stably transfected with wild-type human XRCC1; EFX shows the resistance of wild-type cells to methyl methanesulfonate; data not shown), WTB (mouse embryonic fibroblast of wild-type), 12–7B (mouse Nth1–), Parp1– (from mouse embryonic fibroblast), MB38Δ (mouse Polβ–), and MB36.3 (MB38Δ expressing wild-type human POLβ). All of the above cell lines were propagated in DMEM (Nissui Pharmaceutical, Tokyo) supplemented with 10% FBS at 37°C, in a 5% CO2/95% air atomosphere. Polβ cells (MB38Δ and MB36.3) were grown at 34°C, in a 10% CO2/90% air atomosphere. Cells were plated on glass-bottom dishes (Matsunami Glass, Osaka) at 50% confluence 24 h before the transfection (Fugene-6, Roche) and irradiated with laser light under the microscope 48 h after transfection.
RNA Interference. Short interference RNA (siRNA) was synthesized by using the Silencer siRNA construction kit (Ambion, Austin, TX). The sense sequence, 5′-GACCATCTCTGTGGTCCTA-3′, which codes nucleotides 123–141 relative to the start codon, was used for designing siRNA for hXRCC1. A final concentration of 20 nM siRNA was used for transfection. Immunoblot assay and RT-PCR were performed 48 h after transfection as described in ref. 13. The following RT-PCR primers of XRCC1 were used: sense, 5′-CAGACACTTACCGAAAATGGC-3′, and anti-sense, 5′-TCTCGGAAGGGGACATGAAAG-3′, which amplify the fragment of XRCC1 from nucleotides 81–295 relative to the start codon.
Results and Discussion
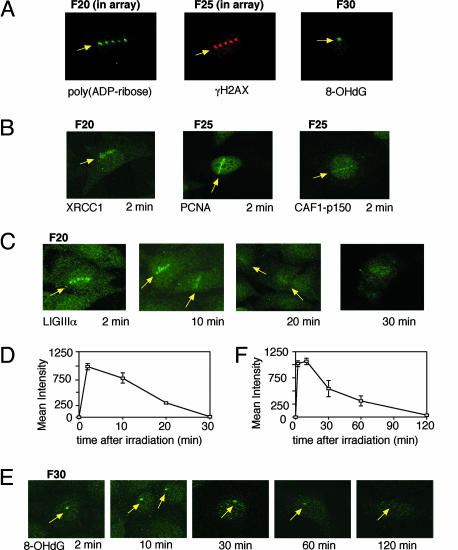
We have developed an experimental system consisting of a confocal laser scanning microscope and other laser-light equipment for 365-nm UVA irradiation. In contrast to the previously reported laser-light irradiation systems (14, 15), light was delivered from the additional UVA laser as pulses through the lens of the microscope, and irradiation conditions of the UVA laser were regulated to produce different types and amounts of DNA damage at the irradiated site (see below). With a fixed amplification scale of ×40 for the lens yielding an irradiated area of ≈1 μm in diameter, the intensity of UVA laser light at the irradiated site was regulated with a filter placed before the lens. We mainly used two scales of this filter, F20 and F25, through which the laser-light energy of 0.19 and 0.49 μJ per pulse was delivered to the irradiated site, respectively. By using this system, we have detected the following products by immunostaining at the irradiated site in the nucleus of HeLa cells immediately after irradiation (Fig. 1A): poly(ADP-ribose), which is produced by PARP at SSBs after 1-pulse irradiation in an array through an F20 filter; phosphorylated histone H2AX (γH2AX), which shows double-strand break formation after 1-pulse irradiation in an array through an F25 filter; and oxidized base damage, 8-OHdG after 10 pulses of irradiation through an F30 filter delivering 1 μJ per pulse. Thus, the laser light produces various types of DNA damage at the irradiated site.
Fig. 1.
Kinetics of repair intermediates measured by using antibodies. (A) Immunochemical detection of immediate products poly(ADP-ribose) (Left), γH2AX (Center), and 8-OHdG (Right) after laser-light irradiation of HeLa cells through F20 (1-pulse irradiation in array), F25 (1-pulse irradiation in array), and F30 (10 pulses) filters, respectively. (B) Immunohistochemistry of XRCC1 after irradiation through the F20 filter and PCNA and CAF1-p150 after irradiation through the F25 filter. (C) Time-dependent detection of repair protein with antibody against LIGIIIα after irradiation through the F20 filter. (D) Time course of the amount of accumulated LIGIIIα.(E) Time-dependent detection of 8-OHdG with its antibody after irradiation through the F30 filter with 10 pulses. (F) Time course of the amount of 8-OHdG after irradiation through the F30 filter with 10 pulses. Mean values with error bar obtained from more than three irradiated cells are shown.
In accordance with the damage production, we have detected accumulation of the following DNA repair proteins by antibodies very early (2 min) after irradiation: XRCC1; PCNA; CAF1-p150, which has been shown to be involved in SSB repair (10); and LIGIIIα (Fig. 1 B and C). XRCC1 and LIGIIIα were detected after irradiation through the F20 filter, whereas detection of PCNA and CAF1-p150 by the antibodies necessitated irradiation through the F25 filter. In the case of LIGIIIα, we could follow a time-dependent decrease of the amount of LIGIIIα at sites after irradiation. Because LIGIIIα is involved in the final step of the XRCC1-dependent process for SSB repair (16), the time course of the amount of accumulated LIGIIIα (Fig. 1 C and D) suggests the time course for the completion of the ligation step in SSB repair. The data suggest that one-half of the initial amount of SSBs was repaired within 15 min after irradiation through the F20 filter. To learn the fate of base damage after irradiation, the amount of 8-OHdG at irradiated sites was followed by antibody against the damage after irradiation with 10 pulses through the F30 filter (Fig. 1E). Despite the presumably large amount of 8-OHdG produced by this irradiation with a high dose, the amount of 8-OHdG rapidly decreased to one-half of the initial amount within 30 min after irradiation (Fig. 1F). This report presents previously undescribed time courses for DNA repair processes of SSBs and base damage in cells. These methods will be useful for determining the repair activity of SSBs and 8-OHdG in primary human cells for diagnostic purposes.
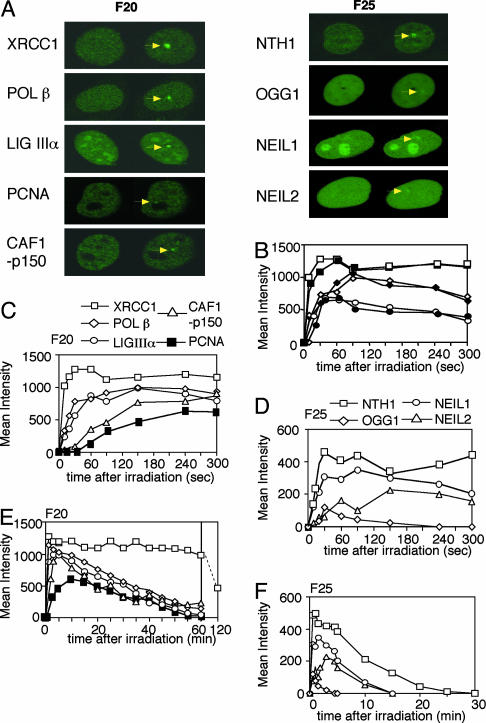
Having shown the production of various types of oxidative DNA damage by irradiation with UVA laser light, we next wanted to know what kinds of proteins accumulated at SSBs and oxidative base damage. We, therefore, expressed various human DNA-repair proteins tagged with GFP in human cells and observed the accumulation of the proteins at the irradiated site. GFP was fused at the appropriate terminus of the proteins to ensure the activity (see Methods). We found that the following GFP-tagged proteins involved in DNA repair, XRCC1, POL β, LIGIIIα, PCNA, and CAF1-p150, accumulated at irradiation sites after one-pulse irradiation through the F20 filter (Fig. 2A Left). Accumulation of various GFP-tagged DNA glycosylases for repair of oxidative base damage, NTH1, OGG1, and the human homologues of Escherichia coli endonuclease VIII, NEIL1 and NEIL2 (12), was achieved after irradiation through the F25 filter (Fig. 2 A Right) but not through the F20 filter (data not shown). Proteins involved in repair of double-strand breaks, such as NBS1, BRCA1, and RAD52, accumulated after irradiation through the F25 filter as well (data not shown). Because irradiation with 10 successive pulses through the F20 filter accumulated a much smaller amount of NTH1 than that obtained by irradiation with 1 pulse through the F25 filter (data not shown), the production of oxidative base damage by the laser light may be influenced by the number of photons in a pulse. Glycosylases for the repair of alkylated-base damage (AAG) or MUTYH that recognizes adenine base paired with 8-OHdG after replication, or GFP-tagged foreign photolyases for UV-induced DNA lesions, cyclobutane pyrimidine dimers and 6-4 photoproducts, did not accumulate after irradiation through the F20 or F25 filters (data not shown), indicating that alkylated base damage and UV-induced damage are hardly produced by the laser light. Although we have observed accumulation of GFP-tagged XPC protein involved in nucleotide excision repair after irradiation through the F20 filter, we did not see any accumulation of other components of nucleotide excision repair, including XPA, ERCC1, and XPG (data not shown), suggesting that XPC accumulated at SSBs, which were not further processed by nucleotide excision repair. These data indicate that the laser light produces mainly oxidative DNA damage of SSBs, double-strand breaks, and base damage in a dose-dependent manner.
Fig. 2.
Repair kinetics of SSBs and base damage in living cells. (A Left) Expression and accumulation of GFP-tagged XRCC1, POLβ, LIGIIIα, PCNA, and CAF1-p150 before and after irradiation through the F20 filter. (A Right) Expression and accumulation of GFP-tagged NTH1, OGG1, NEIL1, and NEIL2 before and after irradiation through the F25 filter. Before irradiation (left side of each image) and maximum accumulation after irradiation (right side of each image) are shown. (B) Accumulation patterns of GFP-tagged proteins in the proficient and deficient rodent cell lines. XRCC1 after irradiation through the F20 filter in wild-type (AA8, □) or deficient (EM9, ▪) cells; POL β after irradiation through the F20 filter in wild-type (MB36.3, ⋄) or deficient (MB38Δ4, ♦) cells; and NTH1 after irradiation through the F25 filter in wild-type (WTB, ○) or deficient (12–7B, •) cells. (C) Accumulation kinetics of GFP-tagged XRCC1 (□), POL β (⋄), LIGIIIα (○), CAF1-p150 (▵), and PCNA (▪) after irradiation through the F20 filter in HeLa cells. (D) Accumulation kinetics of GFP-tagged NTH1 (□), NEIL1 (○), NEIL2 (▵), and OGG1 (⋄) after irradiation through the F25 filter in HeLa cells. (E) Dissociation kinetics of GFP-tagged XRCC1 (□), POL β (⋄), LIGIIIα (○), CAF1-p150 (▵), and PCNA (▪) after irradiation through the F20 filter in HeLa cells. (F) Dissociation kinetics of GFP-tagged NTH1 (□), NEIL1 (○), NEIL2 (▵), and OGG1 (⋄) after irradiation through the F25 filter in HeLa cells.
By using a computer-aided analysis system, the amount of the accumulated GFP-tagged proteins can be quantified, and the kinetics for accumulation, as well as dissociation, of repair proteins can be analyzed. To address the influence of endogenous protein on the accumulation of the expressed GFP-tagged protein, we compared the accumulation kinetics of GFP-POL β, -NTH1, and -XRCC1 at irradiated sites between cells proficient and deficient in these proteins and found no difference in the accumulation kinetics of the proteins (Fig. 2B). These data indicate that the amounts of the induced substrates for these proteins were in excess so that the accumulation of expressed protein at the irradiated site was not influenced by the presence of endogenous protein. Among the proteins tested, XRCC1 accumulated the most rapidly and abundantly at the irradiated site after irradiation through the F20 filter (Fig. 2C). This accumulation is because of abundant production of poly(ADP-ribose) at SSBs by PARP activation. POL β and LIGIIIα are known to interact with XRCC1, and the accumulation kinetics of these proteins at the irradiated sites suggested that the accumulation was indeed by means of interaction with endogenous XRCC1 accumulated at SSBs (see below). PCNA and CAF1-p150 accumulated more slowly than POL β and LIGIIIα. Because PCNA is involved in the late step of the long-patch BER pathway and CAF1 has a role in chromatin assembly in the late steps of SSB repair in vitro (17, 18), this order of accumulation is consistent with the previously proposed model based on the data from cell-free extracts and purified proteins (5). Most importantly, these data indicate the involvement of both POL β and PCNA in SSB repair processes in living cells. Fig. 2D shows the accumulation of four human GFP-tagged glycosylases (NTH1, NEIL1, NEIL2, and OGG1) after irradiation through the F25 filter. Comparison of the maximum intensities of the focus for each glycosylase indicates that laser light produced more substrates for NTH1 and NEIL1, which are oxidized pyrimidines such as thymine glycol, than for OGG1, which are oxidized purines such as 8-OHdG. Oxidized cytosine has been reported to be a substrate for NEIL2 in vitro (19), and it indeed accumulated here more than OGG1 did.
The mean intensity of the fluorescence derived from the accumulated GFP-tagged proteins gradually decreased after the maximum accumulation had been achieved. All of the proteins involved in the repair of SSBs except XRCC1 had dissociated by ≈1 h after one-pulse irradiation through the F20 filter (Fig. 2E). These dissociation kinetics are in good agreement with the data obtained by the antibody against LIGIIIα after irradiation through the same filter (Fig. 1D) and indicate that detection of the GFP molecule at the irradiated site is a sensitive assay to visualize and analyze repair processes in vivo. Fig. 2E shows that, in contrast to their sequential accumulation, POL β, LIGIIIα, PCNA, and CAF1-p150 dissociated almost simultaneously, suggesting cooperative activities of these proteins until the end of the process. However, XRCC1 may play some additional role(s) after DNA damage (SSBs) was repaired. In contrast to the proteins at SSBs, the time for dissociation of DNA glycosylases from the irradiated site was rapid and depended on the amounts of substrates for each glycosylase (Fig. 2F). Thus, analysis of expressed GFP-tagged DNA repair proteins is a simple but powerful method to obtain information about the time course for repair and the protein–protein interactions built at the damaged site.
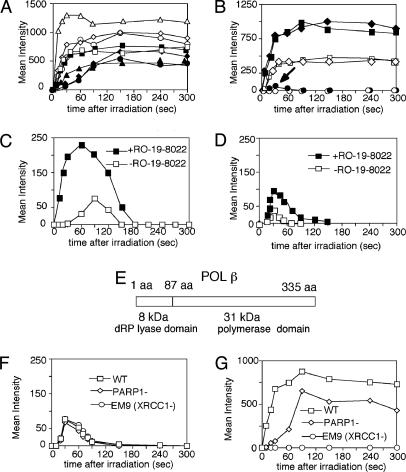
Accumulation of a protein at DNA damage in a sequential process depends on the presence of its interacting protein(s) involved in the preceding process. Therefore, we analyzed the repair pathways for SSBs and oxidative base damage in real time by using expressed GFP-tagged repair protein in cells with different repair capacities. We first examined our previous conclusion that the accumulation of XRCC1 at SSBs depends on PARP activation at the SSB (10) and further extended the analysis to other proteins. As shown in Fig. 3A, accumulation of XRCC1 and the XRCC1-binding proteins POL β and LIGIIIα at the irradiated site was significantly suppressed by DIQ, a potent inhibitor of PARPs, in HeLa cells, whereas DIQ had no influence on the accumulation of the glycosylase, NTH1, and any other accumulated glycosylase (data not shown), demonstrating that accumulation of the glycosylases at base damage is PARP-independent. To analyze the step after XRCC1 accumulation, we examined the CHO cell line EM9, which is deficient in XRCC1. In contrast to the parental wild-type cells, AA8 and EFX cells (EM9 expressing wild-type hXRCC1), POL β failed to accumulate at irradiated sites after F20 filter irradiation, indicating that POL β accumulation at UVA-induced SSBs is fully dependent on XRCC1 (Fig. 3B). However, we found that, despite the presence of XRCC1, POL β did not accumulate at UV damage endonuclease (UVDE) induced SSBs with blocked 5′ end (data not shown). Thus, accumulation of POL β at SSBs depends not only on the presence of XRCC1 at the site but also on the nature of the SSB.
Fig. 3.
POL β accumulation at SSBs and base damage. (A) Accumulation of GFP-tagged XRCC1 after irradiation without (▵) and with (▴) DIQ treatment, POL β after irradiation without (⋄) and with (♦) DIQ treatment, LIGIIIα after irradiation without (○) and with (•) DIQ treatment, and NTH1 after irradiation without (□) and with (▪) DIQ treatment in HeLa cells. XRCC1, POL β, and LIGIIIα are visualized after irradiation through the F20 filter, and NTH1 is visualized after irradiation through the F25 filter. (B) POL β accumulation in XRCC1-deficient and -proficient CHO cells. Accumulation of GFP-tagged POL β in XRCC1-deficient EM9 after irradiation through the F25 (•) or F20 (○) filter; wild-type AA8 after irradiation through the F25 (♦) or F20 (⋄) filter; and wild-type hXRCC1-expressing EM9, EFX, after irradiation through the F25 (▪) or F20 (□) filter. (C) Influence of photosensitizer RO-19-8022 on the accumulation of GFP-tagged OGG1 in EM9 cells after irradiation through the F25 filter. OGG1 accumulation without (□) or with (▪) photosensitization is shown. (D) Influence of RO-19-8022 on the accumulation of GFP-tagged POL β in EM9 cells after irradiation through the F25 filter. POL β accumulation without (□) or with (▪) photosensitization is shown. (E) Two domains of POL β, N-terminal 8-kDa domain with 5′-deoxyribose phosphate lyase activity and C-terminal 31-kDa polymerase domain, are shown. (F) Accumulation of the GFP-fused 8-kDa domain of POL β at irradiated sites in wild-type mouse (□), PARP1-mouse (⋄), and EM9 (XRCC1–)(○) CHO cells after irradiation through the F25 filter. (G) Accumulation of GFP-tagged 31-kDa domain of POL β in wild-type mouse (□), PARP1-mouse (⋄), and EM9 (XRCC1–)(○) CHO cells after irradiation through the F25 filter.
However, even in EM9 cells, there was a rapid but low-level POL β recruitment immediately after F25-filter irradiation producing both SSBs and base damage (arrow, Fig. 3B). We thought that this POL β recruitment might have reflected the involvement of POL β in the BER of oxidized base damage. To test this hypothesis, we used a photosensitizer, RO-19-8022, which has been shown to produce more 8-OHdG after treatment of cells and irradiation with ≈400-nm light (20). By pretreating the cells with RO-19-8022, the expected increase in the accumulation of OGG1 at the irradiated site was observed (Fig. 3C). Similar data were obtained for accumulation of NTH1, NEIL1, and NEIL2 (data not shown), indicating that photosensitization with RO-19-8022 increased the production of various types of oxidized base damage. Because the photosensitizing treatment did not change the accumulation of XRCC1 (data not shown), the treatment enabled us to test proteins that are accumulated at oxidative base damage and involved in BER pathways. In response to this photosensitization, the low-level, short-period accumulation of POL β was enhanced (Fig. 3D), indicating that the accumulation of POL β in EM9 cells represents its rapid response to base damage in BER.
POL β has two distinct domains: one domain is the N-terminal 8-kDa domain providing the enzymatic activity required for removal of the 5′-deoxyribose phosphate group in single-nucleotide BER (i.e., the 5′-deoxyribose phosphate lyase domain), and the other domain is the C-terminal 31-kDa DNA polymerase catalytic domain, where the XRCC1-binding site is located (Fig. 3E) (21, 22). We fused GFP to each of the POL β domains and analyzed their accumulation at irradiated sites in wild-type (HeLa, AA8, and EFX), PARP1-defective, and XRCC1-defective mammalian cells. The 8-kDa protein showed a rapid and short-term accumulation at the irradiated site in each of the cell lines after F25 filter irradiation (Fig. 3F), but the protein did not show any accumulation after F20 filter irradiation (data not shown), indicating that POL β is recruited to oxidative base damage by means of the 8-kDa domain in a PARP- and XRCC1-independent manner. Conversely, the 31-kDa domain is involved in a fully XRCC1-dependent accumulation at SSBs after irradiation through both the F20 (data not shown) and F25 filters (Fig. 3G). PARP1 deletion only partially suppressed the accumulation of the 31-kDa domain because of the presence of alternative PARP activity (23). These results indicate that POL β plays dual roles in base damage- and SSB-repair pathways by its domain-dependent accumulation at the substrates in cells. These data are consistent with the previous results that POL β plays an important role in BER (24, 25), and 5′-deoxyribose phosphate lyase activity of POL β provides mouse fibroblast cells with resistance to methyl methanesulfonate (26). Judging from the accumulation and dissociation kinetics of POL β and various glycosylases, POL β plays a rapid and initial role in the repair of base damage, and a major part of the laser-light-induced base damage, including those recognized by NTH1 and NEIL glycosylases, may be processed by PCNA-dependent long-patch repair of BER in cells.
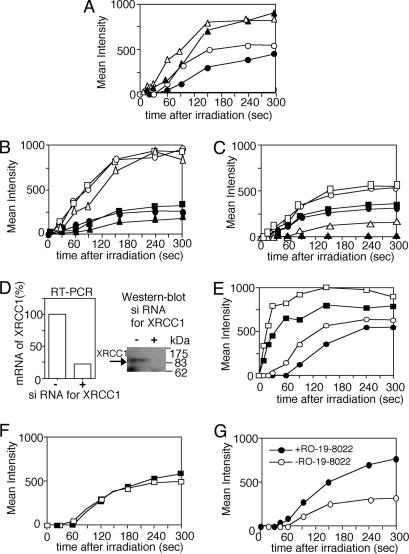
Repair processes for SSBs have not been well understood, mainly because of the absence of an experimental system to produce SSBs alone in cells. It is not known how PCNA-dependent long-patch repair operates for SSBs. We, therefore, analyzed accumulations of PCNA and CAF1-p150 after F20 or F25 filter irradiation and determined how PARP activation and the presence of XRCC1 influence long-patch repair of SSB and base damage. The accumulations of PCNA and CAF1-p150 after F20 filter irradiation were mildly suppressed by DIQ treatment in HeLa cells (Fig. 4A). In the absence of XRCC1 (EM9 cells), the accumulation of CAF1-p150 was slightly delayed or suppressed after irradiation through F20 and F25 filters as compared with those in cells expressing wild-type XRCC1 or wild-type cells (Fig. 4B). Surprisingly, PCNA did not accumulate at the irradiated site in EM9 cells after the F20 filter irradiation producing SSB, and its accumulation was significantly suppressed after the F25 filter irradiation as compared with the accumulation in wild-type cells (Fig. 4C).
Fig. 4.
Accumulation of PCNA at SSBs and base damage. (A) Accumulation of GFP-tagged PCNA after irradiation in HeLa cells through the F20 filter without (○) or with (•) DIQ treatment and CAF1-p150 in HeLa cells after irradiation through F20 filter without (▵) or with (▴) DIQ treatment. (B) Accumulation of CAF1-p150 in CHO wild-type AA8 cells after irradiation through F25 (□) or F20 (▪) filters, EFX cells after irradiation through F25 (○) or F20 (•) filters, and XRCC1-defective EM9 cells after irradiation through F25 (▵) or F20 (▴) filters. (C) Accumulation of PCNA in CHO wild-type AA8 cells after irradiation through F25 (□) or F20 (▪) filters, EFX cells after irradiation through F25 (○) or F20 (•) filters, and XRCC1-defective EM9 cells after irradiation through F25 (▵) or F20 (▴) filters. (D) Influence of RNA interference by siRNA for XRCC1 on the suppression of hXRCC1 expression at mRNA (Left) and protein (Right) levels in HeLa cells. (E) Influence of RNA interference by siRNA for XRCC1 on the accumulation of PCNA after irradiation through the F20 filter in HeLa cells with mock (□) or siRNA (▪) treatment and POL β after irradiation through the F20 filter in HeLa cells with mock (○) or siRNA (•) treatment. (F) Accumulation of PCNA in POL β+ (□) and POL β– (▪) cells through the F20 filter. (G) Influence of RO-19-8022 on the accumulation of PCNA in CHO EM9 cells after irradiation through the F25 filter. Accumulation of GFP-tagged PCNA without (○) or with (•) photosensitization is shown.
To confirm the relationship between the presence of XRCC1 and the accumulation of PCNA at SSBs in human cells, we suppressed the expression of XRCC1 by RNA interference in HeLa cells. mRNA and protein levels of XRCC1 were significantly lower (≈20% of wild type) in the cells treated with siRNA designed for hXRCC1 than those in cells with mock treatment (Fig. 4D). The suppression of XRCC1 expression reduced the accumulations of POL β and PCNA after F20 filter irradiation to similar extents (Fig. 4E), indicating that XRCC1 influenced the accumulation of POL β and PCNA in human cells as well. In contrast to EM9 cells, suppression of XRCC1 only mildly reduced the accumulation of the two proteins, suggesting that the residual amount (approximately one-fifth of the normal amount) of XRCC1 molecules is sufficient to recruit both proteins at the irradiated site. This result is consistent with data that even a low expression of XRCC1 (<10% of normal) could rescue the lethality of XRCC1 knockout mice and methyl methanesulfonate sensitivity of cells derived from the mice to the level of wild-type cells (27). We found that accumulation of PCNA was not influenced by the absence of POL β (Fig. 4F), suggesting that there are two distinct, POL β-dependent and PCNA-dependent (and its interacting polymerase δ/ε) pathways for SSB repair and BER. Because accumulation of PCNA at the irradiated site takes more time than that of POL β, additional factors and steps may be necessary for PCNA accumulation at the site. A recent report showing a physical interaction between XRCC1 and PCNA and their colocalization at replication foci (28) may explain their interaction in SSB repair as well. The accumulation of PCNA in EM9 cells after irradiation through the F25 filter represents the PCNA-dependent repair of base damage, because the accumulation could be enhanced by RO-19-8022 treatment (Fig. 4G), indicating the presence of XRCC1-independent long-patch repair of base damage of BER in cells.
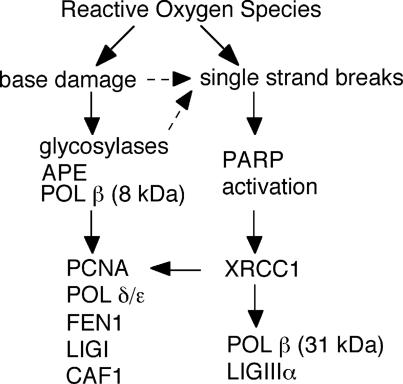
Fig. 5 is a schematic description of repair pathways for oxidative DNA damage suggested by our in situ analysis. XRCC1 plays the important scaffold role in organizing SSB repair in cells. Complete XRCC1-dependency of POL β and PCNA accumulation at SSB may explain severe phenotypes of XRCC1-deficient mice and XRCC1 polymorphisms leading to cancer proneness. Because PARP is activated by SSBs produced during BER (29), XRCC1 could be involved in the repair pathway of base damage initiated by glycosylases as well, as indicated by dashed arrows in Fig. 5. Cooperation between XRCC1 and OGG1 in BER by direct interaction has been reported (30), suggesting more involvement of XRCC1 in base-damage repair, which, however, remains to be identified in vivo. Oxidative base damage is partly processed by POL β by means of its 8-kDa domain independent of XRCC1 or processed by PCNA-dependent long-patch repair, which might be a predominant pathway in BER and might explain the phenomenon that POL β-null cells are proficient in repairing oxidative damage in early passages (6). Thus, the data presented here indicate distinct and interactive repair pathways for oxidative base damage and SSBs in living cells.
Fig. 5.
Schematic description of repair pathways for oxidative DNA damage suggested by in vivo analysis.
In summary, by introducing local UVA laser irradiation coupled with a confocal laser microscope, we addressed some fundamental questions raised in the beginning of this paper. We showed the time scale for the repair of oxidative DNA damage and characterized kinetics for the repair processes of oxidative DNA damage in living cells. These data will help further understanding of mechanisms of genome instability induced by oxidative DNA damage. The experimental system described here has a broad potential for characterizing dynamics of cellular responses to oxidative DNA damage, including temporal and functional interactions among various proteins operating at the site of DNA damage in cells.
Acknowledgments
We thank M. Satou and Y. Watanabe of KS Olympus and Olympus, respectively, for setting up the laser equipment; Dr. B. Epe (University of Mainz, Mainz, Germany) for photosensitizing chemicals; Dr. S. H. McCready for editing the text; and Drs. M. A. Resnick, T. A. Kunkel, and B. Van Houten for reading the manuscript and for valuable comments. This work was supported in part by Grants-in-Aid for Scientific Research 12143201 and 13480162 from the Ministry of Education, Science, Sports, and Culture of Japan (to A.Y.).
Abbreviations: BER, base excision repair; CAF1-p150, chromatin assembly factor 1 p10 subunit; CHO, Chinese hamster ovary; DIQ, 1,5-dihydroxyisoquinoline; 8-OHdG, 8-hydroxy-2′-deoxyguanosine; F, filter transparency; LIGIIIα, ligase IIIα; PARP, poly(ADP-ribose) polymerase; PCNA, proliferating cell nuclear antigen; POL β, DNA polymerase β; siRNA, short interference RNA; SSB, single-strand break.
References
- 1.Lindahl, T. & Wood, R. D. (1999) Science 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 2.Slupphaug, G., Kavli, B. & Krokan, H. E. (2003) Mutat. Res. 531, 231–251. [DOI] [PubMed] [Google Scholar]
- 3.Sugo, N., Aratani, Y., Nagashima, Y., Kubota, Y. & Koyama, H. (2000) EMBO J. 19, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tebbs, R. S., Flannery, M. L., Meneses, J. J., Hartmann, A., Tucker, J. D., Thompson, L. H., Cleaver, J. E. & Pedersen, R. A. (1999) Dev. Biol. 208, 513–529. [DOI] [PubMed] [Google Scholar]
- 5.Caldecott, K. W. (2003) DNA Repair 2, 955–969. [DOI] [PubMed] [Google Scholar]
- 6.Horton, J. K., Baker, A., Berg, B. J., Sobol, R. W. & Wilson, S. H. (2002) DNA Repair 1, 317–333. [DOI] [PubMed] [Google Scholar]
- 7.Loizou, J. I., El-Khamisy, S. F., Zlatanou, A., Moore, D. J., Chan, D. W., Qin, J., Sarno, S., Meggio, F., Pinna, L. A. & Caldecott, K. W. (2004) Cell 117, 17–28. [DOI] [PubMed] [Google Scholar]
- 8.Dianov, G. L., Sleeth, K. M., Dianova, I. I. & Allinson, S. L. (2003) Mutat. Res. 531, 157–163. [DOI] [PubMed] [Google Scholar]
- 9.El-Khamisy, S. F., Masutani, M., Suzuki, H. & Caldecott, K. W. (2003) Nucleic Acids Res. 31, 5526–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okano, S., Lan, L., Caldecott, K. W., Mori, T. & Yasui, A. (2003) Mol. Cell. Biol. 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin, M. A., Zerkowski, M. P., Camp, R. L., Kuefer, R., Hofer, M. D., Chinnaiyan, A. M. & Rimm, D. L. (2004) Am. J. Pathol. 164, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takao, M., Kanno, S., Kobayashi, K., Zhang, Q. M., Yonei, S., van der Horst, G. T. & Yasui, A. (2002) J. Biol. Chem. 277, 42205–42213. [DOI] [PubMed] [Google Scholar]
- 13.Lan, L., Hayashi, T., Rabeya, R. M., Nakajima, S., Kanno, S., Takao, M., Matsunaga, T., Yoshino, M., Ichikawa, M., Riele, H., et al. (2004) DNA Repair 3, 135–143. [DOI] [PubMed] [Google Scholar]
- 14.Lukas, C., Falck, J., Bartkova, J., Bartek, J. & Lukas, J. (2003) Nat. Cell Biol. 5, 255–260. [DOI] [PubMed] [Google Scholar]
- 15.Celeste, A., Fernandez-Capetillo, O., Kruhlak, M. J., Pilch, D. R., Staudt, D. W., Lee, A., Bonner, R. F., Bonner, W. M. & Nussenzweig, A. (2003) Nat. Cell Biol. 5, 675–679. [DOI] [PubMed] [Google Scholar]
- 16.Tomkinson, A. E., Chen, L., Dong, Z., Leppard, J. B., Levin, D. S., Mackey, Z. B. & Motycka, T. A. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 151–164. [DOI] [PubMed] [Google Scholar]
- 17.Verreault, A., Kaufman, P. D., Kobayashi, R. & Stillman, B. (1996) Cell 87, 95–104. [DOI] [PubMed] [Google Scholar]
- 18.Moggs, J. G., Grandi, P., Quivy, J. P., Jonsson, Z. O., Hubscher, U., Becker, P. B. & Almouzni, G. (2000) Mol. Cell. Biol. 20, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazra, T. K., Kow, Y. W., Hatahet, Z., Imhoff, B., Boldogh, I., Mokkapati, S. K., Mitra, S. & Izumi, T. (2002) J. Biol. Chem. 277, 30417–30420. [DOI] [PubMed] [Google Scholar]
- 20.Will, O., Gocke, E., Eckert, I., Schulz, I., Pflaum, M., Mahler, H. C. & Epe, B. (1999) Mutat. Res. 435, 89–101. [DOI] [PubMed] [Google Scholar]
- 21.Idriss, H. T., Al-Assar, O. & Wilson, S. H. (2002) Int. J. Biochem. Cell Biol. 34, 321–324. [DOI] [PubMed] [Google Scholar]
- 22.Marintchev, A., Robertson, A., Dimitriadis, E. K., Prasad, R., Wilson, S. H. & Mullen, G. P. (2000) Nucleic Acids Res. 28, 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schreiber, V., Ame, J. C., Dolle, P., Schultz, I., Rinaldi, B., Fraulob, V., Menissier-de Murcia, J. & de Murcia, G. (2002) J. Biol. Chem. 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 24.Allinson, S. L., Dianova, I. I. & Dianov, G. L. (2001) EMBO J. 20, 6919–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton, J. K., Prasad, R., Hou, E. & Wilson, S. H. (2000) J. Biol. Chem. 275, 2211–2218. [DOI] [PubMed] [Google Scholar]
- 26.Sobol, R. W., Prasad, R., Evenski, A., Baker, A., Yang, X. P., Horton, J. K. & Wilson, S. H. (2000) Nature 405, 807–810. [DOI] [PubMed] [Google Scholar]
- 27.Tebbs, R. S., Thompson, L. H. & Cleaver, J. E. (2003) DNA Repair 2, 1405–1417. [DOI] [PubMed] [Google Scholar]
- 28.Fan, J., Otterlei, M., Wong, H. K., Tomkinson, A. E. & Wilson, D. M., III (2004) Nucleic Acids Res. 32, 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allinson, S. L., Sleeth, K. M., Matthewman, G. E. & Dianov, G. L. (2004) DNA Repair 3, 23–31. [DOI] [PubMed] [Google Scholar]
- 30.Marsin, S., Vidal, A. E., Sossou, M., Menissier-de Murcia, J., Le Page, F., Boiteux, S., de Murcia, G. & Radicella, J. P. (2003) J. Biol. Chem. 278, 44068–44074. [DOI] [PubMed] [Google Scholar]