Abstract
The ResT protein, a telomere resolvase from Borrelia burgdorferi, processes replication intermediates into linear replicons with hairpin ends by using a catalytic mechanism similar to that for tyrosine recombinases and type IB topoisomerases. We have identified in ResT a hairpin binding region typically found in cut-and-paste transposases. We show that substitution of residues within this region results in a decreased ability of these mutants to catalyze telomere resolution. However, the mutants are capable of resolving heteroduplex DNA substrates designed to allow spontaneous destabilization and prehairpin formation. These findings support the existence of a hairpin binding region in ResT, the only known occurrence outside a transposase. The combination of transposase-like and tyrosine-recombinase-like domains found in ResT indicates the use of a composite active site and helps explain the unique breakage-and-reunion reaction observed with this protein. Comparison of the ResT sequence with other known telomere resolvases suggests that a hairpin binding motif is a common feature in this class of enzyme; the sequence motif also appears in the RAG recombinases. Finally, our data support a mechanism of action whereby ResT induces prehairpin formation before the DNA cleavage step.
Bacteria from the genus Borrelia are important pathogens that cause Lyme disease and relapsing fever (see ref. 1 for a recent review). These bacteria defy the paradigm that prokaryotic replicons are circular in nature and that those in eukaryotes are linear (2, 3). The genome of the prototype strain of Borrelia burgdorferi, the Lyme disease spirochete, has a segmented genome with a linear chromosome of 911 kb and at least 12 linear and 9 circular extrachromasomal elements (4, 5). The linear replicons all contain covalently closed hairpin ends called telomeres (6–9). Previous work done on B. burgdorferi suggests bidirectional replication from an internal origin leading to the formation of a circular head-to-head, tail-to-tail dimer intermediate (10). Processing of the dimer junctions to generate linear hairpin ends in Borrelia occurs through a DNA breakage-and-reunion event referred to as telomere resolution (3, 11–13). This process also occurs in the linear phages N15, PY54, and φKO2, which also contain hairpin telomeres (14–20).
The enzymes that promote telomere resolution are most appropriately termed telomere resolvases but have also been referred to as protelomerases (12). The telomere resolvases from phage N15 (14, 15) and B. burgdorferi (13, 21) perform a unique reaction in which a phosphodiester bond on each strand in a single DNA molecule is broken and then joined to its partner strand to form a covalently closed hairpin telomere. This reaction is in sharp contrast to the reactions performed by either type I topoisomerases (breakage and reunion of a single DNA strand) or site-specific recombinases (generation of a recombinant product through the cleavage and joining of four DNA strands) and thus defines telomere resolvases as a new class of DNA breakage-and-reunion enzymes. The telomere resolvases purified to date display limited sequence homology to the tyrosine recombinase family of site-specific recombinases (13, 19) and show some mechanistic similarity to these enzymes (22, 23) as well as to the type IB topoisomerases (24). The reaction chemistry used by ResT appears to be a two-step transesterification involving nucleophilic attack by Y335 that results in the formation of a covalent protein–DNA intermediate (ref. 13; J. Deneke, A. B. Burgin, S. L. Wilson, and G.C., unpublished work). Other features of the reaction related to that of the tyrosine recombinases include a 6-bp overlap region between nicks introduced in the substrate during catalysis and the lack of a requirement for divalent metal ions or high-energy cofactors (13).
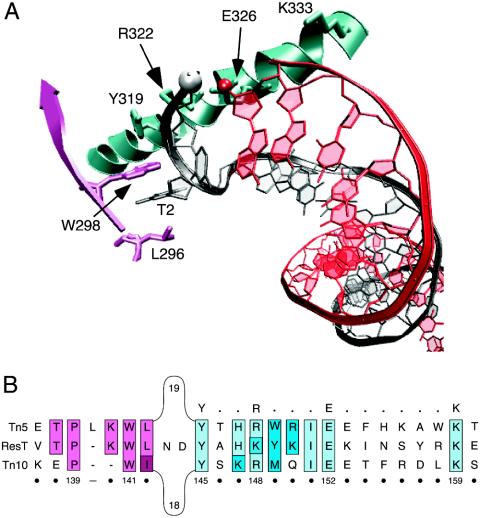
The process of cut-and-paste transposition by the bacterial elements Tn10 (25) and Tn5 (26) has been shown to involve the formation of a hairpin intermediate. Structural analysis of the Tn5 transposase (27, 28) coupled with biochemical and genetic experiments on both Tn5 (29, 30) and Tn10 (31) has defined a hairpin binding module in these transposases (32). Because telomere resolution by ResT also involves the formation of a DNA hairpin, it seemed reasonable to examine the ResT sequence for the hairpin binding region found in the Tn5 and Tn10 transposases. This motif contains a hydrophobic binding pocket that stacks with the flipped-out thymine base (Fig. 1A) at the penultimate position on the nontransferred strand (27, 28, 32). The hairpin binding domain also contains the conserved Y-(2)-R-(3)-E-(6)-K signature found in the transposases of IS4 family members (33). This region of the transposase is involved in a series of interactions with the ends of the transposon DNA that distort the DNA and facilitate hairpin formation (Fig. 1A) (27, 28, 32). Some of the mutations in this region result in a transposase that cannot form the hairpin intermediate and therefore results in the accumulation of nicks at the transposon ends (29–31). Between the hydrophobic binding pocket and the YREK motifs is an intervening stretch of 18 or 19 intervening amino acids that are not part of the hairpin binding domain (Fig. 1B).
Fig. 1.
The hairpin binding motif of cut-and-paste transposases Tn5 and Tn10. (A) The hairpin binding domain bound to Tn5 hairpin DNA. The structural schematic is from the Tn5 crystal structure (Protein Data Bank ID code 1MUS) and was generated by using the vmd molecular graphics program (46). The 5′ phosphate and 3′ hydroxyl group that will take part in forming the interstrand bond are shown in the white and red spheres, respectively. The residue numbering indicated on the structure corresponds to that of the Tn5 transposase amino acid sequence. T2 is the flipped-out thymine base at position 2. (B) Amino acid sequence alignment of ResT (4, 13) with the cut-and-paste transposases of Tn5 and Tn10. Sequence analysis reveals that ResT contains a putative hairpin binding region similar to that found in the Tn5 (27, 28, 32) and Tn10 (31) transposases. The conserved Y-(2)-R-(3)-E-(6)-K signature found in the transposases of IS4 family members (33) is indicated in bold above the alignment. The residues that constitute the hydrophobic binding pocket are colored pink, and those contained in the Y-(2)-R-(3)-E-(6)-K signature are colored blue. Sequence identity is indicated by lighter shades; amino acids with similar properties are denoted by darker shades. The numbering below the alignment corresponds to positions in the ResT sequence.
ResT from B. burgdorferi has what appears to be a hairpin binding region that contains both a hydrophobic binding pocket and a YREK motif but with a lysine substituting for the conserved arginine (YKEK) (Fig. 1B). An alignment of the ResT sequence with that of Tn5 and Tn10 argues for similarity of function as a hairpin binding region. Interestingly, there is more sequence identity in this region between Tn5 and ResT than between Tn5 and Tn10 (Fig. 1B). ResT does not contain the intervening 18- or 19-aa linker but instead contains a 2-aa bridge that connects the two parts of the putative hairpin binding region (Fig. 1B).
To test the role of the putative hairpin binding region in ResT, we have generated a number of point mutants in the protein and assayed their ability to catalyze telomere resolution. Our findings support the existence of a hairpin binding region in ResT and define the only known occurrence of this region outside a transposase. Furthermore, the combination of transposase-like and tyrosine-recombinase-like domains found in ResT indicates the use of a composite active site and helps explain the unique breakage-and-reunion reaction observed with this protein. Our results also further define the molecular details of the reaction mechanism of ResT.
Materials and Methods
In Vitro Reaction Conditions. All reactions for telomere resolution contained 25 mM Tris·HCl (pH 8.5), 100 mM NaCl, 1 mM EDTA, 5 mM spermidine, 100 μg/ml BSA, and 82 μg/ml (2 nM) substrate DNA. When present, the final concentration of ResT in reactions was 4.6 μg/ml (80 nM). Telomere resolution reactions were performed in 20 μl of total volume and incubated at either 20°C or 30°C as specified. Reactions were stopped after 30 min by the addition of 0.5 μl of 10% SDS. Samples were loaded onto 8% native polyacrylamide gels and electrophoresed in 1× Tris acetate buffer at 185 V for 2 h at 4°C. Cleavage assays using the modified phosphorothiolate oligo substrate were performed at 20°C as described above. Samples were loaded onto 8% polyacrylamide Tris/SDS gels and electrophoresed in Tris/Tricine/SDS buffer at 40 mA of constant current for 5 h. Vacuum-dried gels were analyzed with a Packard Cyclone, and band intensities were quantified by using imagequant software (Molecular Dynamics). The percentage of telomere resolution was determined by dividing the net counts corresponding to end products (products minus background) by the total counts (substrate plus net products). The percent relative covalent protein–DNA complex (CPD) was determined by calculating the percent CPD formation by the mutant ResT proteins relative to the cleavage level exhibited by wild-type ResT, defined as 100%.
Oligonucleotide Substrates. The wild-type telomere DNA substrate contained the following annealed oligonucleotides: 5′-GATCGGCGGCACTCTATACTAATAAAAAATTATATATATAATTTTTTATTAGTATAGAGTGGCGGC-3′ and 5′-GATCGCCGCCACTCTATACTAATAAAAAATTATATATATAATTTTTTATTAGTATAGAGTGCCGCC-3′. The heteroduplex DNA substrate consisted of the following annealed oligonucleotides: 5′-GATCGGCGGCACTCTATACTAATAAAAAAT TATATATATA ATTTTTTAT TAGTATAGAGTGGCGGC-3′ and 5′-GATCGCCGCCACTCTATACTAATAAAAAAT TATA AT TATA ATTTTTTAT TAGTATAGAGTGCCGCC-3′. The wild-type asymmetric DNA substrate contained the following annealed oligonucleotides: 5′-CCGGCGCCGCGGCACTCTATACTAATAAAAAATTATATATATAATTTTTTATTAGTATAGAGT-3′ and 5′-TACTCTATACTA ATAAAAAAT TATATATATA AT TTTTTATTAGTATAGAGTGCCGCGGCGCCG-3′. The 5′ bridging phosphorothiolate substrates were synthesized by A. B. Burgin (deCode Biostructures, Bainbridge Island, WA) as described (35, 36) and were assembled by ligation of the following annealed oligonucleotides: 5′-GATCACTCTATACTAATAAAAAATTA*TATAT-3′, where * indicates the position of the bridging phosphorothiolate, and 5′-ATAATTTTTTATTAGTATAGAGTG-3′. Annealing of oligomers was carried out by heating equimolar amounts of each oligonucleotide at 95°C for 5 min, followed by slow cooling to room temperature. Assembled oligomers were 5′-end-labeled by using T4 polynucleotide kinase (NEB) and [γ-32P]ATP (NEN) and then gel-purified on 8% native polyacrylamide gels to separate the annealed substrate from unannealed oligomer and unincorporated [γ-32P]ATP. Bands corresponding to the annealed substrate were excised and crush-soak-eluted in 10 mM Tris·HCl/0.1 mM EDTA (pH 8.0). Further details on the use of these substrates can be found in published work by J. Deneke, A. B. Burgin, S. L. Wilson, and G.C.
Enzyme. Purification of ResT is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
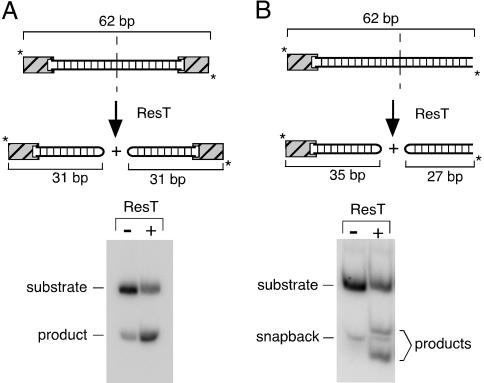
An Oligonucleotide-Based Telomere Resolution Assay. Previously reported in vitro telomere resolution assays have used plasmid DNA substrates carrying replicated telomere targets (13–15, 21, 34). Although plasmid substrates are efficient, replicated Borrelia telomeres are unstable when cloned in Escherichia coli (11), making them inconvenient substrates. Plasmid substrates are also unsuitable for incorporation of modified nucleotides or for generation of heteroduplex substrates. We have, therefore, used oligonucleotide substrates for the work reported here. A 62-bp oligo substrate containing a 50-bp replicated telomere (21) as well as two 6-bp GC clamps was synthesized (Fig. 2A Upper). Wild-type ResT converted ≈50% of our telomeric substrate into 31-bp hairpin products after 30 min at 30°C (Fig. 2A Lower). Comparable telomere resolution activity was observed by using a 62-bp oligonucleotide substrate with asymmetrically located cleavage sites in which the products of spontaneous denaturation of the A-T-rich substrate migrated differently than the products of telomere resolution (Fig. 2B). For the studies reported below, all proteins and substrate variants were tested by using both symmetric and asymmetric oligonucleotides, which yielded very similar results. For reasons not understood at present, oligonucleotide substrates of varying lengths were all processed at a slower rate (×2) and with a reduced yield (×2) compared with plasmid substrates (data not shown; J. Deneke, A. B. Burgin, S. L. Wilson, and G.C., unpublished work).
Fig. 2.
In vitro telomere resolution by ResT. (A Upper) Schematic of the telomere resolution reaction on a double-end-labeled (*), 62-bp, DNA-replicated telomere substrate. Addition of ResT leads to the production of 31-bp resolution products that contain a covalently closed hairpin end. The striped boxes at the ends of the substrate denote different GC clamps, which were required to maximize annealing of the two strands and to minimize snapback of the individual strands on themselves to form hairpins. (A Lower) Autoradiograph of an acrylamide gel showing bands corresponding to the unreacted substrate and the resolution products. A band running at the same position as the product is visible in the unreacted substrate because of some denaturation and subsequent snapback of the very A-T-rich substrate during the 30°C reaction incubation for 30 min. Controls without ResT were run for all substrates, and the background amount of 31-bp hairpin (typically ≈7%) was subtracted from ResT reaction signals. Reactions contained 2 nM labeled substrate and 80 nM ResT and were stopped by the addition of SDS. Products were analyzed on an 8% polyacrylamide gel (see Materials and Methods). (B Upper) Schematic of the telomere resolution reaction on a double-end-labeled (*), 62-bp, asymmetric, DNA-replicated telomere substrate. Addition of ResT leads to the production of 35- and 27-bp resolution products that migrate differently than the unreacted snapback DNA fragment. (B Lower) Autoradiograph of an acrylamide gel showing bands corresponding to the unreacted asymmetric substrate and resolution products.
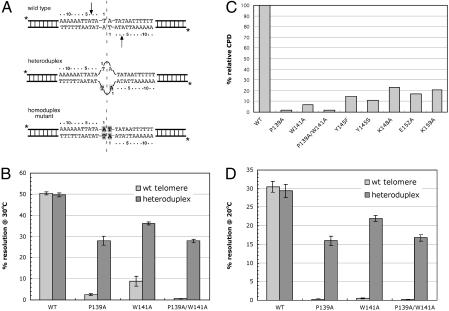
ResT Mutants in the Hydrophobic Binding Pocket of the Hairpin Binding Region. To investigate the suggested presence of a transposase-like hairpin binding region in the telomere resolvase ResT, we changed to alanine the hydrophobic residues P139 and W141, corresponding in position to Tn5 L296 and W298. The mutant ResT proteins were purified and assayed for telomere resolution activity on a labeled 62-bp replicated telomere substrate (Fig. 3A Top).
Fig. 3.
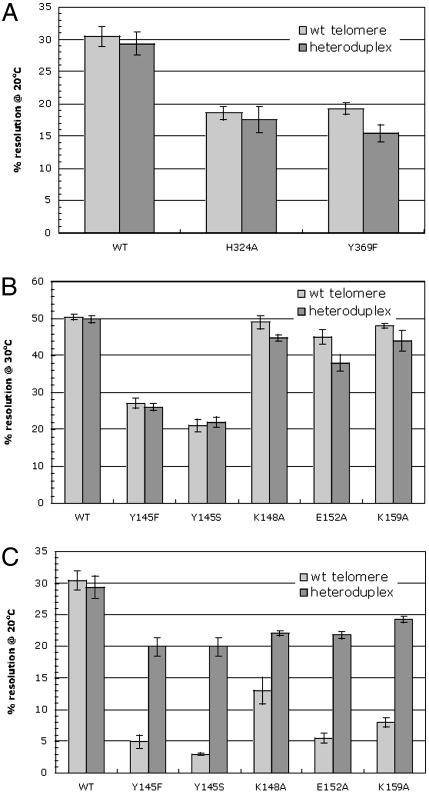
Telomere resolution activity by wild-type and putative hydrophobic binding pocket mutants of ResT. In vitro assays involving resolution of either wild-type or heteroduplex DNA substrates are shown. (A) The symmetric, double-end-labeled, 62-bp wild-type (Top), heteroduplex (Middle), and homoduplex (Bottom) mutant substrates used are shown. The vertical dashed line indicates the axis of symmetry, and the arrows in the wild-type telomere indicate the positions of cleavage. In the heteroduplex DNA substrate, the top strand sequence is wild type throughout, and the bottom strand contains mutations at position 1 flanking the axis of symmetry on both sides. Note that the design of this substrate inhibits base pairing between position-1 nucleotides but allows prehairpinning within each individual strand. The homoduplex mutant substrate contains the same mutation on the bottom strand as in the heteroduplex but also with the complimentary mutation at the top strand to allow base pairing. Mutated bases are shown in bold and boxed in gray. (B) Telomere resolution activity of wild-type ResT, P139A, W141A, and the P139A/W141A double mutant at 30°C. Reactivity levels were determined as the percentage of substrate converted into products. The data shown represent the average of three separate experiments, and the error bars indicate the standard error. (C) DNA cleavage activity of wild-type ResT and ResT with mutations in the putative hairpin binding region. A wild-type substrate carrying 5′ bridging phosphorothiolates at the cleavage positions was used. Cleavage products were detectable as CPDs (see Materials and Methods). The absolute amount of CPD observed with wild-type ResT was 55% of the input substrate. (D) Resolution activity as in B but at 20°C.
The phenotype expected for ResT proteins carrying a mutation in the hairpin binding region was uncertain at the start of these studies, mainly because of a lack of information on the mechanism of ResT action. The expected phenotype of these mutant proteins depended on whether cleavage precedes hairpin formation during the telomere resolution reaction (Scheme 1) or whether hairpinning precedes DNA cleavage (Scheme 2).
Scheme 1.
Scheme 2.
In Scheme 1, one would expect that the mutant proteins would be unable to catalyze the reaction beyond DNA cleavage, thereby leading to an accumulation of covalent protein–DNA intermediate with little or no final product generated. In Scheme 2, where helix destabilization and hairpin nucleation precede cleavage, little or no cleavage would be expected.
As shown in Fig. 3B, a 20- and 6-fold decrease in telomere resolution activity was observed for the P139A and the W141A mutants, respectively, as well as a >80-fold decrease in the double mutant. However, the end-product assay does not allow a reliable assessment of cleavage activity, because reversibility of the reaction could lead to rapid conversion of the cleaved intermediate back into substrate. Therefore, to directly assay the cleavage ability of the mutant proteins, we used a modified DNA substrate containing 5′ bridging phosphorothiolates (the 5′ bridging oxygen is replaced by a sulfur) at the cleavage sites (35, 36). With this substrate, cleavage intermediates become trapped in a CPD because the resulting 5′ sulfhydryl group is a very poor nucleophile and is unable to promote either the forward or reverse ligation step. As shown in Fig. 3C, the mutant ResT proteins did not accumulate the cleaved intermediate, indicating a defect in the mutant proteins at the cleavage step of the reaction.
The combined results suggested that Scheme 2 might be the operative mechanism for telomere resolution by ResT and that disruption of residues presumably involved in stacking with a flipped-out base resulted in a decrease in telomere resolution. However, the recovery of mutant ResT proteins with a general down phenotype was also consistent with a global disruption of protein structure and function. It was therefore necessary to further test whether hairpin nucleation was a requirement at an early stage of the reaction and whether our mutant ResT proteins were, in general, structurally and functionally compromised. The experiment that accomplished both of these goals involved using a heteroduplex substrate where the two central base pairs in the telomere resolution substrate were disrupted (Fig. 3A Middle). Such a substrate would facilitate hairpin nucleation or “prehairpin” formation and might be expected to rescue mutations in the hydrophobic binding pocket. Because the overall structure and extent of the destabilized region in the protein–DNA complex is unknown (see Discussion), we refer to this process as “prehairpinning” throughout the remainder of this paper. As shown in Fig. 3B, use of the heteroduplex substrate resulted in an 11-, 4-, and 50-fold stimulation of ResT activity by P139A, W141A, and the double mutant, respectively.
To rule out the possibility that the ResT mutations conferred a change in sequence specificity, a control experiment was conducted by using a homoduplex substrate in which both DNA strands carried the same mutation as found at position one in the heteroduplex (see Fig. 3A Bottom). The homoduplex mutant substrate did not rescue the activity of the ResT mutants and, in fact, showed a 40% decrease in telomere resolution from that observed with a wild-type telomere (data not shown). The rescue of the mutant proteins by the heteroduplex substrate therefore supports the contention that prehairpinning precedes the cleavage step and that the mutated hydrophobic binding pocket residues play a role in this process.
As a further test of Scheme 2 as the reaction mechanism for ResT action, we also performed telomere resolution assays on the hydrophobic binding pocket mutants at a reduced temperature (20°C). The rationale for this experiment was that a decrease in reaction temperature would make helix destabilization and prehairpin formation more difficult. Indeed, at 20°C, the three hydrophobic binding pocket mutants displayed almost no telomere resolution activity. Moreover, P139A, W131A, and the double mutant were rescued by the heteroduplex substrate with 50-, 44-, and 55-fold stimulations, respectively (Fig. 3D). These results again demonstrate the effectiveness of helix destabilization in rescuing the hydrophobic binding pocket mutants and further support the hydrophobic binding pocket as part of a functional hairpin binding region in ResT.
To determine whether the observed phenotype was specific to mutations within the presumed hairpin binding region, we tested whether mutations outside this region of ResT that negatively affect telomere resolution could be rescued with the heteroduplex DNA substrates. ResT mutants H324A and Y369F (J. Deneke, A. B. Burgin, S. L. Wilson, and G.C., unpublished work) displayed reduced activity on wild-type substrates but showed no increase in activity when heteroduplex DNA substrates were used (Fig. 4A) and no temperature sensitivity relative to wild-type protein (data not shown). This finding indicates that the temperature sensitivity and the beneficial effects conferred by the heteroduplex substrate on the hydrophobic pocket mutants is not a general feature of ResT mutants.
Fig. 4.
Telomere resolution activity by wild-type and YREK mutants of ResT. (A) ResT mutants H324A and Y369F were tested for telomere resolution on symmetric wild-type and heteroduplex substrates to assess specificity of the observed phenotype by hairpin binding mutants of ResT. Reactions at either 30°C (B) or 20°C (C) were performed on YKEK mutants as described in the legend of Fig. 3.
ResT Mutants in the YKEK Region of the Hairpin Binding Module. The conserved Y-(2)-R-(3)-E-(6)-K signature of cut-and-paste transposases (37) is also associated with the hairpin binding motif of Tn5 (38) and functions to facilitate hairpin formation and/or resolution during transposition of Tn5 and Tn10 (29–31). As noted above, ResT has what appears to be a YREK motif but with a lysine substituting for the conserved arginine (Fig. 1B). Single alanine point mutants at the YKEK positions were generated and subjected to telomere resolution assays involving both wild-type and heteroduplex telomere substrates. Again, because of the expectation that these residues were important for helix destabilization and prehairpin formation, it was predicted that any defects observed with the wild-type telomeres would be rescued with a heteroduplex substrate. However, the initial results showed that substitution of the K148, E152, and K159 residues had little effect on telomere resolution efficiency, with only mutation of the Y145 residue having any significant defect (Fig. 4B). Both Y-to-F and Y-to-S mutations at position 145 were generated because previous studies with the Tn10 transposase involved analogous mutations (31). The results with ResT showed that both the Y145F and Y145S mutants had reduced telomere resolution activity (Fig. 4B). Although very modest effects were observed with the remaining YKEK mutant proteins, kinetic assays revealed reduced reaction rates relative to wild-type protein (data not shown).
As performed for the hydrophobic binding pocket mutants, the incubation temperature for the YKEK mutant reactions was reduced from 30°C to 20°C. At the lower temperature, all of the YKEK mutants of ResT show reduced telomere resolution of the wild-type substrate (Fig. 4C). As for the hydrophobic binding pocket mutants, the YKEK mutants did not accumulate cleaved intermediate on the phosphorothiolate substrate (Fig. 3C), again supporting the reaction mechanism in Scheme 2. Finally, the observed defects were rescued when the YKEK mutants were tested with a heteroduplex substrate (Fig. 4C). In general, the YKEK mutants of ResT retained higher activity with both wild-type and heteroduplex DNA substrates than did the hydrophobic binding pocket mutants. This finding may be an indication that the P139 and W141 residues implicated in base-flipping in Tn5 are individually more important for prehairpin nucleation than the YKEK residues we mutated in ResT. Nevertheless, these findings indicate that the YKEK residues are important for efficient telomere resolution and that, as expected, these residues appear to be involved in DNA helix destabilization and prehairpin formation before the cleavage step of the reaction.
Discussion
A Hairpin Binding Module in ResT: Mixing Active-Site Components. We identified a putative hairpin binding region in ResT comprising a hydrophobic binding pocket and a YREK motif. The phenotype of mutations in both of these regions supports the existence of a cut-and-paste transposase-like hairpin binding region in ResT, which plays a role in DNA helix destabilization and prehairpin formation. A hairpin binding module has not been previously identified outside of the cut-and-paste transposase family of proteins. We propose that this region is an essential component of a composite active site in ResT, along with the catalytic residues similar to those of tyrosine recombinases and type IB topoisomerases, which perform the reaction chemistry. The mixing of these two functionally different active-site components helps to explain the unique activity of ResT as a telomere resolvase: prehairpin formation occurs by means of a cut-and-paste transposase hairpin binding module (residues 139–159), whereas the transesterifications to cleave the substrate and seal the hairpins occur by means of a constellation of basic residues (198–299) that activate the tyrosine nucleophile Y335 (ref. 13; J. Deneke, A. B. Burgin, S. L. Wilson, and G.C., unpublished work). The position and movement of these two active-site components must therefore be aligned and well choreographed.
A Hairpin Binding Module in Other Telomere Resolvases and in the RAG Recombinases? It is interesting to note that an alignment of the B. burgdorferi ResT protein with other known telomere resolvases (TelN of phage N15 and TelK of phage φKO2) has identified a putative hairpin binding region in both of these enzymes. A small, conserved signature motif encompassing both parts of the hairpin binding module exists in all of these telomere resolvases (including ResT). This motif, P-X-W-(linker)-Y-X-X-K corresponds to the putative base-stacking residues PW and the YK residues of the YKEK motif of ResT and is found at the corresponding position in each of the telomere resolvases. The YK residues correspond to the YR residues of Tn5, which are involved in phosphate interactions with the nucleotide carrying the flipped-out base. The size of the linker or bridge region, which corresponds to the loop in Tn5 and Tn10 transposases, appears to vary from protein to protein, with only three amino acids in ResT and 16 residues in TelN and TelK. Secondary structure predictions in this region show a striking similarity between the above telomere resolvases and the known structure of the hairpin binding domain in the Tn5 transposase cocrystal structure (27, 28). Moreover, the residues corresponding to the YREK motif are predicted to reside on the same face of an α-helix in all these enzymes, similarly displaced for interaction of these or neighboring residues with DNA. Five other putative telomere resolvases also contain this same signature sequence, but each differs by a single change, suggesting limited flexibility in the motif. These findings suggest that a hairpin binding region may be a general feature of all or many telomere resolvases and that an underlying similarity of mechanism may be found in members of this new class of DNA breakage-and-reunion enzyme.
In addition to the telomere resolvases, we have also identified the P-X-W-(linker)-Y-X-X-K motif within the eukaryotic RAG1 and RAG2 proteins. These proteins promote V(D)J recombination in higher organisms and generate hairpin ends during the cleavage step of the site-specific recombination reaction (39). The motif is highly conserved throughout a range of species, despite large differences in protein size, and is always found on truncated RAG proteins that are active. Mutagenesis adjacent to the P-X-W (K890 and R894) or in the Y-X-X-K (K938) results in a defect in hairpin formation (40), suggesting that the function of this region is a hairpin binding module. As with the telomere resolvases, the size of the linker region is not fixed, with 41 amino acids for RAG1 proteins and 38 residues for RAG2 proteins. Experimental tests on the other telomere resolvases and the RAG proteins will be required to confirm the prediction made by our sequence alignment.
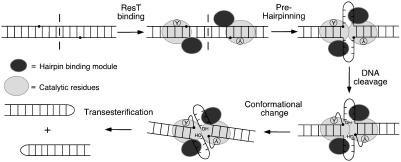
“Prehairpinning-First” Mechanism of Telomere Resolution by ResT. The data presented here support a reaction mechanism in which the catalytic domain of ResT depends on the activity of the hairpin binding region. A model for our current thinking on the reaction mechanism is shown in Fig. 5. Before initiation of the reaction chemistry, the central region of the substrate is destabilized, resulting in prehairpin formation. This structural change is followed by DNA cleavage in the first transesterification, which covalently links ResT to the 3′ DNA end through a phosphotyrosine linkage. A conformational change then occurs to position the 5′ OH terminus from the opposite strand in the active site for subsequent nucleophilic attack of the phosphotyrosine linkage and sealing of the DNA backbone. As shown in Fig. 5, the small prehairpins most likely do not contain intrastrand base pairing because of their short length. In the absence of a solved cocrystal structure, the precise nature of the unpaired region is not currently known. Based on homology with Tn5 transposase, it seems likely that it may approximate a hairpin turnaround. Attempts to directly study prehairpin formation by ResT in vitro have been hampered by our inability to identify bona fide ResT–DNA complexes that can be resolved in a band-shift assay and used for subsequent investigations.
Fig. 5.
Model of the mechanism of the telomere resolution reaction showing the predicted prehairpinning step before the first transesterification. The composite active site is represented by the large gray oval (catalytic residues) and the smaller, darker oval (hairpin binding module). The axis of symmetry is indicated by the dashed line bisecting the telomeric DNA substrate. Prehairpinning is predicted to result in the formation of two small non-base-paired hairpin turnarounds. Although the cis or trans nature of ResT catalysis is not yet determined, the reaction chemistry is depicted in cis by ResT monomers for simplicity.
It has not escaped our attention that localized melting in the prehairpin would involve helix rotation and would also facilitate bending or kinking in the DNA substrate. It may be that such an altered structure in the DNA would play an important role in establishing communication between adjacent ResT monomers or in activating the scissile phosphates for cleavage. A sharp kink has been observed in loxP sites in cocrystal structures of the Cre recombinase, and it has been suggested that this bend plays an important role in coordinated catalysis by Cre and other tyrosine recombinases, such as XerC/D and Flp (23, 41–43).
The formation of prehairpins before cleavage is expected to kinetically favor reaction completion. Upon DNA cleavage, the covalent intermediates would already exist as a prehairpin and would need only undergo nucleophilic attack by the free 5′ hydroxyl groups to produce final products. In addition, the cleavage sites, positioned on opposite sides of the axis of symmetry, would be brought into closer proximity in preparation for forming the interstrand phosphodiester bond. Prehairpinning before cleavage would also move ResT monomers into closer contact, facilitating communication for what is likely a concerted reaction at the two cleavage sites. An intimate association between protein and DNA would also be required for catalysis in trans, a feature observed for the Flp recombinase (44), as well as for Mu (45) and Tn5 (27, 28) transposases. Whether ResT catalysis occurs in cis or in trans is not yet known.
In conclusion, our results identify a composite active site in ResT, explaining the unique breakage-and-reunion activity of this telomere resolvase and perhaps of all members of this enzyme family; the hairpin binding motif sequence has also been found in the eukaryotic RAG proteins. Our findings also provide details on the mechanism of ResT action and offer evidence for a prehairpinning step that precedes DNA cleavage. Further biochemical and structural studies of ResT and other telomere resolvases should shed further light on this fascinating pairing of active-site components and the unique reaction mechanism of this class of DNA breakage-and-reunion enzymes.
Supplementary Material
Acknowledgments
We thank Alex B. Burgin for synthesis of the phosphorothiolate-containing oligonucleotide, Kerri Kobryn (University of Calgary) for participating in the construction of the His-tagged ResT overexpressing plasmid, and Kerri Kobryn, Jan Deneke, and Linda Lee for comments on the manuscript. This research was undertaken, in part, thanks to funding from the Canadian Institutes of Health Research, the Canada Research Chairs Program, and the Alberta Heritage Fund for Medical Research. G.C. was supported by a Scientist Award from the Alberta Heritage Fund for Medical Research and a Canada Research Chair in the Molecular Biology of Lyme Disease.
Abbreviation: CPD, covalent protein–DNA complex.
Noted Added in Proof. Based on work presented here and sequence comparison of hAT transposases with Tn5, a role in hairpinning of a conserved W in hAT transposases and RAG-1 recombinase has been independently suggested by Nancy Craig (personal communication).
References
- 1.Barbour, A. G. (2001) in Emerging Infections 5, eds. Scheld, M. W., Craig, W. A. & Hughes, J. M. (Am. Soc. Microbiol., Washington, DC), pp. 153–173.
- 2.Stewart, P., Rosa, P. A. & Tilly, K. (2004) in Plasmid Biology, eds. Phillips, G. & Funnell, B. (Am. Soc. Microbiol., Washington, DC), pp. 291–301.
- 3.Chaconas, G. & Chen, C. W. (2004) in The Bacterial Chromosome, ed. Higgins, N. P. (Am. Soc. Microbiol., Washington, DC), in press.
- 4.Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., White, O., Ketchum, K. A., Dodson, R., Hickey, E. K., et al. (1997) Nature 390, 580–586. [DOI] [PubMed] [Google Scholar]
- 5.Casjens, S., Palmer, N., van Vugt, R., Huang, W. H., Stevenson, B., Rosa, P., Lathigra, R., Sutton, G., Peterson, J., Dodson, R. J., et al. (2000) Mol. Microbiol. 35, 490–516. [DOI] [PubMed] [Google Scholar]
- 6.Barbour, A. G. & Garon, C. F. (1987) Science 237, 409–411. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch, J. & Barbour, A. G. (1991) J. Bacteriol. 173, 7233–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S., Murphy, M., DeLange, M., Sampson, L., van Vugt, R. & Huang, W. M. (1997) Mol. Microbiol. 26, 581–596. [DOI] [PubMed] [Google Scholar]
- 9.Casjens, S. (1999) Curr. Opin. Microbiol. 2, 529–534. [DOI] [PubMed] [Google Scholar]
- 10.Picardeau, M., Lobry, J. R. & Hinnebusch, B. J. (1999) Mol. Microbiol. 32, 437–445. [DOI] [PubMed] [Google Scholar]
- 11.Chaconas, G., Stewart, P. E., Tilly, K., Bono, J. L. & Rosa, P. (2001) EMBO J. 20, 3229–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobryn, K. & Chaconas, G. (2001) Curr. Opin. Microbiol. 4, 558–564. [DOI] [PubMed] [Google Scholar]
- 13.Kobryn, K. & Chaconas, G. (2002) Mol. Cell 9, 195–201. [DOI] [PubMed] [Google Scholar]
- 14.Deneke, J., Ziegelin, G., Lurz, R. & Lanka, E. (2000) Proc. Natl. Acad. Sci. USA 97, 7721–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deneke, J., Ziegelin, G., Lurz, R. & Lanka, E. (2002) J. Biol. Chem. 277, 10410–10419. [DOI] [PubMed] [Google Scholar]
- 16.Ravin, N. V. (2003) FEMS Microbiol. Lett. 221, 1–6. [DOI] [PubMed] [Google Scholar]
- 17.Ravin, N. V., Kuprianov, V. V., Gilcrease, E. B. & Casjens, S. R. (2003) Nucleic Acids Res. 31, 6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravin, N. V., Strakhova, T. S. & Kuprianov, V. V. (2001) J. Mol. Biol. 312, 899–906. [DOI] [PubMed] [Google Scholar]
- 19.Rybchin, V. N. & Svarchevsky, A. N. (1999) Mol. Microbiol. 33, 895–903. [DOI] [PubMed] [Google Scholar]
- 20.Huang, W. M., Joss, L., Hsieh, T. & Casjens, S. (2004) J. Mol. Biol. 337, 77–92. [DOI] [PubMed] [Google Scholar]
- 21.Tourand, Y., Kobryn, K. & Chaconas, G. (2003) Mol. Microbiol. 48, 901–911. [DOI] [PubMed] [Google Scholar]
- 22.Grainge, I., Buck, D. & Jayaram, M. (2000) J. Mol. Biol. 298, 749–764. [DOI] [PubMed] [Google Scholar]
- 23.Van Duyne, G. D. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 87–104. [DOI] [PubMed] [Google Scholar]
- 24.Shuman, S. (1998) Biochim. Biophys. Acta 1400, 321–337. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, A. K., Guhathakurta, A., Kleckner, N. & Haniford, D. B. (1998) Cell 95, 125–134. [DOI] [PubMed] [Google Scholar]
- 26.Bhasin, A., Goryshin, I. Y. & Reznikoff, W. S. (1999) J. Biol. Chem. 274, 37021–37029. [DOI] [PubMed] [Google Scholar]
- 27.Davies, D. R., Goryshin, I. Y., Reznikoff, W. S. & Rayment, I. (2000) Science 289, 77–85. [DOI] [PubMed] [Google Scholar]
- 28.Lovell, S., Goryshin, I. Y., Reznikoff, W. R. & Rayment, I. (2002) Nat. Struct. Biol. 9, 278–281. [DOI] [PubMed] [Google Scholar]
- 29.Ason, B. & Reznikoff, W. S. (2002) J. Biol. Chem. 277, 11284–11291. [DOI] [PubMed] [Google Scholar]
- 30.Naumann, T. A. & Reznikoff, W. S. (2002) J. Biol. Chem. 277, 17623–17629. [DOI] [PubMed] [Google Scholar]
- 31.Allingham, J. S., Wardle, S. J. & Haniford, D. B. (2001) EMBO J. 20, 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, P. A. & Baker, T. A. (2001) Nat. Struct. Biol. 8, 302–307. [DOI] [PubMed] [Google Scholar]
- 33.Rezsohazy, R., Hallet, B., Delcour, J. & Mahillon, J. (1993) Mol. Microbiol. 9, 1283–1295. [DOI] [PubMed] [Google Scholar]
- 34.Hertwig, S., Klein, I., Lurz, R., Lanka, E. & Appel, B. (2003) Mol. Microbiol. 48, 989–1003. [DOI] [PubMed] [Google Scholar]
- 35.Burgin, A. B., Jr., Huizenga, B. N. & Nash, H. A. (1995) Nucleic Acids Res. 23, 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgin, A. B. (2001) Methods Mol. Biol. 95, 119–128. [DOI] [PubMed] [Google Scholar]
- 37.Mahillon, J., Seurinck, J., van Rompuy, L., Delcour, J. & Zabeau, M. (1985) EMBO J. 4, 3895–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reznikoff, W. S. (2003) Mol. Microbiol. 47, 1199–1206. [DOI] [PubMed] [Google Scholar]
- 39.Gellert, M. (2002) Annu. Rev. Biochem. 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 40.Huye, L. E., Purugganan, M. M., Jiang, M. M. & Roth, D. B. (2002) Mol. Cell. Biol. 22, 3460–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, L., Chu, L. C. & Sadowski, P. D. (2003) J. Biol. Chem. 278, 23118–23129. [DOI] [PubMed] [Google Scholar]
- 42.Hallet, B., Arciszewska, L. K. & Sherratt, D. J. (1999) Mol. Cell 4, 949–959. [DOI] [PubMed] [Google Scholar]
- 43.Chen, Y. & Rice, P. A. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 135–159. [DOI] [PubMed] [Google Scholar]
- 44.Lee, J., Jayaram, M. & Grainge, I. (1999) EMBO J. 18, 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaconas, G. & Harshey, R. M. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 384–402.
- 46.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.