Abstract
Peptide:N-glycanase (PNGase) has been proposed to participate in the proteasome-dependent glycoprotein degradation pathway. The finding that yeast PNGase interacts with the 19S proteasome subunit through the protein Rad23 supports this hypothesis. In this report, we have used immunofluorescence, subcellular fractionation, coimmunoprecipitation, and in vitro GST pull-down techniques for detecting intracellular localization and interactions of PNGase, HR23B, and S4 by using human (h) and mouse (m) homologs. Immunofluorescence studies revealed that hPNGase, hHR23B, and hS4 are present in close proximity to the endoplasmic reticulum (ER) when calnexin was used as an ER marker in HeLa cells. Subcellular fractionation suggests not only cytoplasmic but also ER association of hPNGase in HeLa cells. Immunoprecipitation analysis revealed the interaction of h/mPNGase with the 19S proteasome subunit, hS4, through hHR23B. Using an in vitro GST pull-down assay, we also have shown that recombinant mPNGase requires its N terminus and middle domain for interaction with mHR23B. Finally, using misfolded yeast carboxypeptidase Y and chicken ovalbumin as glycoprotein substrates, we have established that mHR23B acts as a receptor for deglycosylated proteins. Based on this finding, we propose that after deglycosylation of misfolded glycoproteins by PNGase, the aglyco forms of these proteins are recognized by HR23B and targeted for degradation.
The proper folding and assembly of newly synthesized glycoproteins is vital for many cellular functions. Glycoproteins that do not fold into their native state are processed for proteolysis via the endoplasmic reticulum-associated degradation (ERAD) pathway (1–3). Misfolded N-linked glycoproteins destined for degradation are recognized by a highly conserved deglycosylating enzyme, peptide:N-glycanase (PNGase), implicated in proteasomal degradation (4). Very recent findings established that PNGase acts upstream of the proteasome and facilitates subsequent degradation of misfolded N-linked glycoproteins by deglycosylating them (5, 6).
A yeast two-hybrid screen and biochemical studies detected the interaction of cytoplasmic yeast PNGase (yPNGase) with yRpt1 (a 19S proteasome subunit) through yRad23 (a yeast nucleotide excision repair protein), and this complex has been implicated in the ERAD pathway (7). yRpt1 interacts with yRpt2 (8, 9), a protein that has been shown to mediate gating of the proteasome by means of its ATPase domain (10). yRpt1 and yRpt2 have been reported to copurify and form a heterocomplex (11). Another component of the base of 19S proteasome, yRpn1, has been identified as the receptor for yRad23 (12). Moreover, it has been shown that the ubiquitin-like (UBL) domain present at the N terminus in both yRad23 and its mammalian homologs interacts with the proteasome (13) and thereby acts as a shuttle factor by using its ubiquitin-associated (UBA) domain present at its C terminus (14–16) to bind proteins destined for degradation (17).
Mouse PNGase (mPNGase) also has been identified as interacting with mouse HR23B (mHR23B) and mouse S4 (mS4), the mouse homolog of yRpt2, by using both biochemical studies and two-hybrid analysis (18). On the basis of these findings, the existence of a degradation complex formed by mPNGase–mHR23B–mS4 was postulated. Evidence for the presence of a similar proteolytic complex has been provided by the finding of an interaction between human (h) HR23 and S5a (human homolog of yRpn10) (19). All these studies established the role of Rad23 (and its mammalian homologs) and the 19S proteasome subunits in the ERAD pathway. Thus, it is clear that in both yeast and mammals there appears to be a tightly knitted relationship among PNGase, Rad23, and the proteasome.
Our objective was to analyze the function of h/mPNGase in association with h/mHR23B and h/mS4 in the ERAD pathway. We used immunofluorescence, subcellular fractionation, coimmunoprecipitation, and GST-binding assays to analyze the localization and interactions of these proteins. All three proteins were found in close proximity to the ER by using human calnexin (hcalnexin) as an ER marker in HeLa cells. Subcellular fractionation revealed that only human PNGase (hPNGase) was associated with the ER in HeLa cell lysates. mPNGase overexpressed in HeLa cells coimmunoprecipitated with endogenous hHR23B but not with human hS4. However, endogenous hHR23B coimmunoprecipitated with both endogenous hPNGase and hS4. We also found that mPNGase requires not only the N-terminal extension containing the PUB (peptide:N-glycanase/UBA or UBX-containing protein domain) or PUG (peptide:N-glycanase and other putative nuclear UBA or UBX domain) (21), but also the highly conserved middle domain for its interaction with mHR23B. Finally, in vitro studies carried out by using mHR23B and yeast carboxypeptidase Y (yCPY) or chicken ovalbumin as glycoprotein substrates demonstrated that mHR23B recognizes only the deglycosylated form of yCPY and chicken ovalbumin. Therefore, we propose that misfolded protein substrates are deglycosylated by ER-associated or free PNGase, identified by HR23B, and thereby targeted to the nearby proteasome for degradation.
Materials and Methods
Antibodies and Chemicals. mPNGase antiserum (rabbit, polyclonal) was a gift from Tadashi Suzuki (University of Osaka, Osaka). It was affinity-purified by using an N-hydroxysuccinimide-activated column and coupling the N-terminal domain of mPNGase as antigen according to the manufacturer's protocol (Amersham Pharmacia). Anti-CPY (rabbit, polyclonal) was a gift from Randy Schekman (University of California, Berkeley). Monoclonal antibody to hcalnexin was purchased from Abcam (Cambridge, MA). Anti-hHR23B (sheep, polyclonal) and anti-hS4 (rabbit, polyclonal) were obtained from Affiniti (Nottingham, U.K.). Antibodies for secondary detection coupled with AlexaFluor-555, AlexaFluor-488, and glutathione (GSH)-agarose beads were purchased from Molecular Probes. Lipofectamine and normal goat serum were obtained from Invitrogen. DMEM, FBS, and PBS were obtained from GIBCO/BRL. Paraformaldehyde was obtained from Sigma.
Mammalian Cell Culture and Transfection. HeLa cells were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin in a 5% CO2/95% air atmosphere at 37°C. Transfection of HeLa cells was performed with pcDNA-3.1-mPNG1-(HA)3 [hemagglutinin epitope (HA)-tagged-mPNGase] and pcDNA-3.1-mS4-(His)6 (His-tagged-mS4) by using Lipofectamine (Invitrogen) according to the manufacturer's protocol.
Immunofluorescence. Cells were fixed in 3% paraformaldehyde for 20 min at room temperature and permeabilized in 0.5% Triton X-100 on ice for 7 min. The cells were washed in PBS plus 0.5% normal goat serum and incubated with one of the following primary antibodies: mPNGase 1:200, hHR23B 1:100, hS4 1:100, or hcalnexin 1:200. AlexaFluor-488- or AlexaFluor-555-conjugated secondary antibodies were used as required. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were analyzed by using a confocal microscope (Zeiss) to visualize the endogenous level of proteins under study.
Immunoprecipitation, Subcellular Fractionation, and Western Blotting. Immunoprecipitations were conducted in Nonidet P-40 buffer (0.2% Nonidet P-40/150 mM KCl/1 mM EGTA/50 mM Hepes, pH 7.5/5 mM MgCl2/50 mM NaF/1 mM Na3VO4/250 mM sucrose/1 mM PMSF) containing proteasome inhibitor mixture (Pierce). Immunoprecipitation was performed in 500 μl of cell lysate (2 × 107 cells) that was precleared by incubation with protein A or G agarose beads for 1 h, and the resulting supernatant was treated with the indicated antibody and captured on 30 μl of protein A or G agarose beads for 1 h, washed three times, boiled in SDS sample buffer, and processed for SDS/PAGE followed by immunoblotting.
Subcellular fractionation was performed on a sucrose gradient with 108 HeLa cells after lysing in lysis buffer as described (22). The ER membranes sedimenting at the interphase of 1.2 and 1.5 M sucrose were collected, suspended in SDS/PAGE sample buffer, resolved by SDS/PAGE, and transferred to nitrocellulose membranes. Blots were incubated with 1:2,000 dilution of anti-mPNGase, 1:2,000 dilution of anti-hS4, 1:1,000 dilution of anti-hHR23B antibody, or 1:3,000 dilution of anti-hcalnexin antibody followed by 1:3,000 dilution of horseradish peroxidase-conjugated secondary antibody (Roche Diagnostics). Gels were visualized by using chemiluminiscence after exposure to medical x-ray film (Fuji).
In Vitro Binding Assays. A set of deletion constructs of mPNGase was prepared. BL21(DE3)pLysS cells were transformed by using the following plasmids: pET28a-mPNG1(1–651), pET28a-mPNG1(1–171), pET28a-mPNG1(1–470), pET28a-mPNG1(171–470), pET28a-mPNG1(171–651), and pET28a-mPNG1(470–651) [with (His)6 tag at the N terminus]. pGEX-mHR23B and pGEX-mS4 (containing the GST tag at the N terminus) were transformed into DH5α cells. Expression of mPNGase constructs, mHR23B, and GST proteins was carried out as described (18). Proteins were extracted in buffer A (1× PBS, pH 7.2/1% Triton X-100/5 mM DTT/1 mM PMSF/protease inhibitor mix). All of the protein extracts were analyzed on SDS/10% PAGE and used for GST-binding assay as described (18). In brief, GST alone or GST-mHR23B fusion protein extracts (1 ml) in buffer A were incubated with 30 μl (bed volume) of GSH-agarose beads for 1 h, washed five times with buffer A, and incubated with the required cell lysates (1 ml) for 2 h at 4°C. The beads were washed five times with buffer A, followed by elution of the bound proteins in SDS/PAGE sample buffer and immunoblotting with monoclonal anti-His or anti-GST antibody. Similar experiments were performed to detect the binding of yCPY or chicken ovalbumin to mHR23B. yCPY (Roche Diagnostics) or chicken ovalbumin (Sigma) (20 μg/ml) was denatured and deglycosylated with Endo H (New England Biolabs) according to the manufacturer's protocol. Samples of native yCPY or chicken ovalbumin (500 μl of 20 μg/ml) and denatured and deglycosylated yCPY or chicken ovalbumin (500 μl of 20 μg/ml) were then incubated with GSH-agarose beads containing GST-mHR23B or GST alone in buffer B (20 mM sodium phosphate, pH 7.2/500 mM NaCl/5 mM DTT/1% Triton X-100/1 mM PMSF). The beads were washed five times in buffer B, and the bound proteins were eluted in SDS sample buffer, resolved by SDS/PAGE, and immunoblotted with anti-GST, anti-CPY, and anti-ovalbumin antibodies.
Results
Endogenous hPNGase, hHR23B, and hS4 Reside Around the Periphery of the ER. mPNGase and its interacting partners, mHR23B and mS4, have been implicated in the proteasome-dependent degradation pathway (18). mPNGase, mHR23B, and mS4 proteins exhibit high conservancy with the primary sequence of hPNGase, hHR23B, and hS4, respectively. Therefore, we studied the localization patterns of endogenous hPNGase, hHR23B, and hS4 in HeLa cells to shed light on the role of these proteins in the ERAD pathway. Antibodies against mPNGase, hHR23B, and hS4 specifically detected the corresponding proteins in HeLa cells (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). Localization of these proteins also was studied in three other cell lines: Cos1, NIH 3T3, and HT1080. The results for HeLa cells are shown in Fig. 1; similar results were obtained with the other cell lines (S.K. and W.J.L., unpublished data). No labeling was seen after treatment with secondary antibodies alone or after competition of the primary antibody with an excess of the recombinant mPNGase, mHR23B, and mS4 (data not shown).
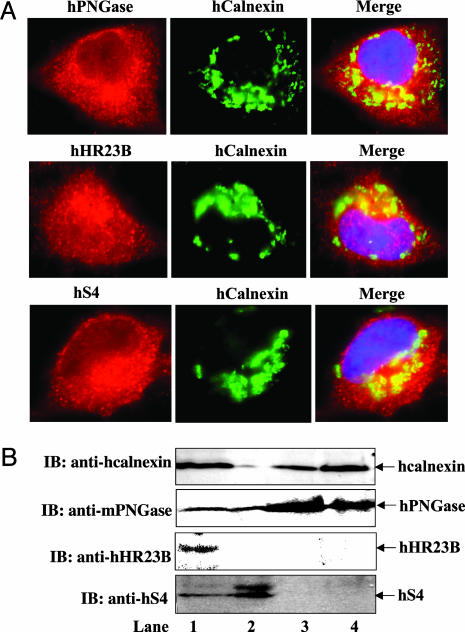
Fig. 1.
Localization of hPNGase, hHR23B, and hS4 in HeLa cells analyzed by immunofluorescence and subcellular fractionation. (A) Localization patterns of hPNGase, hHR23B, and hS4 in HeLa cells. (Top) Colocalization of hPNGase with hcalnexin: hPNGase staining (red), hcalnexin staining (green), and merge of hPNGase and hcalnexin (yellow shows the colocalization of two proteins). (Middle) Colocalization of hHR23B with hcalnexin: hHR23B staining (red), hcalnexin staining (green), and merge of hHR23B and hcalnexin (yellow shows the colocalization of two proteins). (Bottom) Colocalization of hS4 with hcalnexin: hS4 staining (red), hcalnexin staining (green), and merge of hS4 and hcalnexin (yellow shows the colocalization of two proteins). 4′,6-Diamidino-2-phenylindole (blue) shows the nucleus. (B) Subcellular fractionation of HeLa cells. Shown are HeLa cell lysate (lane 1), cytoplasmic fraction (lane 2), microsome-rich pellet (lane 3), and rough ER fraction (lane 4). Shown are immunoblots (IB) with hcalnexin antibody, mPNGase antibody, hHR23B antibody, or hS4 antibody. Nonspecific bands with anti-hS4 were sometimes seen but were distinct from the authentic hS4 band.
As shown in Fig. 1 A, colocalization studies using antibody to the ER marker, hcalnexin, revealed that hPNGase, hHR23B, and hS4 are detectable in the cytosol but are more concentrated around the ER (Fig. 1 A). However, colocalization of hPNGase and hS4 with hcalnexin was observed, whereas a lesser degree of colocalization was observed for hHR23B with hcalnexin. Localization of these proteins around the ER periphery indicates that they could be involved in the ERAD process (23). PNGase has been reported to be present in the ER (24), microsomes (25), and the cytosol (26). Some studies using immunofluorescence suggest the association of proteasomes with the cytosolic face of ER membrane both in mammalian cells (27, 28) and in yeast (29, 30). We found that in HeLa cells, hHR23B was detected in the nucleus (Fig. 1 A) as well as in the cytoplasm, but there was more intense staining around the ER. Nuclear localization of hHR23B is expected, because its function in DNA repair during the cell cycle is well established (31, 32).
Subcellular Fractionation Reveals Association of hPNGase with the ER Membrane. Because the immunofluorescence studies suggested that hPNGase, hHR23B, and/or hS4 might be associated with or anchored to the ER membrane, we carried out subcellular fractionation of a HeLa cell lysate as described in Materials and Methods. The ER membrane protein hcalnexin was used as a marker for the ER. In Fig. 1B, the distribution of hcalnexin, hPNGase, hHR23B, and hS4 after subcellular fraction is shown. Lane 1 shows the endogenous level of all of the proteins in the total cell lysate of HeLa cells (105 cells). Cell lysate (2 ml) obtained from 108 HeLa cells was then processed for subcellular fractionation and three subfractions, i.e., microsome-free cytosol (≈1.75 ml), microsome-rich membrane pellet (≈200 μl), and purified rough ER (RER) (≈50 μl) were obtained. As expected, hcalnexin was not detected in the membrane-free cytoplasmic fraction, whereas hPNGase and hS4 were found in this fraction (lane 2). However, hHR23B could not be detected in this fraction, possibly because of a dilution factor. In addition to hcalnexin, hPNGase was detected in the microsome-rich pellet (lane 3). RER purified by density gradient fractionation also showed the presence of hcalnexin and hPNGase (lane 4). In contrast, neither hHR23B nor hS4 was identified in the microsome-rich pellet or in the RER fraction. These subcellular fractions were not quantified, so the exact protein amount in each fraction could not be estimated. In any case, of the three proteins under study, only hPNGase appears to be associated with the ER. This finding also suggests that although both hHR23B and hS4 may reside near the ER, they are not directly bound to it.
h/mPNGase Interacts with HR23B and S4. Our immunofluorescence studies clearly showed that hPNGase and its partners reside around the ER. In an earlier paper (18), direct interaction of mPNGase with mHR23B was confirmed by GST-binding assays. Moreover, S4 was also found to copurify with PNGase by using gel filtration with mammalian cell lysates obtained from COS1 cells (18). As an extension of this work, we identified in vivo and in vitro interactions among h/mPNGase, h/mHR23B, and h/mS4 by using immunoprecipitation and GST pull-down experiments. After transfection of HeLa cells with HA-tagged-mPNGase, we carried out immunoprecipitation from the HeLa cell lysate by using monoclonal anti-HA antibody. Then, to identify the proteins in the coimmunoprecipitate, SDS/PAGE followed by immunoblotting was carried out with anti-mPNGase, anti-hS4, or anti-hHR23B antibodies (Fig. 2A). The total cell lysate before immunoprecipitation showed the presence of the three proteins under study (Fig. 2 A, lane 1). The supernatant obtained after immunoprecipitation with monoclonal anti-HA antibody and protein G-agarose beads was analyzed. Analysis of immune complex that bound to protein G-agarose beads revealed an enrichment of mPNGase as expected. Moreover, hHR23B, but not hS4, was found to coimmunoprecipitate with mPNGase (Fig. 2 A, lane 2). The failure to detect interaction between overexpressed mPNGase and hS4 by using coimmunoprecipitation could be explained in three ways. (i) Perhaps hS4 dissociates under the conditions used. (ii) Alternatively, the relative abundance of overexpressed mPNGase and hS4 might differ to such a large extent that hS4 could not be detected on the immunoblot. (iii) As a component of 19S proteasome complex, hS4 may interact with another subunit of the proteasome with higher affinity; consequently, it did not coimmunoprecipitate with hPNGase.
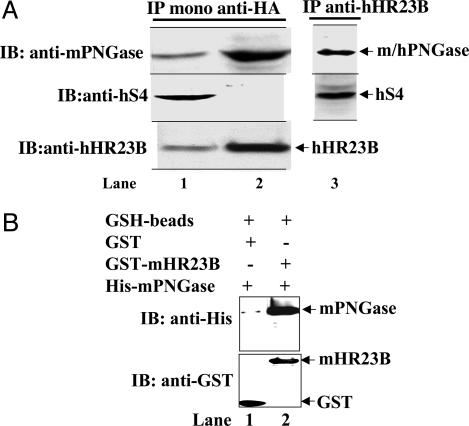
Fig. 2.
In vivo and in vitro interactions among h/m PNGase, h/mHR23B, and h/mS4. (A) Immunoprecipitation (IP) of transiently transfected HeLa cells with HA-tagged-mPNGase expression construct by using monoclonal anti-HA antibody and immunoblotting (IB) with anti-mPNGase, anti-hS4, and anti-hHR23B antibodies. Shown are total cell lysate (lane 1), protein G-agarose beads obtained after immunoprecipitation using mono-HA antibody (lane 2), and protein A-agarose beads obtained after immunoprecipitation using hHR23B antibody (lane 3). (B) In vitro GST-binding assay for interaction between GST-mHR23B and full-length mPNGase. Escherichia coli DH5α cells expressing GST or GST-mHR23B were lysed and subjected to GSH-agarose beads. E. coli BL21(DE3) cell lysates expressing full-length mPNGase with (His)6 tag at the N terminus were used for in vitro binding followed by SDS/PAGE and immunoblotting with anti-His and anti-GST antibodies. Shown are GSH beads carrying GST alone and mPNGase (lane 1) and GSH beads carrying GST-mHR23B and mPNGase (lane 2).
In a second experiment, we asked whether hHR23B associates with hPNGase and hS4 in HeLa cells at the endogenous level. After immunoprecipitation using the hHR23B antibody, we determined whether hPNGase and hS4 were in the immunoprecipitate from the HeLa cell lysate by SDS/PAGE and immunoblotting. Both proteins were found to be associated with hHR23B (Fig. 2 A, lane 3). We confirmed this finding in the reverse coimmunoprecipitation experiment, in which immunoprecipitations were performed with anti-mPNGase or anti-hS4 antibody and immunoblotting was done with anti-hHR23B antibody (data not shown). Based on the immunoprecipitation data, we speculate that hPNGase associates tightly with hHR23B but not with hS4. Perhaps hS4 is more tightly associated with the other subunits of proteasome and therefore could not be detected in the hPNGase–hHR23B complex. Association of hHR23B with both hPNGase and hS4 suggests that hHR23B functions as a bridge between hPNGase and hS4. However, at this point, it is not clear whether the interaction of hHR23B with hS4 is direct or mediated by human S7. In yeast, the interaction of yRad23 with yRpt1 (yeast homolog of human S7) has already been confirmed (7). Moreover, yRpt1 has been shown to form a complex with yRpt2 (8, 9, 11).
Immunoprecipitation experiments detected hPNGase and hHR23B in a complex. Interaction between these proteins was further confirmed by GST-binding assay using recombinant His-mPNGase and GST-mHR23B. Full-length mPNGase was found to interact with mHR23B detected by immunoblotting with anti-His and anti-GST antibodies (Fig. 2B, lane 2). No interaction was observed with GST alone; this result served as a negative control (Fig. 2B, lane 1). GST-binding assay using GST-mS4 and His-mPNGase did not yield reproducible results.
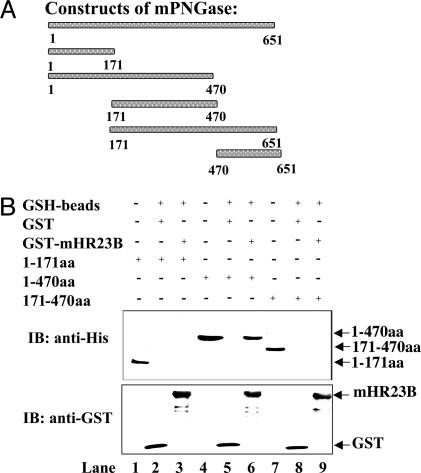
mPNGase Requires the N Terminus and Middle Domains for the Interaction with mHR23B. To identify the region of mPNGase required for interaction with mHR23B, deletion constructs of mPNGase, with His-tag at the N terminus, were prepared (Fig. 3A). To test for a physical interaction between mPNGase constructs and GST-mHR23B, we used a GST pull-down assay. The N terminus of mPNGase contains a PUB/PUG domain implicated in protein–protein interactions; therefore, we expected that only the PUB/PUG domain or the N terminus (1–171aa) containing this domain would be sufficient for protein–protein interactions. In Fig. 3B, the immunoblots with anti-His and anti-GST antibodies are shown. In lane 1, the E. coli cell lysate expressing construct 1–171aa is shown. Lane 2 shows the interaction of GST alone as a control, and lane 3 shows the interaction with GST-mHR23B. No interaction with GST-mHR23B was observed with the 1–171aa construct of mPNGase. We next tested a construct containing the N and the M (middle) domains (1–470aa). The middle domain contains the transglutaminase domain with the catalytic residues, cysteine, histidine, and aspartic acid (33). Lane 4 shows the E. coli cell lysate of mPNGase construct containing 1–470aa, and lane 5 shows the binding with GST alone as a control. The 1–470aa construct of mPNGase showed interaction with GST-mHR23B (lane 6). In contrast, when the mPNGase construct containing only the middle domain (171–470aa, lane 7), the most conserved domain of PNGase in all eukaryotes, was used, no interaction between the two proteins was detected (Fig. 3B, lane 9). These results suggest that neither the N-terminal extension (1–171aa) nor the middle domain alone was sufficient for this interaction. As expected, the other two mouse constructs that contained the middle and the C terminus (171–651aa) or the C terminus alone (470–651aa) failed to show any interaction between mPNGase and mHR23B (data not shown).
Fig. 3.
Deletion constructs of mPNGase and in vitro GST-binding assay for interaction with GST-mHR23B. (A) Various deletion constructs of mPNGase. (B) E. coli DH5α cells expressing GST or GST-mHR23B were lysed and subjected to GSH-agarose beads. E. coli. BL21(DE3) cell lysates expressing mPNGase constructs with a (His)6 tag at the N terminus were used for in vitro binding followed by SDS/PAGE and immunoblotting with anti-His and anti-GST antibodies. Shown are cell lysate mPNGase-1–171aa (lane 1), GSH beads carrying GST alone and mPNGase-1–171aa (lane 2), GSH beads carrying GST-mHR23B and mPNGase-1–171aa (lane 3), cell lysate mPNGase-1–470aa (lane 4), GSH beads carrying GST alone and mPNGase-1–470aa (lane 5), GSH beads carrying GST-mHR23B and mPNGase-1–470aa (lane 6), cell lysate mPNGase-171–470aa (lane 7), GSH beads carrying GST alone and mPNGase-171–470aa (lane 8), and GSH beads carrying GST-mHR23B and mPNGase-171–470aa (lane 9).
mHR23B Interacts with Deglycosylated Protein Substrates. Rad23 and its mammalian homologs (hHR23B and mHR23B) contain a UBL domain that can interact with the proteasome (7, 13, 19). Rad23 and its homologs also contain two UBA domains that interact with ubiquitin (14–16, 34), multiubiquitin chains, and ubiquitinated protein substrates (35). These findings suggest that proteins that contain both UBL and UBA domains represent a previously unrecognized class of proteolytic regulators. Recent findings have provided evidence that Rad23 specifically interacts with multiubiquitin chains in vivo (34) and promotes the targeting of proteolytic substrates to the proteasome (17).
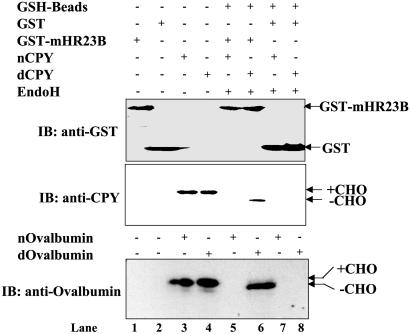
Based on these findings, we asked whether mHR23B could act as a receptor for misfolded, deglycosylated protein substrates and translocate them to the proteasome for degradation. Therefore, we analyzed in vitro binding of two glycoprotein substrates, yCPY and chicken ovalbumin, to mHR23B by using a GST-binding assay. GST-mHR23B and GST (control) were overexpressed in E. coli, and the resulting cell lysates were bound on GSH beads. In Fig. 4, lanes 1 and 2 (anti-GST blot) show the GST-mHR23B and GST cell lysates. Native and denatured yCPY or ovalbumin (lanes 3 and 4, anti-CPY and anti-ovalbumin blots) were subjected to deglycosylation with Endo H and tested for their ability to bind to the GSH beads bound to either GST-mHR23B or GST alone. Neither native yCPY nor native ovalbumin treated with Endo H was bound to GSH beads containing GST-mHR23B (Fig. 4, lane 5), whereas denatured yCPY or denatured ovalbumin that would be deglycosylated with Endo H was bound to GSH beads containing GST-mHR23B (Fig. 4, lane 6). As expected, neither native nor denatured yCPY nor ovalbumin treated with Endo H was detected on GSH beads containing GST alone (Fig. 4, lanes 7 and 8). These findings demonstrate that mHR23B specifically binds to deglycosylated yCPY or to chicken ovalbumin. This finding also suggests that mHR23B acts downstream of PNGase in targeting deglycosylated protein substrates to the proteasome. In contrast, mS4, which is involved in the gating of the proteasome, did not bind to deglycosylated yCPY or ovalbumin directly (data not shown). This analysis supports the idea that the interaction between mHR23B and deglycosylated proteins is specific and not merely because of the hydrophobicity of the misfolded protein. In another study in US11 cells, association of deglycosylated class I MHC heavy chains with Derlin-1 has been reported (36). However, this pathway may be required by a subset of ERAD pathway substrates. This speculation is consistent with the idea that multiple pathways operate to clear misfolded proteins and glycoproteins from the ER.
Fig. 4.
mHR23B binds to deglycosylated proteins yCPY and ovalbumin detected by GST-binding assay. E. coli DH5α cells expressing GST or GST-mHR23B were lysed and subjected to GSH-agarose beads. Native (n) yCPY or chicken ovalbumin treated with Endo H and denatured (d) yCPY or chicken ovalbumin treated with Endo H was used for in vitro binding followed by SDS/PAGE and immunoblotting with anti-GST, anti-CPY, or anti-ovalbumin antibodies. Shown are GST-mHR23B cell lysate (lane 1), GST cell lysate (lane 2), nyCPY or ovalbumin (lane 3), dyCPY or ovalbumin (lane 4), nyCPY or ovalbumin treated with Endo H and bound on GSH beads carrying GST-mHR23B (lane 5), dyCPY or ovalbumin treated with Endo H and bound on GSH beads carrying GST-mHR23B (lane 6), nyCPY or ovalbumin treated with Endo H and bound on GSH beads carrying GST alone (lane 7), and dyCPY or ovalbumin treated with Endo H and bound on GSH beads carrying GST alone (lane 8). In the case of ovalbumin, all other components were added as shown. +CHO and –CHO, proteins with and without carbohydrate, respectively.
Discussion
The current study has established that hPNGase is associated with the ER, based on both biochemical and immunofluorescence analysis. However, in another study using a different cell line, the enzyme activity was found only in the cytoplasmic fraction (6). Recently, PNGase interaction with the ubiquitin complex was detected by two-hybrid analysis (18). Immunoprecipitation and GST-binding assays using two interacting partners, HR23B and S4, revealed that the most likely association of PNGase with S4 is through HR23B. These proteins are present in the cytoplasm but seem to be more concentrated around the ER. Misfolded proteins targeted for proteolysis via the ERAD pathway would be expected to accumulate around the ER. Previous analysis using electron microscopy determined that 14% of the cytoplasmic proteasome is associated with the ER (27, 37), whereas subcellular fractionation methods estimate that only 1% of proteasome is bound to the ER (38). This discrepancy may be due to disruption of the ER–proteasome interaction during subcellular fractionation. Similarly, we presume that during fractionation, the interaction of hHR23B and hS4 with the microsome-bound PNGase is disrupted. Localization of hPNGase and hS4 around the ER supports the hypothesis that deglycosylation and degradation are coupled processes and may occur in close association with the ER for a subset of ERAD pathway substrates.
Mammalian PNGase (mouse and human homologs) exhibits several structural features that differentiate it from yPNGase. First, mPNGase and hPNGase have extended domains at their N and C termini; as a result, mPNGase and hPNGase have a mass that is almost 2-fold greater than that of yPNGase. Second, the N termini of mPNGase and hPNGase contain the PUB/PUG domain, which is conserved among many eukaryotes (20, 21) and has been implicated in protein–protein interactions. However, in this study, we found that the N-terminal domain of mPNGase that contains the PUB/PUG domain is not sufficient to mediate interaction of this enzyme with the proteasome. Instead, both the N terminus and the middle domains are required.
Rad23 and its mammalian homologs already have been shown to play a role in the targeting of ubiquitinated protein substrates to the proteasome (17). Therefore, we asked whether Rad23 also could act as a receptor for misfolded proteins after their deglycosylation. Using an in vitro GST-binding assay, we found that mHR23B specifically binds to two misfolded glycoproteins, yCPY and chicken ovalbumin, but only after they were deglycosylated.
N-linked glycoproteins that are not in their native conformation are deglycosylated and undergo proteasomal degradation in the cytosol. Currently, two models have been proposed for the deglycosylation of misfolded glycoproteins by PNGase (39). In the first model, N-glycanase is present in the cytosol and acts after the translocation (39). In the second model, PNGase is situtated in the membrane and cleaves the glycopeptide bond in a coretrotranslocational manner as it emerges from the ER (39). An increasing number of reports suggest that the N-glycanase is located in or on ER membrane because, in the presence of proteasome inhibitors, the glycosylated protein and deglycosylated intermediates are either exclusively or predominantly associated with the ER rather than with the cytosol (39). This model is supported by various studies using diverse glycoproteins as substrates, e.g., Ig subunits (40), class I MHC heavy chains (41) a ribophorin-I variant (42), cog thyroglobulin mutant (1), cystic fibrosis transmembrane regulator (43), and T cell receptor α-subunit (44). These findings also suggests that N-deglycosylation and proteasome-mediated degradation of proteins may be coupled events. However, PNGase is unlikely to be present in the lumen of the ER; if this had been the case, CPY* in a mutant sec61 strain that cannot retrotranslocate misfolded protein from the ER lumen to the cytosol would have been deglycosylated (45).
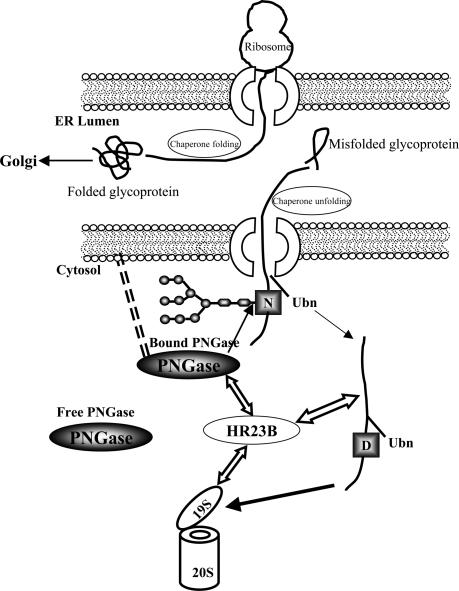
Our current findings support a model in which PNGase is associated with the ER membrane as shown in Fig. 5. This is a working model for the participation of PNGase in the ERAD pathway in mammals. Although the sequence and location of these proteins in the ERAD pathway may vary for different proteins and in different cell types, we propose that PNGase is associated with the surface of the ER and deglycosylates misfolded glycoproteins as they are dislocated into the cytosol (free cytosolic PNGase that could act upon a subset of misfolded glycoproteins also is present). Misfolded deglycosylated protein substrates would then be recognized by HR23B and targeted to the proteasome for degradation. This model prompts several questions for future study. (i) How is PNGase associated with the ER? No hydrophobic domain has been predicted in the primary structure of PNGase that might facilitate its anchoring to the surface of the ER membrane. (ii) Does deglycosylation of misfolded N-glycoprotein occur coretrotranslocationally or after the glycoprotein has completely exited into the cytosol? (iii) After deglycosylation, does the misfolded glycoprotein become ubiquitinated, or does it first bind to HR23B and then become ubiquitinated? (iv) Is this proposed model used by only a subset of ERAD pathway substrates?
Fig. 5.
A working model for the PNGase cascade. As shown, properly folded proteins enter the Golgi complex for further maturation, whereas misfolded glycoproteins exit the ER by means of the retrotranslocon. PNGase, which is associated with the ER surface by an unknown mechanism, deglycosylates a subset of misfolded glycoproteins (free cytosolic PNGase that may be required by another set of glycoproteins also is present). As shown, PNGase interacts with HR23B that, in turn, interacts with the S4 subunit of the 19S proteasome. HR23B binds the deglycosylated (D) protein (CPY and ovalbumin in this study) and delivers it to the proteasome for degradation. The timing of ubiquitination (Ubn) in relation to the deglycosylation process is not clear.
Supplementary Material
Acknowledgments
We thank Dr. Tadashi Suzuki for anti-mPNGase antibody, Dr. Randy Schekman for anti-CPY antibody, and members of the Lennarz laboratory and Dr. Dafna Bar-Sagi for critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM 33814 (to W.J.L.).
Abbreviations: PNGase, peptide:N-glycanase; mPNGase, mouse PNGase; hPNGase, human PNGase; yPNGase, yeast PNGase; hcalnexin, human calnexin; hHR23B, human HR23B; hS4, human S4; mHR23B, mouse HR23B; mS4, mouse S4; ER, endoplasmic reticulum; ERAD, ER-associated degradation; yCPY, yeast carboxypeptidase Y; GSH, glutathione; HA, hemagglutinin epitope.
References
- 1.Tokunaga, F., Brostrom, C., Koide, T. & Arvan, P. (2000) J. Biol. Chem. 275, 40757–40764. [DOI] [PubMed] [Google Scholar]
- 2.Cresswell, P. & Hughes, E. A. (1997) Curr. Biol. 7, R552–R555. [DOI] [PubMed] [Google Scholar]
- 3.Kopito, R. R. (1997) Cell 88, 427–430. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki, T., Park, H., Hollingsworth, N. M., Sternglanz, R. & Lennarz, W. J. (2000) J. Cell Biol. 149, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blom, D., Hirsch, C., Stern, P., Tortorella, D. & Ploegh, H. L. (2004) EMBO J. 23, 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch, C., Blom, D. & Ploegh, H. L. (2003) EMBO J. 22, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki, T., Park, H., Kwofie, M. A. & Lennarz, W. J. (2001) J. Biol. Chem. 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann-Petersen, R., Tanaka, K. & Hendil, K. B. (2001) Arch. Biochem. Biophys. 386, 89–94. [DOI] [PubMed] [Google Scholar]
- 9.Fu, H., Reis, N., Lee, Y., Glickman, M. H. & Vierstra, R. D. (2001) EMBO J. 20, 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohler, A., Cascio, P., Leggett, D. S., Woo, K. M., Goldberg, A. L. & Finley, D. (2001) Mol. Cell 7, 1143–1152. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi, J. & Tamura, T. (2004) FEBS Lett. 565, 39–42. [DOI] [PubMed] [Google Scholar]
- 12.Elsasser, S., Gali, R. R., Schwickart, M., Larsen, C. N., Leggett, D. S., Muller, B., Feng, M. T., Tubing, F., Dittmar, G. A. & Finley, D. (2002) Nat. Cell Biol. 4, 725–730. [DOI] [PubMed] [Google Scholar]
- 13.Schauber, C., Chen, L., Tongaonkar, P., Vega, I., Lambertson, D., Potts, W. & Madura, K. (1998) Nature 391, 715–718. [DOI] [PubMed] [Google Scholar]
- 14.Bertolaet, B. L., Clarke, D. J., Wolff, M., Watson, M. H., Henze, M., Divita, G. & Reed, S. I. (2001) Nat. Struct. Biol. 8, 417–422. [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., Shinde, U., Ortolan, T. G. & Madura, K. (2001) EMBO Rep. 2, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson, C. R., Seeger, M., Hartmann-Petersen, R., Stone, M., Wallace, M., Semple, C. & Gordon, C. (2001) Nat. Cell Biol. 3, 939–943. [DOI] [PubMed] [Google Scholar]
- 17.Chen, L. & Madura, K. (2002) Mol. Cell. Biol. 22, 4902–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, H., Suzuki, T. & Lennarz, W. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiyama, H., Yokoi, M., Masutani, C., Sugasawa, K., Maekawa, T., Tanaka, K., Hoeijmakers, J. H. & Hanaoka, F. (1999) J. Biol. Chem. 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki, T., Park, H., Till, E. A. & Lennarz, W. J. (2001) Biochem. Biophys. Res. Commun. 287, 1083–1087. [DOI] [PubMed] [Google Scholar]
- 21.Doerks, T., Copley, R. R., Schultz, J., Ponting, C. P. & Bork, P. (2002) Genome Res. 12, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker, D., Wuestehube, L., Schekman, R., Botstein, D. & Segev, N. (1990) Proc. Natl. Acad. Sci. USA 87, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodsky, J. L. & McCracken, A. A. (1999) Semin. Cell Dev. Biol. 10, 507–513. [DOI] [PubMed] [Google Scholar]
- 24.Weng, S. & Spiro, R. G. (1997) Biochem. J. 322, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki, T., Kitajima, K., Emori, Y., Inoue, Y. & Inoue, S. (1997) Proc. Natl. Acad. Sci. USA 94, 6244–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki, T., Park, H., Kitajima, K. & Lennarz, W. J. (1998) J. Biol. Chem. 273, 21526–21530. [DOI] [PubMed] [Google Scholar]
- 27.Rivett, A. J., Palmer, A. & Knecht, E. (1992) J. Histochem. Cytochem. 40, 1165–1172. [DOI] [PubMed] [Google Scholar]
- 28.Rivett, A. J. (1998) Curr. Opin. Immunol. 10, 110–114. [DOI] [PubMed] [Google Scholar]
- 29.Enenkel, C., Lehmann, A. & Kloetzel, P. M. (1998) EMBO J. 17, 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enenkel, C., Lehmann, A. & Kloetzel, P. M. (1999) Mol. Biol. Rep. 26, 131–135. [DOI] [PubMed] [Google Scholar]
- 31.van der Spek, P. J., Eker, A., Rademakers, S., Visser, C., Sugasawa, K., Masutani, C., Hanaoka, F., Bootsma, D. & Hoeijmakers, J. H. (1996) Nucleic Acids Res. 24, 2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugasawa, K., Masutani, C., Uchida, A., Maekawa, T., van der Spek, P. J., Bootsma, D., Hoeijmakers, J. H. & Hanaoka, F. (1996) Mol. Cell. Biol. 16, 4852–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katiyar, S., Suzuki, T., Balgobin, B. J. & Lennarz, W. J. (2002) J. Biol. Chem. 277, 12953–12959. [DOI] [PubMed] [Google Scholar]
- 34.Rao, H. & Sastry, A. (2002) J. Biol. Chem. 277, 11691–11695. [DOI] [PubMed] [Google Scholar]
- 35.Ortolan, T. G., Tongaonkar, P., Lambertson, D., Chen, L., Schauber, C. & Madura, K. (2000) Nat. Cell Biol. 2, 601–608. [DOI] [PubMed] [Google Scholar]
- 36.Lilley, B. N. & Ploegh, H. L. (2004) Nature 429, 834–840. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, A., Rivett, A. J., Thomson, S., Hendil, K. B., Butcher, G. W., Fuertes, G. & Knecht, E. (1996) Biochem. J. 316, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyette, J. R. & Mykles, D. L. (1992) Muscle Nerve 15, 1023–1035. [DOI] [PubMed] [Google Scholar]
- 39.Spiro, R. G. (2004) Cell. Mol. Life Sci. 61, 1025–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancini, R., Fagioli, C., Fra, A. M., Maggioni, C. & Sitia, R. (2000) FASEB J. 14, 769–778. [DOI] [PubMed] [Google Scholar]
- 41.Kamhi-Nesher, S., Shenkman, M., Tolchinsky, S., Fromm, S. V., Ehrlich, R. & Lederkremer, G. Z. (2001) Mol. Biol. Cell 12, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitzmuller, C., Caprini, A., Moore, S. E., Frenoy, J. P., Schwaiger, E., Kellermann, O., Ivessa, N. E. & Ermonval, M. (2003) Biochem. J. 376, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong, X., Chong, E. & Skach, W. R. (1999) J. Biol. Chem. 274, 2616–2624. [DOI] [PubMed] [Google Scholar]
- 44.Yu, H., Kaung, G., Kobayashi, S. & Kopito, R. R. (1997) J. Biol. Chem. 272, 20800–20804. [DOI] [PubMed] [Google Scholar]
- 45.Plemper, R. K., Bohmler, S., Bordallo, J., Sommer, T. & Wolf, D. H. (1997) Nature 388, 891–895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.