Abstract
Micrometer-size patterned lipid bilayers containing liganded lipids are used to control the location and size of receptor clusters and enable direct visualization of structural reorganization of cellular components. Subsequent to concentration of Fcε receptor I, the mast cell receptor for IgE, and colocalized tyrosine phosphorylation activity, Lyn kinase and other proteins anchored to the inner leaflet of the plasma membrane redistribute selectively with the receptor clusters in a process that depends on actin polymerization. Surprisingly, outer leaflet components characteristically associated with lipid rafts do not detectably coredistribute with these inner leaflet components. Cell activation using patterned surfaces provides unique insights into cell membrane structural organization, revealing dynamic, large-scale uncoupling of inner and outer leaflet components of lipid rafts.
The functional importance of plasma membrane heterogeneity and compartmentalization has become broadly appreciated since caveolae (1) and detergent-resistant membrane domains (2) were described and the “lipid raft hypothesis” was articulated several years ago. Rafts are considered to be membrane domains of ordered lipids, sphingomyelin, and cholesterol that selectively segregate proteins to participate in a range of cellular functions, including receptor-mediated signaling (3). Despite widespread attention, rafts have been elusive to experimental definition because of their compositional heterogeneity of lipids and proteins and their dynamic nature in the living cell (4). Crosslinking of proteins or lipid-based components that tend to associate with rafts causes their coalescence, allowing visualization in terms of coredistributing markers. Crosslinking in this manner often activates cells, and accumulating data support the view that coalesced rafts enhance selective association of protein and lipid components in the membrane, leading to targeting of the cellular response events (5–7).
A prominent example of a process involving membrane domains is antigen-mediated crosslinking of IgE receptor complexes [IgE–Fcε receptor I (FcεRI)] that initiates signal transduction in rat basophilic leukemia (RBL) mast cells (8). Biochemical studies provide strong evidence that clustered IgE–FcεRI associates with coalesced rafts containing active Lyn tyrosine kinase anchored to the inner leaflet of the plasma membrane (9). As a consequence of this association, cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM) segments of the receptor are phosphorylated, creating new binding sites for Lyn and Syk tyrosine kinase, leading to Syk activation and phosphorylation of additional substrates and downstream signaling events (10). A wide range of questions remain regarding participation of such membrane domains in cell activation. A key issue is the nature of these domains in the outer and the inner leaflets of the plasma membrane and how they may be separately organized or coupled in sequential stages of signaling (2–4).
Previous use of soluble antibodies or antigens to crosslink IgE–FcεRI into patched domains revealed selective coredistribution of characteristic raft components (11, 12). However, these studies are complicated by internalization at physiological temperatures and typically require long-term incubation at cold temperatures to visualize stable coredistribution in patches. A more direct approach is to systematically control the location and size of the receptor clusters to enable observation of responses that are spatially controlled by the cell, including stabilization of membrane domains and targeting of signaling events. For this purpose we recently developed a method for patterning supported lipid bilayers containing liganded lipids on a silicon oxide surface (13, 14). This approach allows both μm-scale definition of the activating ligands and ligand mobility afforded by a fluid bilayer. Moreover, the periodicity of the pattern enables objective, quantitative evaluation of colocalized cellular components. Previously, we demonstrated that these ligand-containing patterned surfaces specifically engage and trigger IgE–FcεRI at room temperature and 37°C, without internalization, as indicated by their redistribution over the patterned features and the consequent plasma membrane ruffling that accompanies cellular activation (14). In the present study, we investigate the redistribution of cellular components with FcεRI that is concentrated at these patterned bilayers.
Materials and Methods
Materials. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[6-[(2,4-dinitrophenyl) amino] hexanoyl] (DNP-cap-DPPE), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (NBD-PE) were purchased from Avanti Polar Lipids. 1,1′-Dioctadecyl-3, 3,3′,3′-tetramethylindocarbocyanine perchlorate [DiIC18 (3)], 3,3′-dihexadecyloxacarbocyanine perchlorate [DiOC16 (3)], Alexa 488-cholera toxin subunit B (recombinant), and latrunculin A were purchased from Molecular Probes. Mouse monoclonal anti-2,4-dinitrophenyl (DNP) IgE was purified as described (15) and fluorescently modified with Alexa 488 according to the instructions of the labeling kit (Molecular Probes). Anti-mouse IgG (Southern Biotechnology Associates) and mAb OX7 specific for glycosylphosphatidylinositol-linked protein Thy-1 (BD Pharmingen) were labeled with Cy3 (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Antiphosphotyrosine, clone 4G10 was purchased from Upstate Biotechnology. PP1 and PP2 were purchased from Biomol (Plymouth Meeting, PA) and Calbiochem, respectively. Actin-enhanced GFP (EGFP) and pleckstrin homology domain (PH)-EGFP constructs were generous gifts from A. Jeromin (Baylor College of Medicine, Houston, TX). Palmitate and myristate-EGFP (PM-EGFP) and EGFP-geranyl geranyl constructs were prepared as described (16). Cytochalasin D was purchased from Sigma.
Parylene Deposition and Patterned Bilayer Formation. On 3- or 4-inch silicon wafers (Silicon Quest, Santa Clara, CA) with thermally grown oxide layer, a pinhole-free conformal layer of parylene C was deposited by using the PDS-2010 Labcoater 2 parylene deposition system (Specialty Coating Systems, Indianapolis). A photoresist layer was then applied and patterned by using standard photolithography. The samples were subjected to oxygen-based reactive ion etching until all of the parylene on the patterns was etched away. The parylene-patterned substrate was plasma-treated by PDC-32G Plasma Cleaner (Harrick Scientific, Ossining, NY) for 30 s just before use. Sonicated small unilamellar vesicles were prepared as described (14). Lipid composition is 10 mol% 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[6-[(2,4-dinitrophenyl) amino] hexanoyl] (DNP-cap-DPPE), 1 mol% fluorescent probe [1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (NBD-PE) or 1,1′-dioctadecyl-3, 3,3′,3′-tetramethylindocarbocyanine perchlorate (DiIC18) (3)], and 89 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). Ten microliters of a small unilamellar vesicle suspension (1 mM) was added to an 8-by-8-mm SiO2 substrate at 22°C for 10 min. The sample was then thoroughly rinsed before and after parylene was mechanically peeled away in one piece.
Cell Culture and Transfection. RBL-2H3 cells and their variant B6A4C1 were maintained in monolayer cultures and harvested with trypsin-EDTA (Life Technologies, Rockville, MD) 3–5 days after passage, as described (17). For IgE localization, Alexa 488 IgE was added to cells (1 × 106 cells per ml) at 1 μg/ml for 1 h at 37°C just before the experiments, otherwise cells were sensitized overnight with unlabeled IgE at 37°C. RBL cells were transiently transfected with various EGFP constructs by using GenePORTER (Gene Therapy Systems, San Diego), or a stable cell line expressing PM-EGFP was used (16). EGFP-expressing RBL cell samples were suspended at 3 × 106 cells per ml in buffered saline solution (BSS: 135 mM NaCl/5.0 mM KCl/1.8 mM CaCl2/1.0 mM MgCl2/5.6 mM glucose/20 mM Hepes, pH 7.4) containing 1 mg/ml BSA. For Alexa 488 cholera toxin B or Cy3-anti-Thy-1 localization, each protein was added to cell aliquots at 5 μg/ml for 30 min at 22°C just before the experiment, then cells were washed twice and resuspended in BSS/BSA at 1 × 106 cells per ml. For experiments using inhibitors, cells were incubated with specific inhibitor (2 μM cytochalasin D, 10 μM PP1, 5 μM PP2, or 1 μM latrunculin A) at 37°C for 10–15 min before stimulation by lipid patterns, and the concentration of inhibitor was maintained during stimulation.
Immunofluorescence of Tyrosine Kinase Activity. After incubation with the patterns for specific periods of time at 37°C, cells were fixed with 3.7% formaldehyde in PBS for 15 min, quenched with 10 mg/ml BSA in PBS, and labeled with primary antibody 4G10 (5 μg/ml) at room temperature for 1 h. After washing by PBS/BSA (1 mg/ml), secondary antibody Cy3-labeled anti-mouse IgG (5.5 μg/ml) was incubated with samples at room temperature for 1 h. Control experiments were carried out with the addition of nonspecific mouse IgG instead of 4G10 as primary antibody. Activity was quantified at each time point by scoring fluorescent cells.
Fluorescence Microscopy. A Bio-Rad confocal head and an Olympus IX 70 inverted microscope were used for confocal microscopy. All experiments were carried out in 35-mm Petri dishes with coverglass bottom (0.16–0.19 mm; MatTek, Ashland, MA). An oil immersion ×40, 1.3 numerical aperature objective was used. For experiments carried out at 37°C, an objective heater and feedback temperature control were used. 585LP and 522/DF35 filter set were used for simultaneous or sequential dual-color image acquisition. A 580/32 filter set was used for reflectance image acquisition. A 300-μl aliquot of suspended cells was carefully added on top of the silicon substrate (8 by 8 mm) in the center of the Petri dish and allowed to settle for at least 2 min. The time adding the suspension was recorded as zero time point. Three milliliters of buffered saline solution/BSA was added into the dish before inverting the chip for microscopy observation.
Patch Identification. Although the periodicity of the pattern facilitates objective judgments, the time point for the first appearance of patchiness could be later than the actual onset of accumulation because of the limitation in visual detection. We used a set of test images to quantify the minimum concentration level needed to be recognized as a patch. Based on a real two-channel image, we divided the field of view into three nonoverlapping areas, on cell/on pattern, on cell/off pattern, and off cell (i.e., background), using average intensity as a threshold value. We then generated a series of test data by using real experimental data for background and arbitrary numbers for on cell/on pattern and on cell/off pattern area, with standard deviations that are twice the square root of the average value. From this set of images (Fig. 4, which is published as supporting information on the PNAS web site) it is apparent that if the patch is 1.2 times brighter than the rest of the cell, it is readily observable. Varying the average intensity of the whole cell to approximate different expression levels for GFP constructs does not significantly diminish this discrimination. Test images were generated and displayed in matlab (Mathworks, Natick, MA).
Results and Discussion
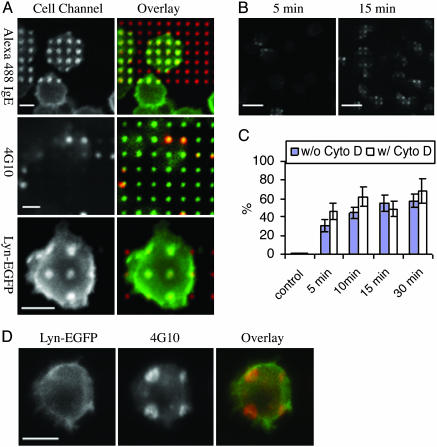
Localized Tyrosine Phosphorylation. RBL cells sensitized with anti-2,4-dinitrophenyl (DNP) IgE were added in buffer at 37°C to substrates patterned with lipid bilayers containing 10% 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[6-[(2,4-dinitrophenyl) amino] hexanoyl] (DNP-cap-DPPE). Cells were allowed to settle and could be viewed at this surface with confocal microscopy 2–3 min after their addition. Within this few minutes, fluorescently labeled IgE–FcεRI clustered over the patterned features for most of the adherent cells (Fig. 1A). The first signaling event activated, tyrosine phosphorylation of IgE–FcεRI clusters, can be monitored with a monoclonal antiphosphotyrosine antibody 4G10 (Fig. 1 A). We observed this activity to concentrate over the patterned bilayers in 30% of the cells within 5 min at 37°C, increasing to >50% of the cells after 15 min (Fig. 1 B and C and Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 1.
Confocal images of RBL cells stimulated by lipid patterns with 10 mol% 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[6-[(2,4-dinitrophenyl) amino] hexanoyl] (DNP-cap-DPPE) at 37°C. (A) Localized IgE, 4G10 indicating tyrosine phosphorylation, and Lyn-EGFP related to the lipid pattern. (B) Micrometer-scale concentration of phosphorylated tyrosine appears as early as 5 min and plateaus at 15 min. (C) Plot of the percentage of activated cells in the absence of cytochalasin D (filled columns) or the presence of cytochalasin D (open columns) as a function of incubation time. For each time points, 350–800 cells were scored. (D) Localized phosphorylated tyrosine (red) appears in the absence of localized Lyn (green), indicating signaling precedes μm-scale concentration of the kinase. (Scale bars: 10 μmin A and D and 30 μmin B.)
Localization of Lyn Kinase. Ligand-dependent concentration of Lyn kinase can be observed directly with transiently expressed Lyn-EGFP (18, 19). Consistent with previous studies carried out at low temperatures, Lyn-EGFP colocalizes with the patterned features in a ligand-dependent manner (Fig. 1 A). However, in contrast to IgE–FcεRI and tyrosine phosphorylation, Lyn-EGFP clustering is delayed and not detectable until ≈15 min after the cells settle on the pattern at 37°C. This difference is illustrated in Fig. 1D, which shows antiphosphotyrosine concentrated over the patterned features, whereas Lyn-EGFP shows little or no patchiness after 10 min of stimulation at 37°C. To assess the basis for this lag time we investigated the effects of Src family tyrosine kinase inhibitors, PP1 and PP2. We found that 10 μm PP1 reduced the percentage of cells with detectable phosphotyrosine at patterned bilayers after 30 min by 36%, whereas the percentage of cells with concentrated Lyn-EGFP was reduced by only 13%. Trends with PP2 were similar, indicating that although tyrosine phosphorylation may contribute to accumulation of Lyn-GFP with clustered IgE–FcεRI and the patterned bilayers, this accumulation is not proportional to the amount of phosphotyrosine detected there. These results suggest that the association of Lyn with clustered IgE–FcεRI is not simply caused by binding to phosphotyrosine residues, but has some additional structural basis in the membrane.
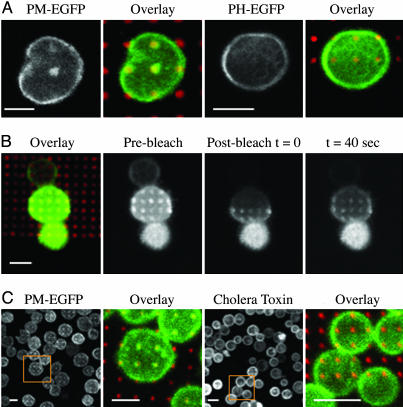
The inner leaflet construct PM-EGFP contains EGFP and the short N-terminal sequence of Lyn necessary for its dual acylation with saturated fatty acids myristate and palmitate, but not its Src homology 2 or other protein-binding domains. Previous studies showed that this probe colocalizes with crosslinked IgE–FcεRI and other raft markers after prolonged incubations at 4°C (16). When IgE-sensitized cells are incubated with the patterned bilayers, optically resolvable patches of PM-EGFP coincident with these patterns are detectable after ≈40 min at 37°C (Fig. 2A), slower than the accumulation of Lyn-EGFP. To investigate the dynamic behavior of PM-EGFP concentrated over the patterned bilayers, half-portions of individual cells were photobleached and fluorescence recovery was monitored (Fig. 2B). Fluorescence reappears over the patterned features on a time scale consistent with lateral diffusion of this lipid-anchored protein (20), confirming the dynamic nature of the coalesced membrane domain that it labels. Photobleaching measurements carried out on patched Lyn-EGFP showed recovery similar to that for PM-EGFP within the range of variations observed for individual cells. Although some differences from PM-EGFP cannot be ruled out, these results indicate that potential additional interactions between Lyn-EGFP and FcεRI or other proteins do not markedly slow down the dynamic exchange of this component in and out of patches that form in response to clustered IgE–FcεRI.
Fig. 2.
Differential colocalization of inner and outer leaflet membrane components with clustered IgE–FcεRI. (A) Confocal images indicate localized PM-EGFP and the absence of localization of PH-EGFP. (B) Fluorescence recovery of PM-EGFP after bleaching half of the cell confirms mobility of the inner leaflet lipid probe PM-EGFP and reveals the dynamic nature of the compartmentalization. (C) Inner leaflet patchiness as represented by RBL cells transfected with PM-EGFP (green) after stimulation with patterned lipid bilayer (red) is shown. Absence of outer leaflet patchiness as represented by RBL cells labeled with Alexa 488 cholera toxin (green) after stimulation with the same patterned lipid bilayer (red) is shown. (Scale bars: 10 μm.)
In contrast to Lyn-EGFP and PM-EGFP, no accumulation over the patterns was observed for PH-EGFP, which is targeted to the inner leaflet of the plasma membrane by the PH of phospholipase C-δ1 that binds selectively to phosphatidylinositol 4,5-bisphosphate (21) (Fig. 2 A). These results are consistent with previous studies in which PH-EGFP remained uniformly distributed at the plasma membrane under conditions where crosslinked IgE–FcεRI coclustered with surface patches of fluorescent cholera toxin B (22). Thus IgE–FcεRI clustering over the μm-size patterns causes selective co-concentration of some inner leaflet components, with rate differences that depend on their structural features.
Localization of Other Membrane Markers. We examined several outer leaflet lipid raft markers that were found previously to redistribute with IgE–FcεRI crosslinked with soluble ligands after several hours at 4°C. We observed that none of those tested colocalize with the surface patterned features that cluster IgE–FcεRI at higher temperatures: Alexa 488-cholera toxin B bound to ganglioside GM1 (Fig. 2C), Cy3-labeled mAb (OX7) specific for the glycosylphosphatidylinositol-linked protein Thy-1, or the lipid probes 1,1′-dioctadecyl-3, 3,3′,3′-tetramethylindocarbocyanine perchlorate (DiIC18) and 3,3'-dihexadecyloxacarbocyanine perchlorate (DiOC16) (data not shown). To confirm accessibility of the antibody probes, we introduced Cy3-anti-Thy-1 after incubating IgE-sensitized cells with the patterned bilayers, and we observed a uniform distribution of this label, independent of the pattern. These findings indicate that inner leaflet components redistribute differently from outer leaflet lipid raft components at this μm scale, and they suggest that the inner leaflet reorganization seen under these conditions may be mediated by the actin cytoskeleton or some other cytoplasmic membrane skeleton.
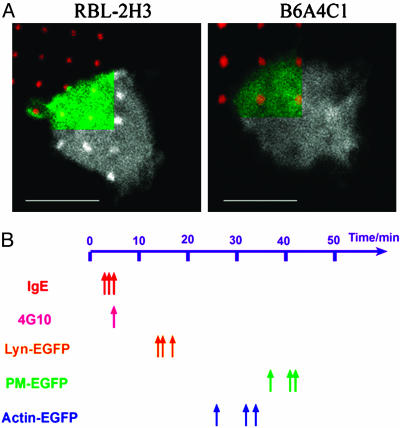
Participation of the Actin Cytoskeleton. Using actin-EGFP transfected into RBL cells we observed that actin accumulates over the patterned bilayers together with clustered IgE–FcεRI after ≈30 min of contact at 37°C (Fig. 3A), somewhat faster than for accumulation of PM-EGFP. We used cytochalasin D to prevent ongoing actin polymerization, and we observed that this reagent slows but does not prevent the accumulation of fluorescent IgE–FcεRI over the patterned bilayers. It enhances the appearance of tyrosine phosphorylation over these patterns (Fig. 1C), consistent with previous evidence that cytochalasin D enhances and sustains the tyrosine phosphorylation response to soluble antigen in these cells (12, 23). However, in the presence of cytochalasin D, no accumulation of Lyn-EGFP, PM-EGFP, or actin-EGFP was detected in any of the cells, and the same results were obtained with latrunculin A that causes actin depolymerization (data not shown). These findings indicate dependence on the actin cytoskeleton for the reorganization of inner leaflet components, and they support the evidence above that structural features in addition to tyrosine phosphorylation are needed, even for Lyn-EGFP.
Fig. 3.
Colocalization of actin with IgE–FcεRI clusters and temporal progression of colocalization events. (A) Actin colocalizes with the patches in RBL-2H3 (Left), but this does not occur in a RBL variant cell line B6A4C1 (Right) with defective activation of Rho family GTPases and downstream signaling. (Scale bar: 10 μm.) (B) Timetable for the first appearance of patchiness of different probes in RBL-2H3. Each arrow corresponds to an independent experiment except in the case of 4G10 where multiple experiments were observed at a single 5-min time point.
For further evaluation of the role of the actin cytoskeleton, we used a mutant RBL mast cell line, B6A4C1, that is defective in IgE–FcεRI-mediated activation of Rho family GTPases Cdc42/Rac and downstream signaling, including stimulated actin polymerization (24). These cells exhibit concentration of both Lyn-EGFP and PM-EGFP with clustered IgE–FcεRI and the patterned bilayers, but no accumulation of actin-EGFP (Fig. 3A). As described for the parental RBL-2H3 cells, actin polymerization that can be inhibited by cytochalasin D is required for the colocalization of these inner leaflet raft components in B6A4C1 cells. Apparently the FcεRI-stimulated signaling leading to concentration of actin over the patterned bilayers is not necessary. Interestingly, Lyn patching in B6A4C1 cells is delayed from an onset of ≈15 to ≈30 min, appearing about the same time at PM-EGFP patches, suggesting a role for stimulated actin polymerization in the accelerated accumulation of Lyn-EGFP in the WT RBL cells.
Implications for Membrane Structural Organization. Collectively, our experiments show that particular membrane and cytoskeletal components of RBL cells redistribute selectively with clustered IgE–FcεRI over the patterned bilayers in a distinctive time sequence (Fig. 3B): the onset for detectable accumulation is ≈3 min for IgE-FcεRI, ≈5 min for tyrosine phosphorylation, ≈15 min for Lyn kinase, and ≈30–40 min for polymerized actin and PM-EGFP. These colocalizations occurring at 37°C and visualized as μm-size patches can be compared to previous studies with soluble crosslinkers of IgE–FcεRI, which showed that these receptors associate in minutes at these higher temperatures with detergent-resistant, ordered regions of the membrane, and tyrosine phosphorylation leading to downstream signaling also proceeds within minutes (9). Our experiments with patterned lipid bilayers do not directly address the nature of lipid rafts as they exist in the plasma membrane of cells before IgE–FcεRI crosslinking or during formation of the small clusters of receptors (tens of nanometers scale) necessary to initiate transmembrane signaling. However, our results are consistent with the view that rafts in the basal state are very dynamic structures that can coalesce after receptors are crosslinked and associate stably with them. The differential colocalization of outer and inner leaflet raft components that we observe with the μm-size receptor patches does not preclude the possibility that both initially coalesce with small receptor clusters at early times, which may be important for the initiation of signaling (9). Our results demonstrate that, in contrast to outer leaflet components, the inner leaflet becomes stabilized such that those raft components preferentially, but dynamically, concentrate with the clustered receptors in larger-scale membrane domains.
The temporal progression of structural rearrangement that we observe with the μm-size clusters of IgE–FcεRI (Fig. 3B) is reminiscent of the assembly that occurs at the interface between T cells and antigen-presenting cells, where T cell receptor (TCR) activation precedes immunological synapse formation (25–26). Membrane reorganization that occurs during the formation of these μm-scale structures has been characterized as a slower process that appears to play a role in regulating the cellular signaling that is initiated by clustered TCR (27). Both TCR and FcεRI are members of the multi-subunit immune recognition receptor family, and the slower, μm-scale membrane reorganization that we observe in the present study with FcεRI may similarly serve a regulatory role in mast cell signaling for cytokine production and other functional responses.
What may be different for IgE–FcεRI clustered on RBL cells with patterned lipid bilayers compared to T cell interactions at the interface with target cells is the selective redistribution we observe for inner leaflet membrane components. Coupling between inner and outer leaflets, particularly within raft structures, is a question of current interest for understanding transmembrane signaling (2–4). Such coupling in cells is suggested by the observation that crosslinking of outer leaflet raft lipids causes increased activity of Src family kinases that associate with inner leaflet rafts (2) and colocalization of both components is observed after crosslinking at low temperatures (16). Furthermore, giant unilamellar vesicles prepared with gel/fluid glycerophospholipid mixtures (28) or cellular lipid mixtures (7) exhibit separated domains that are in register. The striking differences in IgE-mediated accumulation we observe in RBL cells at 37°C for inner leaflet (e.g., PM-EGFP) compared with outer leaflet raft (e.g., GM1) components suggests that coupling between inner and outer leaflets is limited, and stabilization occurs differentially on the cytoplasmic side of the asymmetric plasma membrane. For T cells it has been reported that outer leaflet raft components such as GM1 bound to fluorescent cholera toxin B concentrate in immune synapses (29–31), but, when quantified, this colocalization has been found to be small (29) or attributed to morphology artifacts (32). Furthermore, T cells contain an intracellular pool of GM1 that may undergo stimulated trafficking to the plasma membrane and contribute to the labeled pool observed near the immune synapse (30). Some studies did not detect outer leaflet lipid raft components accumulating with crosslinked TCR and other signaling components (33–35). Thus, the mechanism and significance of selective coredistribution of outer leaflet components with crosslinked TCR that may occur during formation of immune synapses remain to be determined.
In the present experiments with RBL cells, Lyn-EGFP and PM-EGFP accumulation as the result of IgE–FcεRI clustering appears to overlap the patterned bilayers uniformly, but we cannot rule out inhomogeneities on a scale that cannot be resolved optically. Another inner leaflet probe, EGFP-GG, which anchors to the inner leaflet by a geranylgeranyl moiety and a polybasic sequence similar to K-ras, also colocalizes with the patterned bilayers with similar kinetics and efficiency as PM-EGFP (data not shown), even though this lipid-anchored protein has a reduced tendency to associate with ordered lipid domains compared to PM-EGFP (16, 36). Taken together, our results indicate that neither tyrosine phosphorylation nor lipid phase separation alone is sufficient to explain the compartmentalization that we observe. Furthermore, our evidence that coredistribution of lipid-anchored inner leaflet components depends on actin polymerization indicates that the cellular cytoskeleton plays an important role in this process, consistent with other evidence for lipid raft–cytoskeletal interactions (6, 12, 17, 37, 38). However, receptor-mediated signaling downstream of Cdc42 or Rac GTPases is not required, and the molecular basis for the actin dependence remains to be determined.
In summary, patterned lipid bilayers are demonstrated to be a useful tool for visualization of membrane compartmentalization during receptor-mediated signaling. Surprisingly, inner leaflet lipid-anchored proteins are found to redistribute selectively with clustered IgE–FcεRI under conditions of cell activation. This process occurs subsequent to signal initiation and depends on actin polymerization, and its structural basis remains to be fully elucidated. The roles for lipid and cytoskeleton-based membrane reorganization can be further defined with an extended set of probes for selected cellular components. Such studies will be aided by advances in quantitative microscopic analysis and micro/nanofabrication to increasingly smaller dimensions.
Supplementary Material
Acknowledgments
We thank Reid Orth and Parijat Bhatnagar for their assistance in fabrication. This work was supported by the National Science Foundation (Nanobiotechnology Center, a Science and Technology Centers Program under Agreement ECS-9876771), National Institutes of Health Grant AI18306, and the Cornell Nanofabrication Facility.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EGFP, enhanced GFP; FcεRI, Fcε receptor I; PH-EGFP, pleckstrin homology domain-EGFP; PM-EGFP, palmitate and myristate-EGFP; RBL, rat basophilic leukemia; TCR, T cell receptor.
References
- 1.Anderson, R. G. (1998) Annu. Rev. Biochem. 67, 199–255. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. A. & London, E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111–136. [DOI] [PubMed] [Google Scholar]
- 3.Simons, K. & Ikonen, E. (1997) Nature 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 4.Edidin, M. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 257–283. [DOI] [PubMed] [Google Scholar]
- 5.Simons, K. & Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 6.Kusumi, A., Koyama-Honda, I. & Suzuki, K. (2004) Traffic 5, 213–230. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich, C., Volovyk, Z. N., Levi, M., Thompson, N. L. & Jacobson, K. (2001) Proc. Natl. Acad. Sci. USA 98, 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holowka, D. A. & Baird, B. (2001) Semin. Immunol. 13, 99–105. [DOI] [PubMed] [Google Scholar]
- 9.Field, K. A., Holowka, D. & Baird, B. (1997) J. Biol. Chem. 272, 4276–4280. [DOI] [PubMed] [Google Scholar]
- 10.Turner, H. & Kinet, J. P. (1999) Nature 402, B24–B30. [DOI] [PubMed] [Google Scholar]
- 11.Sheets, E. D., Holowka, D. A. & Baird, B. (1999) J. Cell Biol. 145, 877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holowka, D. A., Sheets, E. D. & Baird, B. (2000) J. Cell Sci. 113, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 13.Ilic, B. & Craighead, H. G. (2000) Biomed. Microdevices 2, 317–322. [Google Scholar]
- 14.Orth, R. N., Wu, M., Holowka, D. A., Craighead, H. G. & Baird, B. A. (2003) Langmuir 19, 1599–1605. [Google Scholar]
- 15.Subramanian, K., Holowka, D. A., Baird, B. & Goldstein, B. (1996) Biochemistry 17, 5518–5527. [DOI] [PubMed] [Google Scholar]
- 16.Pyenta, P. S., Holowka, D. A. & Baird, B. (2001) Biophys. J. 5, 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierini, L., Holowka, D. A. & Baird, B. (1996) J. Cell Biol. 134, 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovarova, M., Tolar, P., Arudchandran, R., Draberova, L., Rivera, J. & Draber, P. (2001) Mol. Cell. Biol. 21, 8318–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess, S. T., Sheets, E. D., Wagenknecht-Wiesner, A. & Heikal, A. A. (2003) Biophys. J. 85, 2566–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyenta, P. S., Schwille, P., Webb, W. W., Holowka, D. & Baird, B. (2003) J. Phys. Chem. A 107, 8310–8318. [Google Scholar]
- 21.Varnai, P. & Balla, T. (1998) J. Cell Biol. 143, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stauffer, T. P. & Meyer, T. (1997) J. Cell Biol. 139, 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frigeri, L. & Apgar, J. R. (1999) J. Immunol. 162, 2243–2250. [PubMed] [Google Scholar]
- 24.Field, K. A., Apgar, J. R., Hong-Geller, E., Siraganian, R. P., Baird, B. & Holowka, D. (2000) Mol. Biol. Cell 11, 3661–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wulfing, C., Sjaastad, M. D. & Davis, M. M. (1998) Proc. Natl. Acad. Sci. USA 95, 6302–6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. H., Holdorf, A. D., Dustin, M. L., Chan, A. C., Allen, P. M. & Shaw, A. S. (2002) Science 295, 1539–1542. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. H., Dinner, A. R., Tu, C., Campi, G., Raychaudhuri, S., Varma, R., Sims, T. N., Burack, W. R., Wu, H., Wang, J., et al. (2003) Science 302, 1218–1212. [DOI] [PubMed] [Google Scholar]
- 28.Korlach, J., Schwille, P., Webb, W.W. & Feigenson, G. W. (1999) Proc. Natl. Acad. Sci. USA 96, 8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burack, W. R., Lee, K. H., Holdorf, A. D., Dustin, M. L. & Shaw, A. S. (2002) J. Immunol. 169, 2837–2841. [DOI] [PubMed] [Google Scholar]
- 30.Viola, A., Schroeder, S., Sakakibara, Y. & Lanzavecchia, A. (1999) Science 283, 680–682. [DOI] [PubMed] [Google Scholar]
- 31.Bi, K., Tanaka, Y., Coudronniere, N., Sugie, K., Hong, S., van Stipdonk, M. J. & Altman, A. (2001) Nat. Immunol. 2, 556–563. [DOI] [PubMed] [Google Scholar]
- 32.Glebov, O. O. & Nichols, B. J. (2004) Nat. Cell Biol. 6, 238–243. [DOI] [PubMed] [Google Scholar]
- 33.Harder, T. & Kuhn, M. (2000) J. Cell Biol. 151, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunnell, S. C., Hong, D. I., Kardon, J. R., Yamazaki, T., McGlade, C. J., Barr, V. A. & Samelson, L. E. (2002) J. Cell Biol. 158, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harder, T. & Engelhardt, K. R. (2004) Traffic 5, 265–275. [DOI] [PubMed] [Google Scholar]
- 36.Silvius, J. R. (2002) J. Membr. Biol. 190, 83–92. [DOI] [PubMed] [Google Scholar]
- 37.Harder, T. & Simons, K. (1999) Eur. J. Immunol. 29, 556–562. [DOI] [PubMed] [Google Scholar]
- 38.Gordy, C., Mishra, S. & Rodgers, W. (2004) J. Immunol. 172, 2030–2038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.