Abstract
The crystal structure of the Dps-like (Dps, DNA-protecting protein during starvation) ferritin protein DpsA from the halophile Halobacterium salinarum was determined with low endogenous iron content at 1.6-Å resolution. The mechanism of iron uptake and storage was analyzed in this noncanonical ferritin by three high-resolution structures at successively increasing iron contents. In the high-iron state of the DpsA protein, up to 110 iron atoms were localized in the dodecameric protein complex. For ultimate iron storage, the archaeal ferritin shell comprises iron-binding sites for iron translocation, oxidation, and nucleation. Initial iron–protein interactions occur through acidic residues exposed along the outer surface in proximity to the iron entry pore. This narrow pore permits translocation of ions toward the ferroxidase centers via two discrete steps. Iron oxidation proceeds by transient formation of tri-iron ferroxidase centers. Iron storage by biomineralization inside the ferritin shell occurs at two iron nucleation centers. Here, a single iron atom provides a structural seed for iron-oxide cluster formation. The clusters with up to five iron atoms adopt a geometry that is different from natural biominerals like magnetite but resembles iron clusters so far known only from bioinorganic model compounds.
Keywords: ferritin, iron-binding, inorganic cluster formation, x-ray crystal structure
Iron not only is an essential cofactor of many enzymes but also has been a major threat to cellular life since the advent of atmospheric oxygen 2.8 billion years ago. Despite the abundance of iron, its cellular uptake is compromised, because this element occurs as Fe3+ in a mostly insoluble form under physiological pH and oxidizing conditions. Furthermore, free Fe3+ ions generate highly reactive hydroxyl radicals by Fenton reactions with biogenic oxygen species like superoxide anions or hydrogen peroxide (1). Consequently, the iron present in the cytosol has to be tightly sequestered by complexation to proteins. The protein family of ferritins evolved as such a specialized iron storage container, which incorporates intracellular iron in a bioavailable and nontoxic form. Here, iron is taken up into oligomeric ferritin shells as Fe2+, oxidized therein to Fe3+ by ferroxidase centers, and deposited in the large cavities of the ferritin oligomers as crystalline or amorphous ferrihydrite-like cores (2). Such polymerization processes often occur in nature and usually rely on preorganized bioorganic templates, as provided by proteinaceous surfaces. Several other examples of biological iron oxidation and polymerization processes exist as well, such as the formation of magnetosomes by the magnetic bacterium Magnetospirillum gryphiswaldense or the presence of magnetite-like crystals in unicellular and multicellular organisms such as algae, salmon, pigeons, and humans (ref. 3 and references therein).
Two major subfamilies of ferritins are known that differ in their quaternary structure but share a highly conserved protein fold based on a four-helix bundle. In the eukaryotic and most of the eubacterial ferritins, 24 identical or homologous subunits assemble to ball-like protein shells with 432 symmetry (4, 5). The 432-symmetric ferritin shells have overall molecular masses of 450–500 kDa and outer and inner diameters of 12 and 8 nm, respectively (6, 7). Up to 4,500 Fe3+ ions can be stored per eukaryotic 24-mer apoferritin, whereas eubacterial 24-mer ferritins are able to incorporate up to 2,500 iron atoms (1).
Members of the other subfamily were originally discovered as stress proteins, which protect DNA against oxidative stress during nutrient starvation. Several members of the Dps subfamily, such as Dps from Escherichia coli or the Dps homolog from Bacillus subtilis exhibit a DNA-binding activity that is at least partially linked with iron complexation. DNA binding by these proteins was shown to suffice for protection against oxidative DNA damage and might be mediated by magnesium ions, which bridge the protein surfaces with the polyanionic DNA (7, 8). Functionally, the Dps subfamily is much more diverse, with many members promoting iron incorporation and others acting as immunogens, neutrophile activators (9), cold-shock proteins, or constituents of fine-tangled pili (10). Another mode of protection against reactive oxygen species implies the preferential consumption of hydrogen peroxide instead of oxygen during biomineralization (11).
Due to the absence of the C-terminal fifth helix of 24-mer ferritins, members of the Dps subfamily assemble only to dodecameric protein shells that have cubic 23 symmetry and outer and inner diameters of 9 and 4.5 nm. Consequently, the iron-storage capacity of these proteins is smaller, and ferritin dodecamers from Helicobacter pylori and Listeria innocua were reported to oxidize and sequester up to 500 iron atoms inside their cavity (9, 12, 13). Hereby, the main entry of iron and other ionic species into these members of the Dps subfamily is postulated to occur along pores that penetrate the protein shell at the threefold axes of symmetry (9, 13), and that were also identified as entrance sites in the 24-mer ferritins (2). In the crystal structures of Dps proteins from E. coli (14), L. innocua (13), and Bacillus anthracis (15), these access channels are conserved, besides an interfacial iron-binding site that was assigned in these Dps-like ferritins as a di-iron ferroxidase center for catalytic iron oxidation.
So far, structures of the ferritin family have been described from two of the three domains of life, eukaryotes and eubacteria, but not from archaea (16). Iron uptake and storage by these enzymes were extensively studied (2), but neither the uptake route nor the redox pathway could be analyzed in atomic detail. The lack of x-ray crystallographic data about biomineralization steps in ferritins might be caused by the partial lack of order in their iron cores (2). Here, we report crystal structures of an archaeal Dps-like ferritin from the euryarchaeote H. salinarum at a low- and two high-iron states. This ferritin functionally resembles its eubacterial homologues but exhibits unique structural features for iron uptake and storage pathways. By x-ray crystallography, the sites of initial iron binding, translocation, oxidation, nucleation, and storage in a bioavailable form were delineated at high resolution. Like other processes in nature, the iron storage process here described can be understood on the basis of matrix-controlled biomineralization.
Experimental Procedures
Crystallization, Iron Complexation, and X-Ray Data Collection. The purification of H. salinarum DpsA and its crystallization in crystal forms A–D are described elsewhere (unpublished results). A single nontwinned low-resolution dataset was collected from a form A crystal (space group P3121, a = b = 91 Å, c = 223 Å) at beamline ID14-2 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France, at 2.8 Å (FELR). Later, a low-iron high-resolution dataset (FEHR) was collected at 1.6-Å resolution from crystal form B (space group P321, a = b = 91.11 Å, c = 150.04 Å) as 1° oscillation frames at 100 K on ESRF beamline ID29 by using a charge-coupled device detector (ADSC) quantum detector at λ = 1.0052 Å.
DpsA crystals initiating biomineralization were prepared under transiently anaerobic conditions. Form A crystals were taken from a single drop and transferred into a 2-μl drop containing the precipitant solution plus 20 mM FeCl2 and 2 mM sodium dithionite. The crystals were incubated for 30 (FE30) or 120 min (FE120) against the reservoir solution under aerobic conditions to permit slow reoxidation of iron. As reference served a crystal from the same batch without soaking (FE0). Single anomalous dispersion diffraction data of these crystals differing in iron content were collected at the peak wavelength of iron x-ray absorption (λ = 1.7314 Å) at beamlines ID29 (FE0, FE120), European Synchrotron Radiation Facility, and X06SA, Swiss Light Source, Villigen, Switzerland (FE30). X-ray fluorescence scans were collected at beamline ID29 for the FE0 crystal by using the automated scan procedure. Spectral points were collected in 1-kV steps. Diffraction intensities were integrated by xds, scaled, and merged by and xscale (17). The statistics of the four datasets are summarized in Table 1 and Table 2, which is published as supporting information on the PNAS web site. Because twinning was regularly observed for form A crystals, the absence of twinning in collected diffraction data was tested by using the programs detwin (CCP4, www.ccp4.ac.uk/main.html) and cns (18).
Table 1. Definition of iron sites.
| Data collection | ||||
| Dataset | FEHR | FE0 | FE30 | FE120 |
| Refinement statistics (cns/refmac) | ||||
| Iron content* | Low (0 min) | Low (0 min) | High (30 min) | High (120 min) |
| No. of irons in TS† | 0 | 0 | 3 (T1-T3) | 3 (T1-T3) |
| No. of irons in FOC† | 1 (F1) | 1 (F1) | 3 (F1-F3) | 1 (F1) |
| No. of irons in NI† | 0 | 0 | 3 (N11-N13) | 3 (N11-N13) |
| No. of irons in NII† | 1 (N21) | 1 (N21) | 5 (N21-N25) | 5 (N21-N25) |
Incubation time of crystals with Fe2+ solutions is given in parentheses.
Definition of the subsites used in the text is given in parentheses.
Structure Solution and Refinement. The H. salinarum DpsA structure was solved by molecular replacement by using the nontwinned dataset FELR and a tetrameric model as a search probe that was derived from molecules A, C, I, and K of the Dps dodecamer of E. coli (Protein Data Bank ID code 1DPS), with each chain comprisingresidues26–167. The rotational and translational searches were subsequently performed by molrep (19). The best solution matched a correlation coefficient of 0.164 and an R factor of 0.54. The initial model was further refined by alternative cycles of model rebuilding in o (20) and automatic refinement using cns and refmac with noncrystallographic symmetry restraints (18, 21). When the refinement converged at an R factor/Rfree of 0.206/0.260 for data between 25 and 2.8 Å, the refinement was continued at 1.6 Å with the high-resolution dataset FEHR. The final structure with six Fe3+ atoms bound has a R factor/Rfree of 0.164/0.210 for data between 30 and 1.6 Å (Table 2).
For the FE0, FE30, or FE120 datasets, model phases were generated for the calculation of anomalous difference Fourier maps after initial refinement against the FEHR structure as a starting model. Additional iron atoms were included in the further refinement process of the FE30 and FE120 structures, if anomalous difference peaks >3 I/σ(I) were found at noncrystallographic symmetry-equivalent positions. A few additional irons were added if waters that were originally placed into densities showed remarkably lower B factors than their environment. All magnesium ions from the FEHR structure with its low iron content were replaced by iron in the FE30 and FE120 structures. The final models of the FE30/FE120 datasets consisted of the same number of protein residues, sulfates, and Na+ ions as the FEHR structure but comprised 38 Fe3+/30 Fe3+ and 40/312 waters, respectively (see Table 1).
Results and Discussion
Crystallization and Structural Analysis of H. salinarum DpsA Protein. The crystallization of soluble proteins from halophilic organisms is often hampered by the exceedingly high solubilities these proteins exhibit at saturating salt concentrations (22). Consequently, we pursued a systematic approach to identify crystallizable halophilic proteins to expand the structural repertoire. The cytosol of the halophile H. salinarum was therefore fractionated by conventional chromatography techniques, and concentrated fractions were subjected to an adapted sparse-matrix screen. Crystallized proteins were identified by N-terminal Edman degradation by using single protein crystals and by comparison of these sequences with the genome sequence (see www.halolex.mpg.de). One of them was the archaeal ferritin DpsA protein (ref. 23; 182 aa, Mr = 20,100), which was crystallized in the trigonal crystal forms P3121 (crystal form A) and P321 (crystal form B) by using polyethylene glycol 400 as a precipitant.
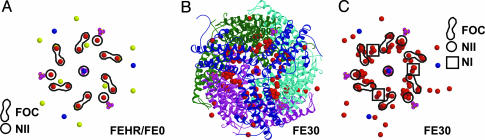
The structure of DpsA from H. salinarum was solved from crystal form B by self-rotation functions and molecular replacement using the structure of E. coli Dps as a search model (Protein Data Bank ID code 1DPS, residues 25–167; for details, see Experimental Procedures). The final structure was refined at 1.6-Å resolution (FEHR low-iron dataset) and comprised four ferritin molecules with 705 protein residues, 6 Fe3+ ions, 4 Mg2+ sites, 2 Na+ ions, 2 sulfates, and 796 water molecules in the asymmetric unit (see Fig. 1A for representation of ions and Table 1 for definition of datasets and iron sites). The traces of the polypeptide chains were clearly defined by electron density from Ala-7 to Leu-181. In addition, the N terminus of chain A in crystal form B extends to Ser-2, because this N-terminal stretch makes crystal contacts with chain A of a symmetry-related dodecamer.
Fig. 1.
Iron-binding sites of low-(FEHR/FE0 data) and high-iron (FE30) content DpsA protein are compared. (A and C) Isolated ion-binding sites of the DpsA ferritin displayed without the protein backbone. The FOC, NI, and NII centers are encircled or boxed. The ion- and sulfate-binding sites only of the H. salinarum ferritin are displayed; iron sites are rendered in red, magnesium in yellow, sodium in blue, and sulfate in yellow/magenta. (B) Ribbon model of the quaternary high-iron structure (FE30) of the ferritin dodecamer as viewed down the threefold axis. Each of the four trimers related by twofold symmetry is in a different color. Figs. 1, 2, 3 were made with molscript (33), raster3d (34), and dino (http://cobra.mih.unibas.ch/dino/intro.php).
X-ray fluorescence spectra collected from a native crystal independently proved the presence of iron (data not shown). Accordingly, anomalous difference Fourier syntheses that were calculated at 2.2-Å resolution from a single anomalous dispersion dataset of the same crystal (FE0) verified the six iron positions of the FEHR dataset with I/σ(I) ratios >4. However, the occupancies of singly occupied ferroxidase centers were subsequently reduced to 0.5 to match the anomalous peak heights and the B factors of the protein environment.
Structural Comparison of DpsA with 24-Mer Ferritins and Dps-Like Orthologues. The archaeal DpsA monomer folds into a four-helix bundle that is commonly found in the 24-mer ferritins and members of the Dps subfamily. One hundred eighteen of 182 residues (65%) belong to α helices (Fig. 1B). Helices 2 and 5 (α2 and α5 in Fig. 2B) form the inner wall of the protein shell, whereas helices α1 and α4 line the cytoplasmically exposed outer protein surface (Figs. 1B and 2B). Helices 2 and 4 are connected by a long irregular stretch that is intersected in its middle by the short helical segment 3 (α3, residues Pro-96-Ala-103). A superposition of the H. salinarum monomer on the subunits of the Dps-like ferritin of L. innocua and Dps of E. coli yields root-mean-square deviations (rmsd) of 1.34 and 1.27 Å for 158 and 149 Cα positions, respectively (13, 14). Structural differences to the eubacterial orthologs are mainly found for the α4–α5 loop region, which comprises three additional residues in the haloarchaeal ferritin. Furthermore, the halobacterial ferritin comprises an elongated N-terminal tail enriched in negatively charged residues and a C-terminal “sickle-like” region (Asp-172-Leu-181). This sickle region is clearly different from the C-terminal region (helix α5) in the structurally related 24-mer ferritins of eukaryotes, e.g., horse spleen L-chain (rmsd of 1.68 Å for 169 Cα positions), where helix 5 was found to stabilize the 24-mer particle along fourfold symmetric contacts (6, 24).
Fig. 2.
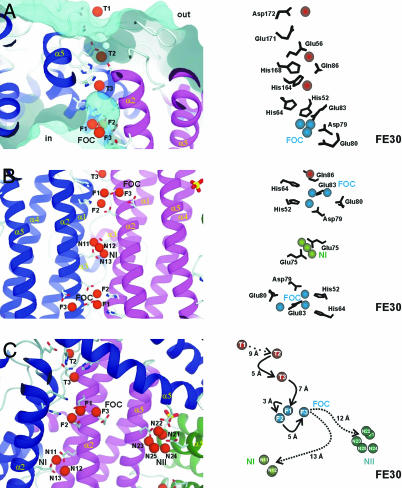
Overview on the iron-binding sites in H. salinarium ferritin based on the FE30 data set. (Left) The general environment of the iron atoms; (Right) detailed analysis of the residues involved and some distances following the iron entry to the final storage center are given. (A) Iron translocation over the DpsA protein shell from the outside (out) to the inner cavity (in). A surface representation of the outer and inner surface is shown in cyan. Side view of the iron translocation channel (from the FE30 dataset) with the initial iron-binding site T1 and the successive iron-binding sites T2 and T3, which occur in the iron translocation channel. The important side chains guiding the iron atoms are shown. (B) Two ferroxidase centers of symmetry-related protein molecules (colored in blue and red) are viewed along the twofold axis. The helices α1–α5, important residues, and the three iron-binding subsites F1–F3 of the FOC and nucleation center NI are depicted. Nucleation center NI, including the two symmetry-related iron atoms N11 and N13 and a third iron atom as well as the symmetry-related liganding residues Glu-72 and Glu-75, is shown. (C) View almost perpendicular to the threefold molecular axis. Overview of the ferroxidase and the two nucleation centers NI and NII. Possible routes for iron atom transfers from the FOC to NI and NII, respectively, are marked. Distances among iron centers and possible storage possibilities are indicated.
Accordingly, the H. salinarum ferritin assembles to a 23-symmetric homododecamer. Helix 5 faces the inner compartment of the dodecamer like helix 2, but the latter resides at almost identical relative locations in the ferritin dodecamers from E. coli, H. salinarum, and L. innocua. The functional role of helix 2 is underlined by at least one iron-binding site [ferroxidase center (FOC) site, Fig. 2 A and B] that is conserved among eubacterial Dps homologues and a cluster of carboxylate residues [nucleation site 1 (NI) center, Fig. 2B] that resembles the iron core nucleation site of mammalian ferritin L chains (1, 25).
Iron-Binding Sites in Low- and High-Iron State DpsA Complexes. The crystallized halobacterial ferritin DpsA was purified as an intact and iron-comprising complex from the cytosol of H. salinarum. In the low-iron-state crystals FEHR (1.6-Å resolution, low iron) and FE0 (2.2-Å resolution, single anomalous dispersion, low iron), the presence of 16 iron-binding sites per protein dodecamer was clearly confirmed by anomalous difference Fourier maps. All of these sites were attached along the interior protein surface, of which 12 iron atoms were placed at the dimeric FOC and four at the trimeric subunit interfaces, nucleation site 2 (NII) (see Fig. 1A for all sites and Fig. 3A for the anomalous density of NII).
Fig. 3.
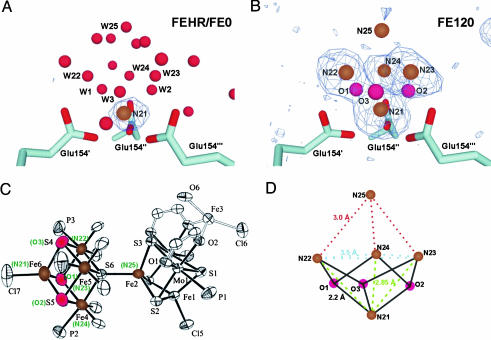
Detailed structural analysis of nucleation center NII of Dpsa from H. salinarium. (A) View perpendicular to the threefold axis on the nucleation center NII of the low-iron FEHR structure. Anomalous difference Fourier maps at 4σ indicating the positions of individual iron atoms are shown in blue. Subsite N21 of nucleation center NII is shown in brown, whereas selected water molecules are depicted in red. Some important solvent molecules resembling atom positions of the high-iron structure FE120 (B) form a crown-like structure (W22–W25 represent N22–N25 and W1–W3 represent O1–O3). The waters are labeled according to iron and oxygen atoms of the FE120 iron cluster. (B) The same view toward the NII iron cluster of the FE120 structure. Anomalous difference Fourier maps at 3.2σ indicating the positions of individual iron atoms are shown in blue. Four strong (N21–N24) and one additional weak (N25) iron peaks are visible. The iron atoms N21–N24 are triply bridged by three OH– or O2– atoms (O1–O3). (C) Molecular structure of a polynuclear inorganic model compound with similar arrangement of iron atoms (Fe2–Fe6 resemble N21–N25), whereas the sulfurs of the structure replace oxygen O1–O3 from FE120. For clarity, only the O donors of the Cl4-cat ligand and the C atoms attached to them are shown. Similarly, only the O atoms of the DMF molecule bound to Fe (3) are shown (29). Equivalent atomic positions of the NII cluster are marked in green. (D) Schematic view of the NII iron cluster. Symmetry equivalent distances between iron–iron and iron–oxygen atoms are marked, and the respective distances are given.
Recently, DpsA of H. salinarum was shown to complex up to 100 iron atoms per dodecamer, which are at least partly constituents of large ferric iron clusters according to EPR spectroscopy (23). To locate iron-binding sites still missing in the native structure of H. salinarum DpsA, we incubated crystals for 30 (FE30) and 120 min (FE120) with Fe2+ under conditions that may promote biomineralization. In the FE30 crystals, a total number of 110 irons per dodecameric complex could be identified (Fig. 1C), whereas in the FE120 crystals, the number of irons decreased to 86 irons, because only the F1-binding sites of the 12 ferroxidase centers were occupied (data not shown). Overall, the iron sites can be subdivided into four classes according to their locations and presumptive function in the DpsA complex (see also Table 1): (i) iron access route (three subsites T1–T3 in FE30 and FE120); (ii) the ferroxidase center FOC (three subsites F1–F3 in FE30, F1 in FE120); (iii) nucleation center NI (three subsites, N11–N13 in FE30 and FE120; N11 and N13 are symmetry related); (iv) nucleation center NII (five subsites, N21–N25, in FE30 and FE120, N22, N23, and N24 are related by symmetry). It is noteworthy that the overall numbers agree quite well with the amount of bound iron determined in isolated samples of DpsA (23).
Translocation Sites T1–T3: An Iron Access Route. Significant structural differences between the meso- and halophilic homologues of the Dps subfamily are observed along the threefold axes of the dodecamers. In the Listeria ferritin and Dps from E. coli, a hydrophilic pore with a diameter of ≈8 Å runs between the α4α5 loops of three symmetry-related subunits and is exclusively formed by acidic residues. Because these features are well conserved among 12- and 24-mer ferritins, it was suggested that the pore region around this threefold axis represents the major entrance site for Fe2+ into the ferritin protein shell (4, 13, 26). In the halophilic ferritin dodecamer, this pore is plugged by a row of four residues, Glu-141, His-150, Arg-153, and Glu-154 (not shown). Only the threefold related Glu-154 residues serve as ligands of the NII nucleation center (see Figs. 2C and 3 A and B). Moreover, a sulfate ion that may be introduced during the purification process is observed 6.7 Å apart from the N21 iron atom in the midst of the plugged pore, where it makes numerous hydrogen bonds with the threefold symmetry-related His-150 and Arg-153 residues (Fig. 2C, side chains of liganding residues are not shown). Hence in the H. salinarium ferritin, the passage of ions along this pathway would be feasible only if major conformational changes transiently open the channel.
In contrast to other structurally characterized 12- and 24-mer ferritins, the DpsA structure reveals a pathway for iron binding and translocation. At the outer protein periphery, an initial translocation site (T1) is found ≈20 Å distant from the catalytic FOC (Fig. 2A). This T1 site is close to the entrance of a water-filled translocation pore leading to the inner compartment of the ferritin shell. The iron of the T1 site is coordinated by Glu-171 and Asp-172. The T1 site is placed on top of a shallow depression, which is surrounded by N-(Arg-8-Asp-18) and C-terminal (Glu-167-Asp-173) stretches derived from two different monomers. These stretches contribute to a cluster of six acidic residues (Glu-13, Glu-15, Asp-18, Glu-167, Glu-171, and Asp-173), which might together act as an electrostatic guide for incoming Fe2+. An analogous electrostatic arrangement was also observed around the threefold axis of the L. innocua ferritin (13). A second binding site (T2) is buried inside the pore, 13 Å away from the FOC. The iron in the T2 site is bound to the residues Glu-56, Gln-86, and His-168, which are recruited from two adjacent monomers. It is noteworthy that in the FEHR and FE0 structures, a magnesium ion replaces the T2 site iron of the FE30 and FE120 structures. Between the T2 site and the FOC, a third binding site (T3) is about halfway between these. Here, weak interactions are formed to residues His-164 and Gln-86. The distances between the four iron sites are all in the range of 5–9 Å (T1–T2, 9 Å; T2–T3, 5 Å; T3–F1, 7 Å, see Fig. 2 A and C).
From the structural data, one might conclude that a hydrated Fe2+ atom is initially bound to the outer surface of DpsA and progressively guided through the pore by hopping in discrete steps from the initial binding site T1 via T2 and T3 to the ferroxidase site. This pathway is obviously unique to the haloarchaeal ferritin. For example, in the Listeria ferritin, two bulky aromatic residues tightly plug the corresponding entrance channel (13). Only the minimal pore diameter in the DpsA complex of ≈5 Å and the distance between the pores and the catalytic FOCs resembles the pores of mesophilic ferritins, which are supposed to be exclusively formed along the threefold symmetry axes (2). Instead, the diversion from the threefold symmetric pores of mesophilic ferritins might be dictated by the hypersaline environment. First, due to the usage of a nonsymmetric site, the H. salinarium ferritin provides 12 entrance pores for iron to the inner compartment pores instead of only four, as in the mesophilic Dps-like ferritins. The increased permeability of the DpsA protein shell might circumvent the problem that electrostatic guidance of iron ions as described for other ferritins is not feasible for a halophilic ferritin ortholog, because long-range electrostatic interactions are efficiently shielded by the almost saturating salt content of the haloarchaeal cytosol (≈5 M KCl). Second, the participation of histidine residues in the iron translocation pathway (His-164, His-168, and His-172) differs significantly from other mesophilic Dps orthologs. Coordination of Fe3+ to imidazole groups is known to proceed with similar dissociation constants (<1 mM) as to carboxylates, whereas potassium ions form ion pairs only with the latter. The competition of iron and alkali ions for coordination to the ferritin shell hence might be better overcome by the more covalent liganding to histidine residues, which are distributed along the inner and outer surfaces of DpsA.
The Ferroxidase Center FOC. The ligands of the ferroxidase center are conserved throughout the dodecameric ferritins, including ferritin from L. innocua and Dps from E. coli. In the low-iron state structures FE0 and FEHR, only a single iron atom (F1 subsite) is bound to the FOC. The iron is coordinated in a hexagonally distorted manner to two water molecules and the conserved residues Asp-79 (A), Glu-83 (A), His-52 (B) from two monomers, which are related by a twofold symmetry axis (data not shown). Such a geometry is similar to the FOCs found in the structures from L. innocua, B. anthracis, and E. coli (13–15).
In the high-iron state dataset FE30, two additional subsites, F2 and F3, were detected at the FOC (Fig. 2B). The F2 subsite is 3.2 Å (averaged value of four symmetry-related monomers) apart from F1 and almost exactly replaces a water molecule, which was observed in the low-iron-state structures. The ion is coordinated to residues Glu-83 (A) and the His-64 (B) of the adjacent monomer. The averaged distance calculated for the four protein molecules related by noncrystallographic symmetry of 3.2 Å between F1 and F2 reflects the canonical distance of a Fe-O-Fe μ-oxo-bond. Surprisingly, a third iron (F3) was found at an averaged distance of 3.9 Å to F2 and 4.9 Å to F1. This F3 iron is coordinated to Glu-80 and Glu-83 of the same monomer. Interestingly, the F2 and F3 subsites are apparently transiently occupied only during the iron charging of DpsA crystals, because these sites were missing in the FE0/FEHR and FE120 and other DpsA structures where crystals were soaked longer than 120 min (data not shown). Such a loose coordination of iron in the F2 subsite was also noted for the Bacillus brevis Dps structure, where a water ligand bridges the F2 iron to the histidine ligand (26). Consequently, the F2 and F3 subsites might be occupied only by Fe2+, which is continuously oxidized to Fe3+ during the prolonged incubations in the presence of oxygen. However, due to the low resolution of the FE30 dataset, we cannot yet prove the water and oxygen coordination geometries of the F2 and F3 subsites. Although the F2 site participates in the oxidation of hydrogen peroxide, the functional role of the F3 subsite is still elusive. Interestingly, such a third iron-binding site was also located in the E. coli ferritin (EcFtnA) next to the nonhomologous di-iron ferroxidase center (27). In the E. coli ferritin, the third iron site assists in the complete reduction of O2 to water, which avoids the generation of reactive oxygen species. Accordingly, Dps-like ferritins are highly efficient with hydrogen peroxide as oxidant but may also accept O2 with much lower turnover (11). Furthermore, the third site of the E. coli ferritin was also reported to stabilize the di-iron FOC, such that the proportion of protein-bound iron was larger than in ferritins without this site (28). Such a slowed incorporation of iron into iron cores was suggested to allow better bioavailability to the microorganism compared to the highly biomineralizing mammalian ferritins.
In any case, if the oxidation of F2 and F3 site irons leads to Fe3+ ejection from the di- or tri-iron FOC, these irons would have to migrate ≈12–13 Å to the closest NI or NII nucleation sites (Fig. 2C). The distance between the F3 subsite and the NII nucleation center might be overcome by intervening residues like Glu-158 and Glu-161, which are both in 6-Å distance to subsite F3 and the nucleation center NII, respectively.
Nucleation Center NI. Two acidic amino acids, which reside next to the twofold symmetry axis of the dodecamer, were supposed to form the iron core nucleation site along helix 2 (α2 in Fig. 2B) in the ferritin from L. innocua (13). It was suggested that, after oxidation at the ferroxidase site, one Fe3+ is displaced by Fe2+ and migrates to the proposed helix 2 iron nucleation site. However, the acidic residues Glu-44 and Asp-47 of L. innocua ferritin (E. coli, Glu-64, Asp-67; B. subtilis, Glu-47, Glu-50) are replaced by leucine and glycine in H. salinarum DpsA (Leu-65, Gly-68). As an alternative iron nucleation site, the H. salinarium ferritin provides the four acidic residues Glu-72 (A, B) and Glu-75 (A, B) at similar positions along the dimeric interface, which are related by twofold symmetry. Three iron peaks (N11–N13) are observed there in anomalous difference maps, two of which are protein-bound (N11 and N13). A short distance of 2.2 Å between two peaks (N11 and N13) can be explained only by their proximity to the twofold axis and the exclusive occupancy of this site by a single iron atom. Because the iron ligands Glu-72 and Glu-75 are not conserved among Dps-like ferritins and a further polynuclear nucleation center (NII) was identified, the mechanism of Ilari et al. (13) for iron accumulation in Listeria ferritin (12, 13) is not directly applicable to the halophilic DpsA.
Nucleation Center NII: An Iron-Oxide Nanocluster. The anomalous difference maps of FE0, FE30, and FE120 revealed a new nucleation center (NII) for the deposition of iron oxides (Fig. 3 A and B). In DpsA in its low-iron state (FEHR and FE0), a single iron atom (subsite N21) is bound with full occupancy along the threefold axis and seems to serve there as a seed for iron cluster formation (Fig. 3A). In the FEHR structure, the Fe3+ of this N21 subsite is prismatically coordinated to three symmetry-related Glu-154 (Fe-OE1, 2.35 Å) residues and three symmetry-related water molecules (Fe-W1/2/3, 2.1 Å). At the N21 subsite, strong ordering of the solvent environment is observed, because 13 water molecules form an unusual three-layered crown around the central Fe3+ ion (see Fig. 3A). In the FE30 and FE120 structures, some of these water molecules are replaced by threefold symmetry-related iron or oxygens atoms (Fig. 3B; subsites N22, N23, N24, O1, O2, and O3), which ligand like the N21 subsite to Glu-154 (Fe-OE2, 2.1 Å). Together with subsite N21, these subsites form a distorted tetrahedron with a distance of 2.9 Å to N21, 2.15 to a bridging oxygen, and distances of 3.5 Å between each other (Fig. 3 B and schematically presented in C). In the better-resolved FE120 structure, three water-derived ligands, presumably OH– or O2–, were observed as triply bridging ligands between N21 and pairs of the other subsites.
The structure of the [4Fe-3O] cluster in the NII nucleation center is clearly different from the cubane-like [4Fe-4S] clusters found in many redox-active iron–sulfur proteins with typical iron–iron distances of ≈2.8 Å. The 4-iron-3-oxo cluster resembles a site-differentiated cubane with C3v–symmetry, although additional OH–/O2– ligands could not be resolved as missing corners between subsites N22 and N24 of the FE120 structure. Such a geometry with widened iron–iron interatomic distances between subsites N22 and N24 is not found among the 588 entries of the protein database (as of December, 2003), which comprise [4Fe-4S] clusters. Likewise, a search in the Inorganic Crystal Structure Database (http://icsd.ill.fr/icsd) demonstrated that the [4Fe-3O] cluster is not observed as a substructure of other iron–oxide/hydroxide minerals such as ferrihydrite, magnetite, or lepidocrocite. Therefore, the unique arrangement of the iron atoms in the four subsites N21–N24 appears to be enforced by the unique threefold symmetric protein environment.
Recently, a synthetic model compound of a singly bridged double cubane was described [Inorganic Crystal Structure Database entry PUWTEG (29)], in which a [4Fe-4S] cluster adopts almost exactly the same geometric arrangement as the [4Fe-3O] cluster of the nucleation site (Fig. 3D). The cluster of this model compound is linked to a [2Fe-2Mo-4S] cluster via the fourth sulfido ligand, whose oxygen analog is missing in the FE120 structure. Consequently, one might infer that the observed cluster in the NII site of H. salinarum DpsA might serve as a center for the further outgrowth of larger iron-oxo clusters. Indeed, the FE30 and FE120 structures show a fifth iron-binding site (N25) with low occupancy that protrudes further into the inner compartment of the DpsA protein shell. This subsite is equidistant from subsites N22–N24 (3.0 Å) and lacks any interactions with the protein. From this study, it is not yet clear whether the observed NII cluster further increases in size on prolonged iron incubations as larger iron-oxo clusters or even as nanocrystals, which may generally be disordered inside the protein shell and hence not trackable by x-ray crystallography (30).
Biological Function. So far, DpsA from H. salinarum is the only known example of an archaeal ferritin. Because the haloarchaeon H. salinarum is an aerobic chemoorganotrophically growing organism (31), the function of DpsA, iron storage and protection again reactive oxygen species, presumably resembles its eubacterial relatives. Iron storage, the hallmark of ferritins, was recently described for DpsA (23), whereas direct binding to DNA that is exerted by Dps-like ferritins from E. coli and a few other eubacteria could not yet be demonstrated for DpsA.
The structure of DpsA suggests that its function is at least twofold affected by the hypersaline environment. First, due to the high salt concentrations of the haloarchaeal cytosol, the long-range electrostatic attraction of incoming iron atoms cannot play a prominent role as described for mesophilic ferritins. The outer protein surface of DpsA has a high density of acidic residues (1.0 Asp/Glu nm–1) that is typical for the protein surface of a halophilic protein. However, this density also resembles the inner acidic surfaces of mesophilic 24-mer ferritins and Dps orthologs (on average, 1.3 Asp/Glu nm–1), which act as biotemplates for iron complexation and biomineralization. Apparently, the accumulation of acidic groups on protein surfaces is not per se sufficient for iron binding, because there is no tendency of DpsA to complex iron atoms along its outer surface apart from the translocation site T1. The transport of iron atoms toward the inner compartment of the DpsA complex, as observed in the crystals, might hence be facilitated by the 3-fold increased number of entrance pores (12 instead of 4) and the utilization of histidine residues that allow local but sufficiently strong complexation of iron. Furthermore, the inner surface has a surface density of acidic residues (2.0 Asp/Glu nm–1) that is still high enough to trap intruded cationic iron species.
Although electron microscopy demonstrated nanocrystals of ferrihydrite inside the 24-mer ferritins with particle diameters of up to 8 nm, the molecular mechanism of biomineralization inside the ferritins is still enigmatic. Due to smaller cavity volumes, the overall storage capacity of 12-mer ferritins is smaller. In both cases, a first set of iron atoms is postulated to ligand directly to the protein shell, but further iron atoms should connect with these surface-bound irons via O2– or hydroxide bridges. In contrast, the tetra-/pentanuclear iron-oxide clusters of the NII nucleation sites are comparably small, although the number of complexed irons is for H. salinarum DpsA in the range known from biochemical studies. The utilization of such small iron oxide clusters for storage was not expected, but clusters of three iron atoms were demonstrated for a bacterial iron-trafficking protein (32). Iron reduction and release should proceed faster from such small iron-oxide clusters than from larger crystalline assemblies due to their higher surface to volume ratios. Due to its unique occurrence, one might currently speculate that the improved mobility of iron is also a demand of the halophilic environment. Because larger crystalline iron-oxide assemblies were not yet described among the 12-mer ferritins, it is feasible to assume that nature created alternative ways of iron storage by using an increased number of small iron clusters.
Supplementary Material
Acknowledgments
We are very grateful to J. Kellermann for N-terminal sequencing and S. Monaco, S. Arzt (European Synchrotron Radiation Facility), and C. Schulze-Briese (Swiss Light Source) for beamline assistance.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DpsA, DpsA protein; NCS, noncrystallographic symmetry; TS, translocation site; FOC, ferroxidase center; NI, nucleation site 1; NII, nucleation site 2; FEHR, low-iron high-resolution dataset; FEn, DpsA crystals incubated for n min.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1TJO (FEHR), 1TK6 (FE0), 1TKO (FE30), and 1TKP (FE120)].
References
- 1.Harrison, P. M. & Arosio, P. (1996) Biochim. Biophys. Acta 161–203. [DOI] [PubMed]
- 2.Chasteen, N. D. & Harrison, P.M. (1999) J. Struct. Biol. 126, 182–194. [DOI] [PubMed] [Google Scholar]
- 3.Bauerlein, E. (2003) Angew. Chem. Int. Ed. Engl. 42, 614–641. [DOI] [PubMed] [Google Scholar]
- 4.Lawson, D. M., Artymiuk, P. J., Yewdall, S. J., Smith, J. M. A., Livingstone, J. C., Treffry, A., Luzzago, A., Levi, S., Arosio, P., Cesareni, G., et al. (1991) Nature 349, 541–544. [DOI] [PubMed] [Google Scholar]
- 5.Carrondo, M. A. (2003) EMBO J. 22, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hempstead, P. D., Yewdall, S. J., Fernie, A. R., Lawson, D. M., Artymiuk, P. J., Rice, D. W., Ford, G. C. & Harrison, P. M. (1997) J. Mol. Biol. 268, 424–448. [DOI] [PubMed] [Google Scholar]
- 7.Frenkiel-Krispin, D., Levin-Zaidman, S., Shimoni, E., Wolf, S. G., Wachtel, E. J., Arad, T., Finkel, S. E., Kolter, R. & Minsky A. D. (2001) EMBO J. 20, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf, S. G., Frenkiel, D., Arad, T., Finkel, S. E., Kolter, R. & Minsky, A. (1999) Nature 400, 83–85. [DOI] [PubMed] [Google Scholar]
- 9.Tonello, F., Dundon, W. G., Satin, B., Molinari, M., Tognon, G., Grandi, G., Del Giudice G., Rappuoli, R. & Montecucco, C. (1999) Mol. Microbiol. 34, 238–246. [DOI] [PubMed] [Google Scholar]
- 10.Brentjens, R. J., Ketterer, M., Apicella, M. A. & Spinola, S. M. (1996) J. Bacteriol. 178, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao, G., Ceci, P., Ilari, A., Giangiacomo, L., Laue, T. M., Chiancone, E. & Chasteen, N. D. (2002) J. Biol. Chem. 277, 27689–27696. [DOI] [PubMed] [Google Scholar]
- 12.Bozzi, M., Mignogna, G., Stefanini, S., Barra, D., Longhi, C., Valenti, P. & Chiancone, E. (1997) J. Biol. Chem. 272, 3259–3265. [DOI] [PubMed] [Google Scholar]
- 13.Ilari, A., Stefanini, S., Chiancone, E. & Tsernoglou, D. (2000) Nat. Struct. Biol. 7, 38–43. [DOI] [PubMed] [Google Scholar]
- 14.Grant, R. A., Filman, D. J., Finkel, S. E., Kolter, R. & Hogle, J. M. (1998) Nat. Struct. Biol. 5, 294–303. [DOI] [PubMed] [Google Scholar]
- 15.Papinutto, E., Dundon, W.G., Pitulis, N., Battistutta, R., Montecucco, C. & Zanotti G. (2002) J. Biol. Chem. 277, 15093–15098. [DOI] [PubMed] [Google Scholar]
- 16.Woese, C. R., Kandler, O. & Wheelis, M. L. (1990) Proc. Natl. Acad. Sci. USA 87, 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabsch, W. J. (1988) Appl. Crystallogr. 21, 67–71. [Google Scholar]
- 18.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Vagin, A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- 20.Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg, H., Mevarech, M. & Zaccai, G. (1992) Adv. Protein Chem. 43, 1–62. [DOI] [PubMed] [Google Scholar]
- 23.Reindel, S., Anemüller, S., Sawaryn, A. & Matzanke, B. F. (2002) Biochim. Biophys. Acta 1598, 140–146. [DOI] [PubMed] [Google Scholar]
- 24.Boyd, D., Vecoli, C., Belcher, D. M., Jain, S. K. & Drysdale, J. W. (1985) J. Biol. Chem. 260, 11755–11761. [PubMed] [Google Scholar]
- 25.Levi, S., Santambrogio, P., Cozzi, A., Rovida, E., Corsi, B., Tamborini, E., Spada, S., Albertini, A. & Arosio, P. (1994) J. Mol. Biol. 238, 649–654. [DOI] [PubMed] [Google Scholar]
- 26.Ren, B., Tibbelin, G., Kajino, T., Asami, O. & Ladenstein, R. (2003) J. Mol. Biol. 329, 467–477. [DOI] [PubMed] [Google Scholar]
- 27.Treffry, A., Zhao, Z., Quail, M. A., Guest, J. R. & Harrison, P. M. (1998) FEBS Lett. 432, 213–218. [DOI] [PubMed] [Google Scholar]
- 28.Levi, S., Santambrogio, P., Corsi, B., Cozzi, A. & Arosio, P. (1996) Biochem. J. 317, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han, J. & Coucouvanis, D. (2001) J. Am. Chem. Soc. 123, 11304–11305. [DOI] [PubMed] [Google Scholar]
- 30.Ilari, A., Ceci, P., Ferrari, D., Rossi, G. L. & Chiancone, E. (2002) Biol. Chem. 277, 37619–37623. [DOI] [PubMed] [Google Scholar]
- 31.Oesterhelt, D. & Krippahl, G. (1983) Ann. Microbiol. 134B, 137–150. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, H., Alexeev, D., Hunter, D. J., Campopiano, D. J. & Sadler, P. J. (2003) Biochem. J. 376, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraulis, P. J. (1991) Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 34.Merrit, E. A. & Murphy, M.E.P. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.