Abstract
Simple (i.e., nonstratified) epithelial cells use two different routes to target their newly synthesized luminal plasma membrane proteins to the cell surface: a direct route from the Golgi complex, as in the kidney-derived MDCK cell line, or an indirect route that involves a intermediate stop at the ab-luminal (basolateral) membrane, as in hepatocytes. The mechanisms or proteins responsible for these different protein targeting strategies are not known. Here, we show that increased expression of EMK1, a mammalian ortholog of Caenorhabditis elegans Par-1, in MDCK cells promotes a switch from a direct to a transcytotic mode of apical protein delivery and other trafficking changes typical of hepatocytes. These results, together with our recent demonstration that PAR-1 promotes morphological features of hepatocytes in MDCK cells, indicate that Par-1 modulates the developmental decision to build a columnar versus a hepatic epithelial cell. To our knowledge, Par-1 is the first gene assigned to this task in epithelial morphogenesis.
Most epithelial cell types in adult and developing multicellular organisms establish a columnar shape with a vertical polarity axis, a single lumen at the cell apex, and vertically arranged microtubules (MTs) (e.g., urinary, respiratory, glandular, reproductive, and vascular systems and neuroepithelia) (1). The “apical” lumen is separated by a junctional complex from the lateral membrane, involved in cell–cell adhesions, which is contiguous with the basal membrane, involved in substrate attachment. Alternative organizations of epithelial cells are those found in stratified epithelia (e.g., skin, cornea, and esophagus) and in the liver. Although stratified epithelia are characterized by the detachment from the substrate of maturing cells, liver epithelial cells differ from columnar epithelial cells in that they establish a horizontal polarity axis with horizontal MT arrays and multiple luminal domains (the bile canaliculi, where bile is secreted), interrupting the lateral membrane (1). Columnar and hepatic epithelial cell lines are exemplified by MDCK (2) and WIF-B (3), respectively. These cell lines reproduce the typical morphological and polarity features of native columnar and hepatic epithelia (4). Columnar and hepatic epithelial cells also display fundamental differences in their biosynthetic trafficking of luminal proteins. Columnar cells use, to various degrees, a direct route from the Golgi complex to the lumen, with MDCK cells the paradigm of this vectorial transport mode (5, 6). By contrast, liver cells use predominantly an indirect biosynthetic route to target their luminal plasma membrane proteins initially to the basolateral membrane and associated endosomes, followed by transcytosis to the lumen (4, 7). In a few cases, e.g., dipeptidyl peptidase IV (DPPIV), it was shown that a protein that utilizes a direct pathway in MDCK cells (8, 9) uses a transcytotic pathway in liver cells (10). These differences in trafficking to the lumen apply to most proteins studied to date; however, some luminal proteins use a transcytotic route in MDCK cells (11) and some use a direct route in liver cells (12). Even though the direct and transcytotic trafficking phenotypes were described >20 years ago, the underlying molecular mechanisms remain unknown.
We have recently demonstrated (13) that increased expression of EMK1, the mammalian ortholog of the Caenorhabditis elegans polarity gene PAR-1, in MDCK cells that are undergoing polarization induces striking morphological changes to a hepatic epithelial phenotype. These changes include the formation of lateral lumina resembling bile canaliculi and the rearrangement of MTs from a predominantly vertical organization with apical negative ends, typical of columnar epithelia (14), to a horizontal organization, with negative ends facing the lateral lumen, typical of liver cells (15). PAR-1 is a serine/threonine kinase identified as a polarity determinant in the C. elegans embryo (16) and as a MT-regulating kinase in mammalian cells (17). Inhibition of EMK1 expression by RNA interference prevents the reorganization of MT from a centrosomal array to a vertical array characteristic of MDCK cells (13). These findings prompted us to investigate whether increased EMK1 expression in MDCK cells results in a transcytotic trafficking phenotype similar to that of the hepatocyte. To this end, we used assays to determine the trafficking of luminal proteins in EMK1-MDCK cells.
Our results indicate that indeed EMK1 promotes the establishment of a robust transcytotic route for luminal proteins in MDCK cells. In addition, experiments with nonpolarized MDCK cells showed that increased EMK1 expression promotes the establishment of a recycling route to an intracellular storage compartment for luminal proteins recently described in nonpolarized liver cells (18).
Methods
Antibody Uptake and Immunofluorescence Analysis. Maintenance of EMK1-MDCK TET-OFF cells and immunofluorescence analysis were performed as described (13). Polarized monolayers were generated by Ca-switch assays as described (13). In brief, cells were plated at confluency in Suspension MEM (SMEM) containing 5–10 μM Ca2+ (“low Ca-medium”) and subsequently switched to DMEM medium containing 2 mM Ca2+ (“normal Ca-medium”) for 24 h. Microinjection of cDNA encoding p75 neurotrophin receptor-GFP (NTRp75-GFP) were carried out as described (19, 20). The EMK1-MDCK TET-OFF cells are derived from the T23 MDCK clone (21). These cells also express pIgR; in some experiments, this protein was induced by overnight incubation in 10 mM sodium butyrate (22). Cells prevented from establishing contacts by incubation for 12 h in 5–10 μM Ca2+ in Suspension MEM (Sigma) (contact-naïve) were transduced with adenoviruses encoding low-density lipoprotein receptor (LDLR) (23) or transferrin (Tf) receptor (provided by I. Mellman, Yale University, New Haven, CT). For ligand-recycling experiments, the appropriate cells were incubated with cy3-conjugated Tf (provided by T. McGraw, Weill Medical College of Cornell University) or dimeric IgA (dIgA) [provided by K. Mostov (University of California, San Francisco) and detected with rhodamine-tagged anti-IgA] for 2 h at 37°C to achieve steady-state distribution. Antibodies against the ectodomain of the following proteins were used: gp135 (clone 3F21D8; provided by G. Ojakian, Downstate Medical Center, Brooklyn, NY); NTRp75 (clone 20.1); and LDLR (clone C7). Confocal and wide-field microscopy was performed as described (13, 23).
Quantification of gp135 fluorescence in experiments represented by Figs. 1B and 5A was performed on 40× wide-field images taken with the same exposure by using the region statistics analysis tool of metamorph (Universal Imaging, Downingtown, PA). Statistical significance was determined by two-paired Student's t test.
Fig. 1.
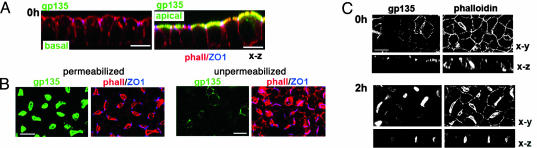
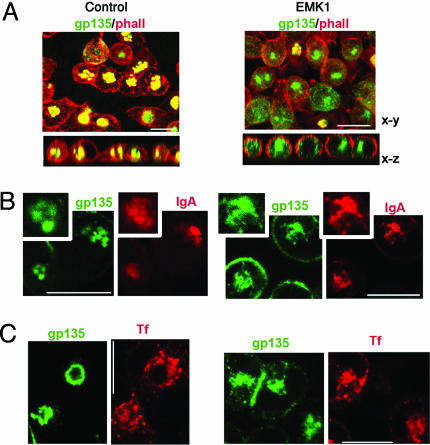
EMK1 promotes indirect targeting of gp135 to lateral lumina. Confluent, contact-naïve EMK1-overexpressing or control MDCK cells were polarized in a Ca-switch assay as described in Methods.(A) gp135 antibody-binding, control MDCK. gp135 antibodies were incubated with the basal (Left) or apical (Right) medium of living cells for 1 h on ice; gp135 (green), phalloidin (red), and ZO-1 (blue). Shown are confocal x-z views. (B) gp135 immunofluorescence, EMK1-MDCK. gp135 (green) in Tx-100 permeabilized and unpermeabilized cells; phalloidin (red) and ZO-1 (blue) labeling was after permeabilization in both cases. Shown are confocal x-y sections. Quantification of gp135 fluorescence indicates that only 11 ± 3% of gp135 labeled in permeabilized cells was accessible to the gp135 antibody in unpermeabilized cells. (C) gp135 antibody binding and uptake, EMK1-MDCK. Shown is the distribution of gp135 antibodies bound to living cells (Left) during a 1-h incubation on ice after a 0-h (Upper) and 2-h (Lower) chase at 37°C. Apical lumina are visualized by phalloidin staining (Right); shown are confocal x-y and x-z sections. (Bars, 10 μm.)
Fig. 5.
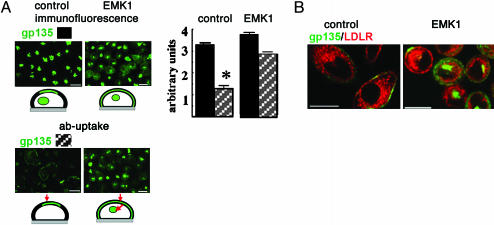
EMK1 promotes “hepatic-type” luminal targeting in nonpolarized cells. Contact-naïve control and EMK1-MDCK monolayers were generated as in Fig. 4. (A) Untransduced cells were incubated with antibodies against the ectodomains of gp135 (green) for 1 h at 37°C. Cell-associated antibodies were detected after fixation and permeabilization with secondary antibodies (ab-uptake). In parallel, samples were fixed, permeabilized, and labeled for total gp135 (immunofluorescence). Shown are 40× wide-field images. (Right) Quantification: Total cell-associated gp135 fluorescence in 30 individual control cells was determined in two separate experiments where gp135 antibodies were added to live cells (striped bars) or after fixation and permeabilization (filled bars). The difference between “total” (filled bars) and “live”(striped bars) fluorescence is statistically significant, P < 0.005. (Schemes in Upper and Lower Left) gp135 immunofluorescence (green, Upper) and gp135 antibody binding and uptake (green and red arrows, Lower) in nonpolarized cells. (B) Cells transduced with an adenovirus encoding LDLR were incubated live with antibodies against the ectodomain of gp135 (green) and LDLR (red) as described in A. Shown are confocal x-y sections. (Bars, 10 μm.) [Image gp135 in A is reprinted from ref. 42 with permission from Elsevier (Copyright 2003, Elsevier).]
Secretion Experiments. EMK1-MDCK cells or EMK1-MDCK cells stably transfected with a soluble form of rat DPPIV (solDPPIV) (9) were cultured for 2 days in the presence or absence of doxycycline before being replated at confluency for 48 h for the secretion assays. Pulse–chase with 35S-sulfate to label trans-Golgi network (TGN) cargo was as described (24). SolDPPIV was immunoprecipitated with rat DPPIV antibody (clone MRC OX-61, Serotec). For the analysis of total secreted proteins, serum-free medium was used for the chase, and proteins were precipitated with 10% trichloroacetic acid.
Results
To determine whether increased EMK1 expression promotes a liver-trafficking phenotype in MDCK cells, we investigated the transport of well characterized luminal markers in EMK1-MDCK cells. Gp135 is an endogenous luminal membrane protein that localizes apically and is delivered directly from the Golgi complex to the apical surface in control columnar MDCK cells (25, 26) (Fig. 1A). Addition of antibody against the ectodomain of gp135 to the basal side of control confluent cultures of MDCK cells on filters did not result in detectable transcytosis of antibody to the apical domain (Fig. 1 A). In EMK1-MDCK, gp135 has a radically different organization. It is present in lateral lumina rich in microvilli that label heavily with FITC-phalloidin, it is not accessible to antibodies due to the presence of functional tight junctions, but it becomes accessible to antibodies upon detergent permeabilization (Fig. 1B). In agreement with these experiments in fixed cells, addition of gp135 antibodies to monolayers of live EMK1-MDCK cells on ice labeled the exposed basolateral surfaces but did not label the lateral lumina (Fig. 1C, 0-h chase). A subsequent 2-h chase at 37°C, however, resulted in antibody transport to the lateral lumina, resulting in colocalization with phalloidin-labeled microvilli (Fig. 1C, 2-h chase). This finding is consistent with the transcytosis of gp135 by EMK1-MDCK cells.
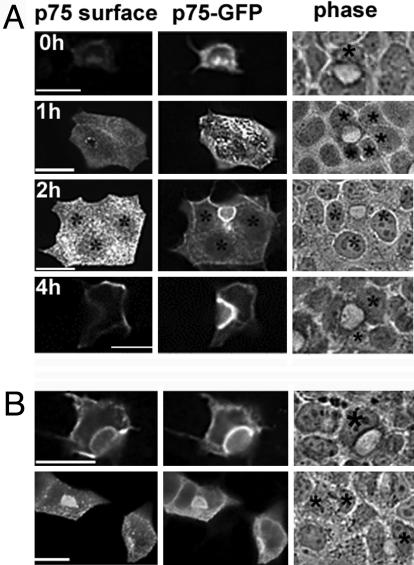
To provide additional evidence for the transcytosis of luminal markers in EMK1-MDCK cells we followed the biosynthetic pool of the recombinant luminal protein p75 neurotrophin receptor coupled to GFP (NTRp75-GFP) by using a pulse–chase live imaging protocol. This protein uses a direct route from the Golgi complex to the apical surface in MDCK cells (11, 27). We microinjected cDNA encoding NTRp75-GFP into the nucleus of EMK1-MDCK cells lying next to a lateral lumen and allowed protein expression for 1 h at 37°C (Fig. 2A). At this time point (0 h), most of NTRp75-GFP is in the endoplasmic reticulum and the Golgi complex and has not yet reached the cell surface, as detected by staining with antibodies to the extracellular domain of NTRp75 in living cells at 4°C. EMK1-MDCK monolayers treated in the same fashion were incubated for an additional 1, 2, and 4 h (chase period) in the presence of cycloheximide to prevent further protein synthesis. At the end of the chase, the monolayers were transferred to ice and probed with antibodies against the extracellular domain of NTRp75 and then fixed with paraformaldehyde. Comparison between NTRp75 surface immunofluorescence and GFP fluorescence revealed that newly synthesized NTRp75-GFP initially appeared at the plasma membrane domain in contact with the medium and was subsequently detected in increasing amounts at lateral lumina (Fig. 2 A, 1 h, 2 h, and 4 h). These kinetic data suggest that a substantial fraction of NTRp75-GFP travels by means of the basolateral cell surface to its final destination at the lateral luminal domain. Antibody-uptake experiments similar to those described above for gp135 confirmed the transcytotic route of NTRp75 in EMK1-MDCK cells (Fig. 2B).
Fig. 2.
EMK1 promotes indirect targeting of NTRp75-GFP to lateral lumina. (A) NTR-p75-GFP transport kinetics. cDNA encoding NTRp75-GFP was microinjected into the nucleus of cells facing an apical lumen and expressed for 1 h at 37°C. At this point (time, 0 h), the addition of cyclohexamide prevented further protein synthesis. After 0, 1, 2, and 4 h of chase, cells were analyzed for GFP fluorescence (Center) and by p75 surface immunofluorescence (Left). *, Microinjected cells facing apical lumina identified by phase (Right). Note that the face and fluorescence images at 0-h and 1-h chase are taken at different focal planes. (B) p75 antibody uptake. After a 1-h chase, cells were incubated with antibodies against the ectodomain of p75 for 1 h on ice, washed, and chased for 2 h at 37°C. GFP fluorescence (Center) was compared with p75 antibody distribution (Left). (Upper and Lower) Representation of different examples of the same experimental treatment. (Bars, 10 μm.)
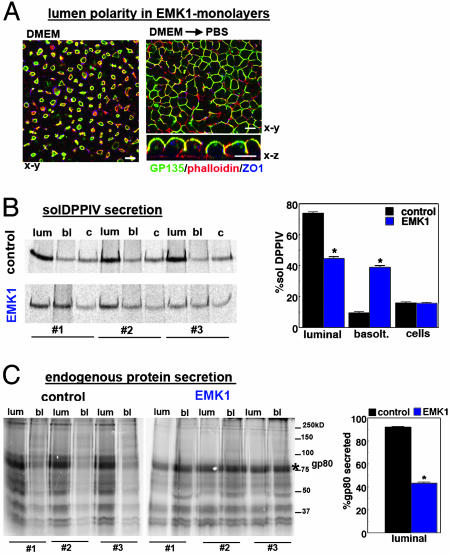
In hepatic cells, most secretory proteins are exocytosed toward the sinusoidal (basolateral) surface, which is in direct contact with the specialized liver capillary system (28). Hence, we investigated whether EMK1-MDCK cells exocytose basolaterally soluble luminal markers, which are released apically in control MDCK cells. DPPIV, a type II luminal membrane protein that is membrane-anchored by its signal sequence, becomes a secretory protein when engineered with a cleavable signal sequence (9). The recombinant protein is secreted into the luminal space in MDCK cells (9) but is targeted basolaterally in the hepatic model cell line WIF-B (10). We exposed EMK1-MDCK clones expressing recombinant solDPPIV to a pulse of 35S-sulfate during a 20°C temperature block [which prevents exit from the TGN and allows labeling of the accumulating proteins by a TGN-specific sulfotransferase (24)] and to a subsequent chase at 37°C to allow release of the protein into the medium. We carried out these biochemical experiments under conditions where >90% of EMK1-overexpressing cells organized with lateral lumen polarity (see Fig. 3A Left). In control MDCK cells, 74 ± 5% of solDPPIV was secreted into the apical medium, representing the luminal fraction (“lum”) and only 9.6 ± 3% into basolateral medium (Fig. 3B, control), with 15.8 ± 3.7% of the protein remaining cell-associated. (In these cells, maximal secretion occurred after a 2-h chase; data not shown.) By contrast, EMK1-MDCK cells secreted 40 ± 7.5% of pulse-labeled solD-PPIV into the medium; this fraction represents the basolaterally secreted pool of solDPPIV (Fig. 3B, lane bl). To gain access to the luminally secreted pool, we subsequently incubated the EMK1-MDCK monolayers in calcium-free PBS for 45 min, which disrupts the tight junctional fence, as reflected by the loss of gp135 polarity (Fig. 3A Right). This procedure led to the release of an additional 44 ± 8% of 35S-labeled solDPPIV, whereas 15 ± 2.5% remained cell-associated (Fig. 3B, lanes lum and c). Similar results were obtained when we analyzed the pattern of endogenous secretory proteins (Fig. 3C). As reported previously, MDCK cells secrete a distinct set of 35S-sulfate-labeled proteins preferentially into the apical medium (29). The most prominent luminal secretory protein of MDCK cells is gp80/clusterin (30), which has an apparent molecular mass of ≈80 kDa in unreduced gels (see asterisks in Fig. 3C). Of secreted gp80, 92 ± 3% appeared in the apical medium in control cells. In EMK1-MDCK cells, by contrast, 56 ± 5.8% of this secreted endogenous protein was recovered from the medium before the opening of tight junctions. This finding represents a substantial increase (from 8% to 44%) in basolateral secretion compared with control MDCK cells (Fig. 3C).
Fig. 3.
EMK1 promotes basolateral targeting of luminal secretory proteins. EMK1-MDCK cells constitutively expressing solDPPIV (B) or untransfected EMK1-MDCK cells (C) were cultured as described for Fig. 1. Cells were labeled for 2 h with 35S-sulfate at 20°C and subsequently chased for 2 h in the presence of 10 mM (Na)2SO4. Note that EMK1-overexpressing cells had ≈50% reduced 35S-sulfate incorporation compared with control conditions. (A) EMK1-MDCK monolayers before and after low Ca-incubation. (Left) Confocal x-y image of EMK1-expressing monolayers similar to those analyzed in B and C labeled with gp135, phalloidin and ZO-1. Note that >90% of cells had lateral lumina organization. (Right) After PBS treatment (2 × 20 min at 37°C), polarity is lost, cells round up, and luminal markers become accessible to the medium; gp135 (green), phalloidin (red), ZO-1 (blue). Presented are confocal x-y and x-z images. (Bars, 10 μm.) (B) SolDPPIV secretion. SolDPPIV was immunoprecipitated from the basal (bl) and luminal (lum) medium and from the cell lysate (c). Shown are triplicate samples (#1–#3) of a representative experiment and quantification by PhosphorImager analysis from three independent experiments. (C) Endogenous apical secretory proteins. Cells were treated as in B, and total protein in apical and basolateral medium was analyzed. Shown are triplicate samples (#1–#3) of a representative experiment and quantification of luminal gp80 (indicated by * in nonreduced gels) from three experiments. Bar graphs in B and C show standard errors. *, Significant difference between control and EMK1 with P > 2 × 10–5.
Our data indicate that, in addition to generating morphological changes typical of the liver phenotype in MDCK cells, EMK1 promotes a transcytotic trafficking route for luminal proteins typical of hepatocytes.
Nonpolarized EMK1-MDCK Cells Generate an Intracellular Luminal Compartment Found in Liver Cells. Earlier work showed that nonpolarized MDCK cells, prevented from establishing cadherin-mediated cell–cell contacts (contact-naïve) by incubation in the presence of low (micromolar) levels of Ca2+, store a large fraction of their luminal proteins in a specialized vacuolar apical compartment (VAC), highly enriched in microvilli and apical markers (31). A similar compartment enriched in apical markers is found during development of columnar epithelia and in epithelial cells undergoing malignant transformation (32). Likewise, nonpolarized hepatic cells (WIF-B cells before polarization or Fao cells, a permanently nonpolarized hepatoma cell line) accumulate apical markers in a specialized intracellular compartment, devoid of basolateral or late endocytic markers (18). The intracellular apical compartment in nonpolarized hepatocytes, however, appears to be distinct from the VACs of columnar epithelia because it is not enriched in actin and shows morphological and functional characteristics of endosomes, whereas VACs resemble vacuoles in shape and size and do not display endocytic markers (18).
Gp135 and phalloidin immunofluorescence labeling of contact-naïve EMK1-MDCK cells revealed that their gp135-enriched intracellular luminal compartment was devoid of actin (Fig. 4, EMK1), in contrast with the VACs of control MDCK cells, which displayed high actin levels (Fig. 4A, control). Other experiments detected differences between control and EMK1-MDCK cells in the accessibility of intracellular lumina to dIgA, a ligand of pIgR that undergoes basolateral to luminal transcytosis in polarized MDCK cells and hepatocytes. In nonpolarized EMK1-MDCK cells dIgA accumulated in the intracellular luminal compartment enriched in the apical marker gp135 (Fig. 4B, EMK1); a similar codistribution with the intracellular apical compartment was previously shown in nonpolarized WIF-B and Fao cells (18). In contrast, in contact-naïve control cells, dIgA accumulated in a perinuclear endocytic compartment that did not overlap with the gp135-labeled VAC structures (Fig. 4B, control). As expected, no colocalization between gp135 and Tf, a basolaterally recycling endocytic marker, was observed in either control or EMK1-MDCK cells (Fig. 4C). Our results apparently contradict previous data by Low et al. (33), who reported colocalization of dIgA with VACs in contact-naïve control MDCK cells. However, Low et al. (33) exposed cells to the ligands for 16 h, a much longer time than the 2-h exposure used in this study, suggesting that prolonged incubation allows inefficient access of dIgA to VACs or that, alternatively, the authors may not have noticed subtle differences in the localization of apical markers and dIgA.
Fig. 4.
The intracellular apical compartment of nonpolarized EMK1-MDCK cells resembles that of hepatocytic cells. EMK1-MDCK cells maintained either in the presence (control) or absence (EMK1) of doxycycline were cultured in Ca2+-free medium for 12 h to generate contact-naïve monolayers. (A) gp135 colocalization with phalloidin. Confocal x-y and x-z views of permeabilized cells colabeled with anti-gp135 (green) and phalloidin (red). (B and C) gp135 colocalization with dIgA and Tf. PIgR expression in EMK1-MDCK cells was induced with 10 mM sodium butyrate overnight. Human Tf receptor was expressed by adenovirus-mediated gene transfer. Cells were incubated for 2 h with either dIgA (B) or cy3-conjugated Tf (C) before fixation; colocalization between gp135 (green) and dIgA (red; detected with rhodamine-coupled anti-IgA) or gp135 (green) and cy3-Tf (red) is shown in individual confocal x-y sections. (Bars, 10 μm.)
In summary, our data demonstrate that the intracellular luminal compartment of EMK1-MDCK cells is different from the VACs of control cells but instead resembles an intracellular compartment enriched in apical markers that was recently described in nonpolarized hepatocytes.
Nonpolarized EMK1-MDCK Cells Traffic Luminal Proteins as Nonpolarized Hepatic Cells. The intracellular luminal compartment of nonpolarized hepatic cells very actively exchanges luminal markers with the plasma membrane; this recycling route may represent the transcytotic route to the lumen that operates in polarized liver cells (18). By contrast, the itinerary of luminal markers in nonpolarized MDCK cells has not been studied. We therefore carried out experiments to test whether nonpolarized EMK1-MDCK cells traffic luminal proteins like nonpolarized hepatocytes.
Exposure of control MDCK monolayers kept in 5–10 μM Ca2+ overnight to gp135 ectodomain antibodies for 1 h at 37°C resulted in labeling of a small pool of gp135 at the cell surface and in no labeling of gp135 in VACs [Fig. 5A, gp135, antibody uptake (ab-uptake)]. In striking contrast, EMK1-MDCK cells allowed ample access of gp135 antibody to the intracellular luminal compartment enriched in gp135 (Fig. 5A, gp135, abuptake, EMK1). Quantification of these results for 30 individual cells is shown as a bar graph in Fig. 5A. Control experiments with antibodies to the extracellular domain of basolaterally recycling LDLR (Fig. 5B, ab-uptake, LDLR) demonstrated labeling of an endocytic compartment in control and EMK1 cells. Similar to Tf-positive endosomes (see Fig. 4C), the LDLR-positive endosomes did not codistribute with the intracellular luminal compartment labeled by gp135 antibody uptake in EMK1-MDCK cells.
Together, our data demonstrate that nonpolarized MDCK and EMK1-MDCK cells establish different trafficking routes for luminal markers. Moreover, they provide further evidence that EMK1 promotes trafficking alterations for luminal markers in MDCK cells that are characteristic of the hepatic trafficking phenotype.
Discussion
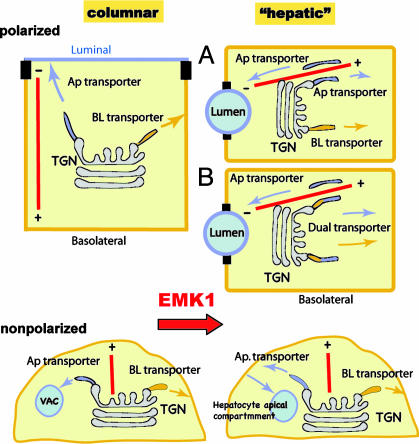
The serine/threonine kinase EMK1 is, to our knowledge, the first signaling molecule that modulates the decision to establish a transcytotic or a direct targeting mode for luminal proteins in epithelial cells. We have used two different assays, gp135 antibody-uptake and -targeting experiments (Fig. 1) and pulse–chase analysis of newly synthesized NTRp75-GFP (Fig. 2), to establish that EMK1 overexpression promotes the establishment of an indirect luminal protein-targeting route, characteristic of hepatocytes, in polarized MDCK cells. In addition, we determined by pulse–chase analysis of metabolically labeled recombinant and endogenous luminal secretory proteins that the basolaterally targeted fraction of this class of proteins increased from ≈10% in control monolayers to ≈40–50% in EMK1-MDCK cultures (Fig. 3). Finally, we established that EMK1 altered luminal protein trafficking in nonpolarized MDCK cells such that it resembled luminal protein trafficking in nonpolarized hepatic cells (Figs. 4 and 5). The change in luminal trafficking mode in EMK1-MDCK cells is accompanied by a change in lumen polarity characterized by the formation of bile canaliculi-like lateral lumina and by establishment of a horizontal MT array. All these features are hallmarks of liver epithelial cells, suggesting that EMK1 is a key regulator of the morphogenetic branching decision that leads to either columnar or hepatic polarity phenotypes (Fig. 6).
Fig. 6.
Model. EMK1 promotes a hepatic-type lumen, MT, and trafficking phenotype in epithelial cells. (Upper) Polarized MDCK and other columnar epithelial cells position their luminal domain at the apex, whereas liver cells position their luminal domains at the lateral surface (bile canaliculi). Columnar cells organize their MTs (red lines) as vertical arrays with the negative ends facing the apical domain, whereas liver cells position their MTs horizontally, with the negative ends facing the lateral lumina. Many luminal proteins in MDCK cells use a direct route, whereas the same proteins use an indirect (transcytotic) route in polarized hepatocytes or liver cell lines (e.g., WIF-B). EMK1 is expressed constitutively by all epithelial cells. Overexpression of EMK1 in MDCK cells at the time they are establishing their polarized phenotype promotes the establishment of all three landmarks of hepatic polarity: lateral lumina, horizontal MTs, and indirect luminal targeting. EMK1 regulates post-Golgi exocytosis by either promoting basolateral targeting of luminal post-Golgi carriers (option A) or by preventing protein sorting at the TGN (option B). (Lower) In contrast to the epithelial-type-specific MT arrays of polarized epithelial cells, nonpolarized MDCK and hepatic epithelial cells maintain a radial MT array. In addition, under certain conditions, they generate an intracellular compartment enriched in luminal proteins (VAC and hepatocyte apical compartment). Although superficially similar, these intracellular compartments have striking morphological and trafficking differences in columnar and hepatic epithelial cells (see text). Whereas in hepatic cells luminal proteins actively recycle between this compartment and the cell surface, in kidney cells the luminal proteins do not exchange with the plasma membrane. EMK1 promotes a hepatic-type mode of luminal protein trafficking in nonpolarized MDCK cells. Ap, apical; BL, basolateral.
Targeting of luminal proteins from the TGN to the basolateral surface in EMK1-MDCK cells could result from inclusion of luminal proteins into basolateral post-Golgi transport carriers (Fig. 6, option B) or, alternatively, from routing luminal transport carriers to the basolateral surface (Fig. 6, option A). In MDCK cells, the glycosylphosphatidylinositol (GPI) moiety is a luminal targeting signal (34). Recent evidence with two recombinant GPI-anchored GFP-fusion proteins has suggested that they undergo indirect luminal targeting in MDCK cells and reach the cell surface in basolateral post-Golgi carriers (11). Whether this is a general feature of indirect luminal protein targeting remains to be seen as the basolateral post-Golgi targeting mechanisms of GPI- or non-GPI-anchored luminal proteins in hepatocytes have not yet been elucidated.
Does EMK1 regulate transcytosis of luminal membrane markers upon their arrival at the basolateral domain? The recent targeting data for GPI-anchored proteins in MDCK cells argue that efficient basolateral-to-luminal transcytosis of luminal marker can occur at endogenous EMK1 levels, suggesting that this leg of the indirect targeting pathway is at least not dependent on EMK1 overexpression. Unlike the basolateral receptor pIgR, which undergoes luminal trancytosis upon ligand binding (21), the signals and mechanisms that govern endocytic and postendocytic events of luminal markers upon their delivery to the basolateral membrane have not been elucidated. pIgR possesses multiple targeting signals in the cytoplasmic domain that promote its clathrin-mediated endocytosis and prevent basolateral recycling and lysosomal targeting (reviewed in refs. 35 and 36). Luminal proteins, by contrast, have only very short (four amino acids in DPPIV) or no (GPI-anchored proteins) cytoplasmic domains, implying a different regulation of their transcytosis (4). In hepatocytes, pIgR meets newly synthesized luminal proteins in a subapical endosomal compartment that is devoid of basolateral proteins (37). We found a colocalization of gp135 with dIgA but not with basolaterally recycling Tf in contact-naïve EMK1-MDCK cells, suggesting that the pIgR transcytotic pathway and the indirect luminal targeting route converge in a similar apical endosomal compartment in these cells.
The only experimental manipulation known to result in redirection of luminal proteins to the basolateral surface in MDCK cells, as reported here for EMK1-overexpression, is the disruption of the MT network. In the latter case, however, no basolateral-to-luminal transcytosis was observed, consistent with a strict dependence of the transcytotic pathway on MTs (reviewed in ref. 38). This finding raises the question whether EMK1 regulates indirect protein targeting by mean of the MT network. We have previously reported that EMK1 is required for the reorganization of the radial MT array of nonpolarized epithelial cells (Fig. 6 Lower) into the horizontal arrays of polarized hepatocytes and the vertical arrays of columnar epithelia (Fig. 6 Upper). The regulation by EMK1 of luminal protein targeting, however, appears to be separable from the effect of the kinase on MT organization, because the apical targeting phenotype was apparent before MT reorganization in contact-naïve EMK1-MDCK cells. In addition to regulating MT organization (13, 39), EMK1 is known to decrease the affinity for MTs of MT-associated proteins (MAP), thereby reducing MT stability (17). Decrease in MT-MAP binding also increases the accessibility of MT motors and is associated with a stimulation of MT-based transport processes (40). Could this result in altered luminal protein targeting? Recent evidence from live imaging studies suggests that MT motors participate in the generation of transport carriers from the TGN in addition to mediating their targeting (19, 41). This sorting role of MT motors could be an explanation of why MT disruption does not simply decrease transport efficiency but also alters the polarity of luminal protein targeting in MDCK cells. Luminal transport carriers contain plus-end and minus-end directed MT motors, implying that their activity is subject to regulation (reviewed in ref. 38). Thus, by regulating the MT-based activity of motors associated with Golgi microdomains enriched in luminal markers, EMK1 could affect the generation of post-Golgi transport carriers and their MT-based targeting; both events that are likely coordinated by the same motors. As MT-plus ends face the cell cortex in nonpolarized cells and the basolateral surface in polarized cells (see MT polarity in Fig. 6), an EMK1-mediated stimulation of plus-end-directed motors at the Golgi is compatible with the alterations in post-Golgi targeting that we observed. Future work should test this hypothesis.
Acknowledgments
We thank Drs. K. Mostov, O. Weisz (University of Pittsburgh, Pittsburgh), A. Hubbard (Johns Hopkins University School of Medicine, Baltimore), T. McGraw, and I. Mellman for providing reagents. This work was supported by National Institutes of Health Grant RO1 GM 34107 (to E.R.-B.) and by American Heart Association National Program Scientist Development Award 0235130N (to A.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MT, microtubule; GPI, glycosylphosphatidylinositol; TGN, trans-Golgi network; DPPIV, dipeptidyl peptidase IV; solDPPIV, soluble form of rat DPPIV; LDLR, low-density lipoprotein receptor; VAC, vacuolar apical compartment; Tf, transferrin; dIgA, dimeric IgA.
References
- 1.Fawcett, D. (1994) in Bloom & Fawcett, A Textbook of Histology (Chapman & Hall, London), pp. 57–81.
- 2.Leighton, J., Brada, Z., Estes, L. W. & Justh, G. (1969) Science 163, 472–473. [DOI] [PubMed] [Google Scholar]
- 3.Ihrke, G., Neufeld, E. B., Meads, T., Shanks, M. R., Cassio, D., Laurent, M., Schroer, T. A., Pagano, R. E. & Hubbard, A. L. (1993) J. Cell Biol. 123, 1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuma, P. L. & Hubbard, A. L. (2003) Physiol. Rev. 83, 871–932. [DOI] [PubMed] [Google Scholar]
- 5.Powell, S. K. & Rodriguez-Boulan, E. (1992) in Epithelial Organization and Development., ed. Fleming, T. P. (Chapman & Hall, London), pp. 89–110.
- 6.Keller, P. & Simons, K. (1997) J. Cell Sci. 110, Part 24, 3001–3009. [DOI] [PubMed] [Google Scholar]
- 7.Hubbard, A. L. (1991) Semin. Cell. Biol. 2, 365–374. [PubMed] [Google Scholar]
- 8.Low, S. H., Wong, S. H., Tang, B. L., Subramaniam, V. N. & Hong, W. J. (1991) J. Biol. Chem. 266, 13391–13396. [PubMed] [Google Scholar]
- 9.Weisz, O. A., Machamer, C. E. & Hubbard, A. L. (1992) J. Biol. Chem. 267, 22282–22288. [PubMed] [Google Scholar]
- 10.Bastaki, M., Braiterman, L. T., Johns, D. C., Chen, Y. H. & Hubbard, A. L. (2002) Mol. Biol. Cell 13, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polishchuk, R., Di Pentima, A. & Lippincott-Schwartz, J. (2004) Nat. Cell Biol. 6, 297–307. [DOI] [PubMed] [Google Scholar]
- 12.Kipp, H. & Arias, I. M. (2000) J. Biol. Chem. 275, 15917–15925. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, D., Brennwald, P. J., Rodriguez-Boulan, E. & Musch, A. (2004) J. Cell Biol. 164, 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacallao, R., Antony, C., Dotti, C., Karsenti, E., Stelzer, E. H. & Simons, K. (1989) J. Cell Biol. 109, 2817–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meads, T. & Schroer, T. A. (1995) Cell Motil. Cytoskeleton 32, 273–288. [DOI] [PubMed] [Google Scholar]
- 16.Guo, S. & Kemphues, K. J. (1995) Cell 81, 611–620. [DOI] [PubMed] [Google Scholar]
- 17.Drewes, G., Ebneth, A., Preuss, U., Mandelkow, E. M. & Mandelkow, E. (1997) Cell 89, 297–308. [DOI] [PubMed] [Google Scholar]
- 18.Tuma, P. L., Nyasae, L. K. & Hubbard, A. L. (2002) Mol. Biol. Cell 13, 3400–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreitzer, G., Marmorstein, A., Okamoto, P., Vallee, R. & Rodriguez-Boulan, E. (2000) Nat. Cell Biol. 2, 125–127. [DOI] [PubMed] [Google Scholar]
- 20.Musch, A., Cohen, D., Kreitzer, G. & Rodriguez-Boulan, E. (2001) EMBO J. 20, 2171–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostov, K. E. & Deitcher, D. L. (1986) Cell 46, 613–621. [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., Kumar, R., Navarre, J., Casanova, J. E. & Goldenring, J. R. (2000) J. Biol. Chem. 275, 29138–29146. [DOI] [PubMed] [Google Scholar]
- 23.Cohen, D., Musch, A. & Rodriguez-Boulan, E. (2001) Traffic 2, 556–564. [DOI] [PubMed] [Google Scholar]
- 24.Yeaman, C., Burdick, D., Muesch, A. & Rodriguez-Boulan, E. (1998) in Cell Biology: A Laboratory Handbook, ed. Celis, J. (Academic, San Diego), 2nd Ed., Vol. 2, pp. 237–245. [Google Scholar]
- 25.Ojakian, G. K., Schwimmer, R. & Herz, R. E. (1990) Am. J. Physiol. 258, C390–C398. [DOI] [PubMed] [Google Scholar]
- 26.Grindstaff, K. K., Bacallao, R. L. & Nelson, W. J. (1998) Mol. Biol. Cell 9, 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreitzer, G., Schmoranzer, J., Low, S. H., Li, X., Gan, Y., Weimbs, T., Simon, S. M. & Rodriguez-Boulan, E. (2003) Nat. Cell Biol. 5, 126–136. [DOI] [PubMed] [Google Scholar]
- 28.Bartles, J. R., Feracci, H. M., Stieger, B. & Hubbard, A. L. (1987) J. Cell Biol. 105, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prydz, K. & Simons, K. (2001) Biochem. J. 357, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban, J., Parczyk, K., Leutz, A., Kayne, M. & Kondor-Koch, C. (1987) J. Cell Biol. 105, 2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vega-Salas, D. E., Salas, P. J. I. & Rodriguez-Boulan, E. (1988) J. Cell Biol. 107, 1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remy, L., Marvaldi, J., Rua, S., Secchi, J. & Lechene de la Porte, P. (1984) Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 46, 297–305. [DOI] [PubMed] [Google Scholar]
- 33.Low, S. H., Miura, M., Roche, P. A., Valdez, A. C., Mostov, K. E. & Weimbs, T. (2000) Mol. Biol. Cell 11, 3045–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisanti, M. P., Sargiacomo, M., Graeve, L., Saltiel, A. R. & Rodriguez-Boulan, E. (1988) Proc. Natl. Acad. Sci. USA 85, 9557–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojas, R. & Apodaca, G. (2002) Nat. Rev. Mol. Cell. Biol. 3, 944–955. [DOI] [PubMed] [Google Scholar]
- 36.Mostov, K., Su, T. & ter Beest, M. (2003) Nat. Cell Biol. 5, 287–293. [DOI] [PubMed] [Google Scholar]
- 37.Barr, V. A., Scott, L. J. & Hubbard, A. L. (1995) J. Biol. Chem. 270, 27834–27844. [DOI] [PubMed] [Google Scholar]
- 38.Musch, A. (2004) Traffic 5, 1–9. [DOI] [PubMed] [Google Scholar]
- 39.Doerflinger, H., Benton, R., Shulman, J. M. & Johnston, D. S. (2003) Development 130, 3965–3975. [DOI] [PubMed] [Google Scholar]
- 40.Bulinski, J. C., McGraw, T. E., Gruber, D., Nguyen, H. L. & Sheetz, M. P. (1997) J. Cell Sci. 110, Part 24, 3055–3064. [DOI] [PubMed] [Google Scholar]
- 41.Polishchuk, E. V., Di Pentima, A., Luini, A. & Polishchuk, R. S. (2003) Mol. Biol. Cell 14, 4470–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen, D. & Musch, A. (2003) Methods 30, 269–276. [DOI] [PubMed] [Google Scholar]