Abstract
The evolutionary history of the largest salamander family (Plethodontidae) is characterized by extreme morphological homoplasy. Analysis of the mechanisms generating such homoplasy requires an independent molecular phylogeny. To this end, we sequenced 24 complete mitochondrial genomes (22 plethodontids and two outgroup taxa), added data for three species from GenBank, and performed partitioned and unpartitioned Bayesian, maximum likelihood, and maximum parsimony phylogenetic analyses. We explored four dataset partitioning strategies to account for evolutionary process heterogeneity among genes and codon positions, all of which yielded increased model likelihoods and decreased numbers of supported nodes in the topologies (Bayesian posterior probability >0.95) relative to the unpartitioned analysis. Our phylogenetic analyses yielded congruent trees that contrast with the traditional morphology-based taxonomy; the monophyly of three of four major groups is rejected. Reanalysis of current hypotheses in light of these evolutionary relationships suggests that (i) a larval life history stage reevolved from a direct-developing ancestor multiple times; (ii) there is no phylogenetic support for the “Out of Appalachia” hypothesis of plethodontid origins; and (iii) novel scenarios must be reconstructed for the convergent evolution of projectile tongues, reduction in toe number, and specialization for defensive tail loss. Some of these scenarios imply morphological transformation series that proceed in the opposite direction than was previously thought. In addition, they suggest surprising evolutionary lability in traits previously interpreted to be conservative.
Keywords: partitioned Bayesian analysis, larvae, tongues, tail autotomy, digits
More than two-thirds of the 522 species of salamanders are members of Plethodontidae (http://amphibiaweb.org), a clade that exhibits both extreme long-term stasis and great adaptive diversity in life history, ecology, and morphology. Morphological evolution in plethodontids is characterized by extensive homoplasy (1). Previous studies examining the causes of this homoplasy identify recurrent morphological transformations and address both their outcomes, or derived character states, and their necessary ancestral preconditions (2, 3). Two plethodontid features figure prominently in shaping morphological evolution: lunglessness, a synapomorphy for the clade, and direct development, present in three of the four major groups.
No well supported molecular phylogenetic hypothesis exists for plethodontids. As a consequence, all analyses of morphological homoplasy are based on phylogenies constructed from many of these same homoplastic characters (4). We present a molecular phylogenetic hypothesis for plethodontids based on 27 complete mitochondrial genomes, 24 of which were sequenced for this study. We explore four strategies for partitioning our dataset in a Bayesian phylogenetic framework and compare those results to maximum parsimony (MP) and maximum likelihood (ML) results. Our mitochondrial phylogeny differs markedly from the morphological phylogenetic hypotheses reflected in current taxonomy; accordingly, we reevaluate plethodontid life history evolution, origins, and historical biogeography. We examine three recurring evolutionary morphological transformations: modification effecting tongue protraction, reduction in toe number, and specialization for defensive tail loss (autotomy). We present scenarios of morphological transformation that will inform future research into the evolutionary history of plethodontid form. These scenarios suggest previously undescribed transformation series for homoplastic characters. Although some are consistent with traditional hypotheses regarding the direction of evolutionary change, others suggest surprising, previously unconsidered reversals in the direction of morphological evolution.
Methods
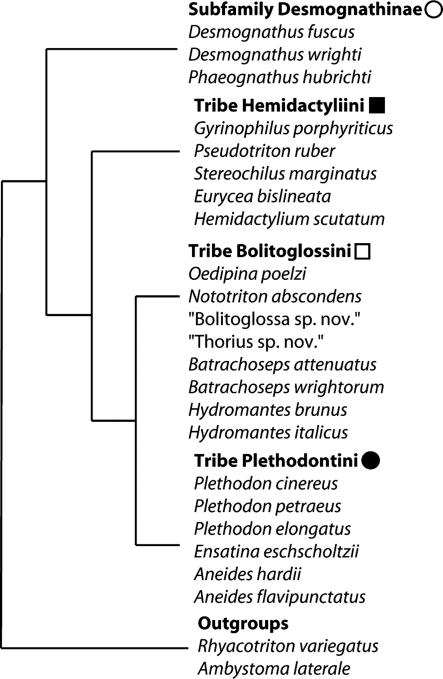
Taxon Sampling. Taxa were selected for sampling across plethodontid taxonomic diversity and to minimize long branches (5). The 24 taxa sequenced represent 17 of 26 plethodontid genera, all four major plethodontid groups, and two outgroups (Fig. 1). We included three complete salamander mitochondrial genomes from GenBank, which represent three additional families (Salamandra luschani, NC 002756; Andrias davidianus, NC 004926; and Ranodon sibiricus, NC 004021) (6–8). Locality information is listed in Table 2, which is published as supporting information on the PNAS web site.
Fig. 1.
Taxa sequenced grouped according to traditional plethodontid clades. Hemidactyliini + Bolitoglossini + Plethodontini = Plethodontinae. Plethodontinae + Desmognathinae = Plethodontidae.
DNA Sequencing. Whole genomic DNA was extracted from frozen tissue in the Museum of Vertebrate Zoology collection. Each mitochondrial genome was PCR-amplified in two to four overlapping fragments by using both universal and specific primers; primer sequences are available from the authors. PCR products were sheared to ≈1.5 kb with a HydroShear device (GeneMachines, San Carlos, CA) and enzymatically repaired to blunt their ends. Products were gel-extracted, ligated into pUC18 vector, and electroporated into competent cells (Invitrogen) by using a Gene Pulser II (Bio-Rad). Plated cells were grown overnight. Colonies were picked using a Qbot robotic colony picker (Genetix, Boston) and processed robotically through the following steps: (i) rolling circle amplification of plasmids, (ii) sequencing reactions using fluorescent dideoxynucleotide terminators, (iii) cleanup, and (iv) loading onto either ABI 3730XL (Applied Biosystems) or Megabace (Amersham Biosciences) 4000 DNA sequencing machines.
Assembly, Annotation, and Alignment. Sequences from each genome were assembled into contigs by using phrap (www.phrap.org) and confirmed visually by using consed, Ver. 13 (9). Genomes were annotated manually or by using dogma (10). Sequences of each gene were aligned by using gcg, Ver. 10.3 (Accelrys, San Diego) (gap creation and extension costs set to the defaults 8 and 2, respectively), adjusted to preserve reading frame and tRNA secondary structure, and concatenated for phylogenetic analysis. Gene-by-gene alignment was necessary because of variation in gene order; these rearrangements are not informative at this phylogenetic level and will be discussed in detail elsewhere. The control region and 1,812 other ambiguously alignable positions (including tRNA loops, beginnings and ends of many protein-coding genes, and rRNA regions with indels) were excluded, resulting in a final alignment of 14,040 bp. For one species, Hydromantes italicus, 384 bp were not sequenced and were coded as missing data for phylogenetic analysis. The alignment is available from treebase (www.treebase.org), study accession no. S1139.
Bayesian Phylogenetic Analysis. Bayesian phylogenetic analyses were implemented by using mrbayes, Ver. 3.04b (11). Flat Dirichlet distributions were used for substitution rates and base frequencies, and default flat prior distributions were used for all other parameters. Metropolis-coupled Markov chain Monte Carlo analyses were run with one cold and three heated chains (temperature set to default 0.2) for 15 million generations and sampled every 1,000 generations. Stationarity was confirmed using converge, Ver. 0.1 [courtesy of D. L. Warren (University of California, Davis), J. Wilgenbusch (Florida State University, Tallahassee), and D. L. Swofford (Florida State University, Tallahassee)] and by examining plots of negative log likelihood scores and parameter values; 8–10 million generations were discarded as burn-in. The tree was rooted with the simultaneous inclusion of five outgroups: A. davidianus, R. sibiricus, S. luschani, Ambystoma laterale, and Rhyacotriton variegatus.
In addition to the unpartitioned dataset, analyses were performed by using four data partitioning strategies designed to improve the fit of the substitution model to the data in light of heterogeneous nucleotide substitution processes (12, 13). The partitioning strategies divided the dataset into 6, 16, 29, and 42 partitions, which will be referred to as 6p, 16p, 29p, and 42p, respectively. Each strategy included a separate partition for each ribosomal RNA and the concatenated tRNAs. Strategies differed in the partitioning of protein-coding genes: 6p defined a separate partition for all first, second, and third codon positions; 16p defined a separate partition for each of the 13 protein-coding genes; 29p defined a separate partition for the first and second codon positions together for each gene and a partition for the third codon position for each gene; and 42p defined a separate partition for each codon position in each protein-coding gene. Alternate partitioning strategies were compared using Bayes factors, the ratios of the marginal likelihoods of two alternate hypotheses (13–16). The 42p analysis was conducted by using an unreleased version of mrbayes modified by J. Huelsenbeck (University of California, San Diego) to accommodate >30 data partitions.
For the unpartitioned dataset and each of the 71 total partitions used in all analyses, the best-fitting nucleotide substitution model was selected by using the hierarchical likelihood ratio test (hLRT) and Akaike Information Criterion (AIC) implemented in mrmodeltest, Ver. 1.1b (modified from modeltest, ref. 17, by J. A. A. Nylander, Uppsala University, Uppsala, Sweden). Likelihood scores were estimated on a minimum-evolution tree (ML distances) of the entire dataset. For 47 of 71 partitions, the two methods selected identical models despite AIC's penalty for increased model complexity. hLRT-selected models were used for all partitioning strategies and the unpartitioned analysis. The 42p analysis was repeated using AIC-selected models.
Comparison of Bayesian Results with Other Phylogenetic Analyses. Equally weighted MP and ML analyses were performed by using paup*4.0b10 (18). MP analyses were performed both including and excluding third codon positions. Heuristic searches were performed with 10 random addition replicates and tree bisection-reconnection branch-swapping. For the ML analysis, the general time-reversible + invariants + gamma and transversion model + invariants + gamma models of nucleotide substitution and parameter values were selected by using the hLRT and AIC, respectively, implemented in modeltest, Ver. 3.06 (17). Both were used in heuristic searches with five random addition replicates. Nonparametric bootstrap proportions (BP) for clades were assessed for MP analyses (1,000 pseudoreplicates) and ML analyses (100 pseudoreplicates). ML analysis of amino acids was performed by using quartet puzzling implemented in tree-puzzle 5.0 (www.nsc.liu.se/software/biology/puzzle5). The mtREV24 substitution model was used with Γ-distributed rates, with amino acid frequencies and α estimated from the data.
Statistical Test of the Monophyly of Traditional Taxonomic Groups. To test whether all possible topologies containing the traditional taxonomic groups Plethodontinae, Hemidactyliini, Plethodontini, or Bolitoglossini are statistically rejected by our Bayesian analyses of mitochondrial genomes, the 95% credible set of trees for each of the four partitioned analyses and the unpartitioned analysis was constructed (12, 19, 20). This is the set of all topologies contained in the cumulative 0.95 posterior probability distribution. All topologies within these sets were examined for the presence of a monophyletic Plethodontinae, Hemidactyliini, Plethodontini, or Bolitoglossini.
Results
Comparisons of Different Analyses and Partitioning Strategies. The results of all ML analyses and the MP analysis excluding third codon positions are largely consistent with the partitioned Bayesian results, although MP and amino acid ML produce phylogenies with fewer resolved nodes. MP analysis with third codon positions included yields a different topology.
All partitioned Bayesian analyses result in increased model likelihoods relative to the unpartitioned analysis, but posterior probabilities (PP) of two nodes in the topologies drops below 0.95. The unpartitioned analysis supports sister group relationships between “Bolitoglossa sp. nov.” and “Thorius sp. nov.” (PP = 0.97) and between Hydromantes and Aneides (PP = 0.99); no partitioned analysis supports these nodes with PP > 0.95. Analyses of the 6p, 16p, 29p, and 42p partitioning strategies yield near-identical topologies and nodal support, although model likelihood scores differ significantly based on Bayes factor comparisons (Table 1). The 42p analyses using models selected with AIC or hLRT yield the same topology and nodal support.
Table 1. Pairwise comparisons of partitioning strategies using twice the natural logarithm of the Bayes factor.
| 42p | 29p | 6p | 16p | Unpartitioned | |
|---|---|---|---|---|---|
| Unpartitioned | 9,708.78 | 8,090.42 | 6,632.44 | 3,034.28 | — |
| 16p | 6,674.5 | 5,056.14 | 3,598.16 | — | — |
| 6p | 3,076.34 | 1,457.98 | — | — | — |
| 29p | 1,618.36 | — | — | — | — |
| 42p | — | — | — | — | — |
Values >10 indicate very strong evidence in favor of the more likely partitioning strategy over the alternate (16). The marginal likelihoods of the unpartitioned analysis and the partitioning strategies, in order of highest to lowest likelihood, are: 42p = -184,509.32; 29p = -185,318.50; 6p = -186,047.49; 16p = -187,846.57; and unpartitioned = -189,363.71.
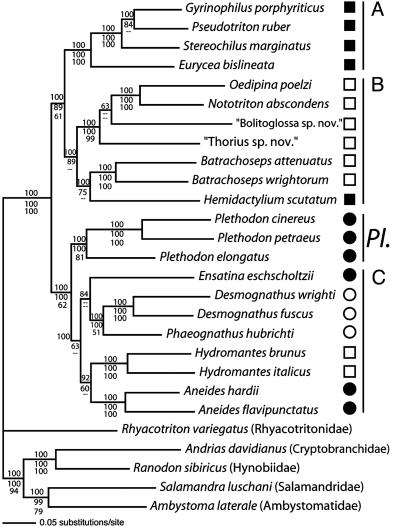
Plethodontid Phylogenetic Hypothesis. The results of the partitioned Bayesian and ML nucleotide analyses, and the MP analysis excluding third codon positions, are shown in Fig. 2. The monophyly of Plethodontidae is supported by all analyses (PP and BP = 1.0). Three previously unnamed clades are identified as follows: Clade A, Gyrinophilus porphyriticus, Pseudotriton ruber, Stereochilus marginatus, and Eurycea bislineata; Clade B, Oedipina poelzi, Nototriton abscondens, “Bolitoglossa sp. nov.,” “Thorius sp. nov.,” Batrachoseps attenuatus, Batrachoseps wrightorum, and Hemidactylium scutatum; and Clade C, Desmognathus wrighti, Desmognathus fuscus, Phaeognathus hubrichti, Hydromantes brunus, H. italicus, Ensatina eschscholtzii, Aneides hardii, and Aneides flavipunctatus. This topology is presented because it results from model-based analyses, appropriate for this dataset which displays saturation, long terminal branches, and short internodes (21, 22). In the MP analysis including third codon positions, H. scutatum is sister to all other plethodontids (BP = 0.89), Clade A is sister to Clade C (BP = 0.56), and “Bolitoglossa sp. nov.” is sister to “Thorius sp. nov.” (BP = 1.0) (not shown). H. scutatum is never sister to Batrachoseps in additional parsimony analyses and is consistently located at the base of the plethodontid tree (BP = <0.50–0.99) (not shown).
Fig. 2.
Consensus phylogram inferred using partitioned Bayesian analysis of 27 complete mitochondrial genomes (42 data partitions). Numbers above the internodes are PP. Numbers below the internodes are BP; upper numbers are MLBP using AIC-selected models, and lower numbers are MP bootstrap proportions excluding third positions. –, relationships not resolved, or resolved differently, by analysis. ML and MP analyses recover “Bolitoglossa sp. nov.” + “Thorius sp. nov.” (BP = 0.74 and 0.95, respectively). Shapes indicate traditional taxonomic groups. Open circles, Desmognathinae; open squares, Bolitoglossini; filled squares, Hemidactyliini; filled circles, Plethodontini. Open squares + closed squares + closed circles = Plethodontinae. Species lacking shapes are outgroup taxa; families are indicated in parentheses. “...” indicates unnamed species. A, B, and C designate clades referred to in the text.
Traditionally Recognized Plethodontid Groups. Desmognathinae and Plethodontinae. The traditional basal dichotomy within plethodontids separates the subfamilies Desmognathinae and Plethodontinae (Fig. 1). Monophyly of Desmognathinae (D. wrighti, D. fuscus, and P. hubrichti) is supported [PP and ML bootstrap proportion (MLBP) = 1.0]; however, the group is nested within Clade C. Monophyly of Plethodontinae is rejected.
Hemidactyliini. Monophyly of Hemidactyliini is rejected. H. scutatum is sister to Batrachoseps in Clade B. The remaining hemidactyliine lineages form Clade A.
Plethodontini. Monophyly of Plethodontini is rejected. The plethodonine lineages (Plethodon cinereus, Plethodon petraeus, Plethodon elongatus, E. eschscholtzii, A. hardii, and A. flavipunctatus) are paraphyletic with respect to the desmognathines and Hydromantes.
Bolitoglossini. Monophyly of Bolitoglossini is rejected, although each of the three clades comprising it is supported (PP and MLBP = 1.0). The tropical plethodontids (O. poelzi, N. abscondens, “Bolitoglossa sp. nov.,” and “Thorius sp. nov.”) are sister to a clade comprised of Batrachoseps and H. scutatum. Hydromantes is a member of Clade C. The basal relationships within Clade C and the position of “Bolitoglossa sp. nov.” within the tropical plethodontids are not supported with PP > 0.95.
Relationships Among Salamander Families. Our mitochondrial phylogeny supports a sister-group relationship between Salamandridae (S. luschani) and Ambystomatidae (A. laterale) and between Cryptobranchidae (A. davidianus) and Hynobiidae (R. sibiricus) (PP and MLBP ≥ 0.99), assuming the root does not fall within either of these two relationships. Similarly, a sister-group relationship between these two clades, to the exclusion of Rhyacotritonidae (R. variegatus), is supported (PP and MLBP = 1.0). Because interfamilial relationships among salamanders remain unresolved, we present our phylogeny rooted along an internal node with a basal polytomy; however, rooting with Crytobranchidae + Hynobiidae supports a sister-group relationship between Rhyacotritonidae and Plethodontidae (not shown) (23).
Statistical Test of the Monophyly of Traditional Taxonomic Groups. The number of topologies in the 95% credible sets ranged from 3 to 11 for each Bayesian analysis. In all cases, the topologies differed only in the position of “Bolitoglossa sp. nov.” within the tropical salamanders or in the relationships within Clade C, both of which are not supported with PP > 0.95 in our phylogeny. None of the topologies in the 95% credible sets contained a monophyletic Plethodontinae, Hemidactyliini, Bolitoglossini, or Plethodontini; therefore, the monophyly of these taxonomic groups is rejected statistically.
Discussion
Different Levels of Nodal Support Among Partitioning Strategies and Analyses. The higher model likelihood scores of the partitioned analyses relative to the unpartitioned analysis indicate that the data are better explained by partitioning the dataset than by applying an average model and parameter values across all genes and codon positions (12, 13). Partitioned analyses yield two fewer nodes with PP > 0.95 than the unpartitioned analysis, suggesting that the high support for these nodes in the unpartitioned analysis may result from model misspecification (24, 25). Partitioning by codon position across the 13 protein-coding genes (6p) is significantly better than partitioning by gene (16p), although it defines fewer partitions.
The most striking results of our phylogenetic analyses are: (i) the inclusion of Hydromantes in Clade C + Plethodon, (ii) the inclusion of the desmognathines in Clade C + Plethodon, and (iii) the inclusion of H. scutatum in Clade B. The first two are supported by PP and MLBP = 1.0, and the third is supported by PP = 1.0 and MLBP = 0.89.
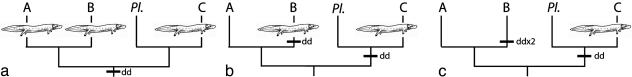
Life History Evolution. Plethodontids exhibit several life history strategies. Some species hatch as aquatic larvae and metamorphose into terrestrial or semiaquatic adults; some retain larval morphology throughout ontogeny; and some are direct developers, hatching from terrestrial eggs as miniature adults. Direct development in salamanders is unique to plethodontids, which suggests a biphasic ancestral life history that includes an aquatic larval stage and metamorphosis. The morphological plethodontid phylogeny (26) implied two appearances of direct development, at the base of Bolitoglossini + Plethodontini and in nested lineages in Desmognathinae, and no instances of the reevolution of a larval stage. Later, a desmognathine mtDNA phylogeny suggested the surprising basal position of direct development and subsequent reevolution of larvae in derived lineages, consistent with three total evolutionary transitions in plethodontid life history strategy (27). Three different transitions had been inferred by Wake (1), but all were from a biphasic life history to direct development. Our results indicate higher levels of homoplasy in life history evolution, necessitating a minimum of four transitions. Fig. 3 shows three different, equally parsimonious life history evolution scenarios. Discrimination among these scenarios will require weighting either the loss or reevolution of larvae in Clades A and B. However, all of these scenarios necessitate at least one instance of a larval stage reevolving from a direct-developing ancestor, a morphological transformation rarely reported and previously considered to be unlikely (28). A direct-developing plethodontid ancestor (Fig. 3a) necessitates reevaluation of several hypotheses, including a stream origin for the clade and the ecological causes of lung loss (29–35).
Fig. 3.
Three equally parsimonious reconstructions of life history evolution on a simplified mitochondrial genome cladogram. Larvae represent transitions from direct development to a biphasic life history that includes a larval stage. “dd” represents transitions from a biphasic life history to direct development. (a) The ancestral plethodontid evolves direct development. Larvae reevolve three times: at the base of Clade A, in H. scutatum (Clade B), and in D. fuscus (Clade C). (b) The ancestral plethodontid has a biphasic life history. Direct development evolves at the base of Clade B and at the base of Clade Plethodon (Pl.) + C. Larvae reevolve in H. scutatum (Clade B) and D. fuscus (Clade C). (c) The ancestral plethodontid has a biphasic life history. Direct development evolves twice within Clade B (at the base of the tropical plethodontids and in Batrachoseps) and at the base of Clade C + Pl. Larvae reevolve in D. fuscus (Clade C).
Morphological Homoplasy. Tongue evolution. Because plethodontids are lungless, the tongue musculoskeletal elements used for ventilation in lunged salamanders are freed from this functional constraint. Consequently, these elements are specialized for tongue protraction (1). Direct-developing lineages are freed from an additional constraint; they no longer require the tongue skeleton for larval suction feeding, a function that may conflict with specialization for extreme tongue protraction (36). Three categories of tongue function are recognized: protrusible, attached projectile, and free. All recent phylogenetic hypotheses for plethodontids require extensive convergence such that none of these tongue types defines a monophyletic group, and different morphological modifications effecting free tongue protraction have evolved in different lineages. Convergence in tongue function represents repeated morphological exploration within different lineages made possible by loss of an ancestral functional constraint (2, 37).
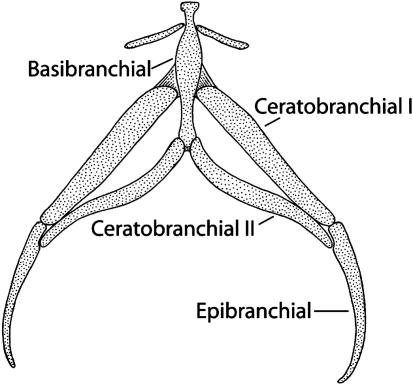
Protrusible tongues are the least modified from the ancestral state. To effect protraction, force is applied by the paired subarcualis rectus I muscles to the epibranchials and transmitted via ceratobranchial I to the basibranchial element in the tongue pad; all of these elements are cartilaginous and comprise the hyobranchial apparatus, or tongue skeleton (Fig. 4). The tongue skeleton folds slightly during protrusion, and the tongue can extend ≈7% snout-vent length beyond the mouth (38, 39). The anterior tip of the tongue is tightly attached to the front of the lower jaw by paired short genioglossus muscles.
Fig. 4.
The hyobranchial apparatus, or tongue skeleton, of a plethodontid salamander with a protrusible tongue (Desmognathus quadramaculatus). The basibranchial element is in the tip of the tongue.
Attached projectile tongues are specialized for directionality and distance, extending ≈15% snout-vent length beyond the mouth (38). The anterior tip is attached by elongated genioglossus muscles whose insertions have moved posteriorly along the lower jaw. Epibranchial length is increased relative to the other elements, and ceratobranchial lengths are decreased. The tongue skeleton folds significantly during projection. Various changes to the associated musculoskeletal components are present.
Free tongues are specialized for extreme long-distance projection, and the muscular connection between the tongue tip and lower jaw has been lost. Morphological modifications are similar to those seen in attached projectile tongues, but tongues are projected 30–80% snout-vent length (40). In two independent cases, projection is truly ballistic; the tongue skeleton leaves the mouth altogether, and the tongue reaches the prey under its own momentum (41, 42).
Free-tongued lineages are characterized by one of two force-transmitting mechanisms to effect tongue projection. The first [Option 1 (37)] is mechanically similar to protrusible tongue protraction. The second (Option 2) differs in that ceratobranchial II, rather than ceratobranchial I, is the force-transmitting pathway from the epibranchials to the basibranchial; this difference significantly alters projection biomechanics. Commitment by a lineage to Option 1 or Option 2 may be irreversible, because an evolutionary transition between the two requires an intermediate tongue skeletal configuration less optimal for tongue projection than either option (37).
The simplest scenario for convergent tongue evolution consistent with our phylogeny requires five total transitions among functional types, similar to the number implied by the morphological phylogeny. Based on comparisons with outgroups, the ancestral state for plethodontids is a protrusible tongue. Free tongues appear three times: at the base of Clade A, at the base of the tropical plethodontids, and in Hydromantes. Attached projectile tongues appear twice: at the base of Batrachoseps + H. scutatum and in Ensatina. The morphological transformation series in this scenario are similar to those suggested by the morphological phylogeny.
Such extensive functional mode homoplasy is expected of lineages exploring a finite set of morphological possibilities (2, 3); the mitochondrial phylogeny is consistent with this interpretation of convergent tongue evolution. However, our finding that Batrachoseps, Hydromantes, and the tropical plethodontids do not form a monophyletic Bolitoglossini demonstrates extensive homoplasy of characters previously thought to have evolved more conservatively (26). Despite their different tongue functional categories, these three groups share many morphological modifications for tongue protraction including: the loss of two different skeletal elements and one muscle; the fusion of several skeletal elements; changes in the proportions of the tongue skeletal elements; Option 2 force transmission and the associated changes in relative size and orientation of additional tongue skeletal elements; loss of a cell type in the motor column of the neck and trunk; and possession of a complex musculoskeletal aiming cylinder to control tongue direction (26). Based on the mitochondrial phylogeny, all of these characters either (i) evolved three times in parallel or (ii) evolved twice in parallel and were regained or reversed in H. scutatum. Again, choosing between these scenarios will require weighting certain morphological transformations; however, in either case, our results necessitate multiple gains and losses of morphological characters not previously thought to be homoplastic.
Toe loss. A reduction from five to four toes characterizes four plethodontid taxa: H. scutatum, Eurycea quadridigitata, Eurycea chamberlaini, and Batrachoseps. Traditional phylogenetic hypotheses imply at least three independent losses, resulting from a developmental constraint imposed by decreasing limb bud size and large cell size (3). The sister-group relationship between Batrachoseps and H. scutatum in the mitochondrial phylogeny decreases the minimum number of toe reductions from three to two.
Tail autotomy. Plethodontids vary in their degree of specialization for defensive tail loss. Some lineages have highly specialized tail morphologies in which shortened vertebrae and musculature and weakened connective tissue form a constriction at the base of the tail and localize intervertebral breakage (constricted-based tails). In addition, the skin breaks one vertebra behind the muscle, forming a sleeve over the wound that facilitates healing, blastema formation, and subsequent tail regeneration (wound healing) (43). Other lineages possess wound healing but do not localize breakage to the base of their uniformly slender tails (slender-based tails). Finally, some lineages have no specializations for loss; their tails simply break mechanically. These thick, often laterally compressed tails are used for aquatic propulsion (thick-based tails). Most lineages comprising the two traditional basal clades, Desmognathinae and Hemidactyliini, are aquatic or semiaquatic with thick-based tails. Close outgroups also lack autotomy, which implies an unspecialized plethodontid ancestral tail morphology. Wound healing was inferred to have arisen in the common ancestor of Plethodontini and Bolitoglossini, after evolution of direct development, and was interpreted as a necessary precondition for convergent and parallel evolution of constricted- and slender-based tails (43). Tail-loss specialization coincident with a shift from aquatic to terrestrial habitat was also noted, to a lesser extent, within desmognathines and hemidactyliines.
We outline a substantially different scenario for the evolution of tail autotomy. We infer the evolution of wound healing in the ancestral plethodontid, because it is present in multiple lineages spanning the basal mitochondrial dichotomy. Across the two main clades, Clade A + B (AB) and Clade Plethodon + C (PC), convergent evolution of all three tail types is present: constricted-based tails appear in the tropical plethodontids and H. scutatum (AB) and in E. eschscholtzii (PC); slender-based tails appear in Batrachoseps and secondarily in several tropical lineages (AB) and in Plethodon and nonarboreal Aneides (PC); and thick-based tails appear in Clade A (AB) and in the desmognathines (PC). In this scenario, the presence of thick-based tails indicates a loss of wound-healing specialization and a new reliance on the tail for aquatic propulsion. Within these largely aquatic groups with thick-based tails, however, there are terrestrial lineages; this terrestriality is associated with the reevolution of a novel, less-efficient tail autotomy specialization that differs from the ancestral specialization. A similar loss of wound healing specialization has occurred in some nonaquatic species as well. Within PC, Hydromantes and some Aneides evolved reliance on their tails for terrestrial and arboreal locomotion, respectively, and tail loss is generally infrequent in these clades. In this scenario for the evolution of tail autotomy, morphological change from one tail type to another proceeds in previously unconsidered directions. Lineages with specializations for autotomy have, in some instances, given rise to lineages with a generalized tail morphology more typical of basal forms.
Plethodontid Origins and Historical Biogeography. Wilder and Dunn (29) first proposed an Appalachian center of origin for plethodontids in 1920. That view is still widely accepted and is supported by three factors: (i) high present-day plethodontid adaptive diversity and species richness in the Appalachians (44); (ii) current Appalachian distributions of the basal groups Desmognathinae and Hemidactyliini, whereas nested clades have both eastern and western North American distributions (Plethodontini), western North American and European distributions (Hydromantes and Batrachoseps, clade Bolitoglossini), or Central and South American distributions (the tropical plethodontids, clade Bolitoglossini); and (iii) existence of Appalachian mountain streams, the inferred ancestral habitat, since the postulated origin of the clade in the Cretaceous (refs. 1, 29, and 31, but see refs. 34 and 35).
The “Out of Appalachia” hypothesis does not receive phylogenetic support from our analysis, although we cannot refute it. The basal split in the mitochondrial phylogeny is into two clades, each of which contains both eastern and western lineages, and we report many associations between eastern and western groups. Our results, coupled with the current plethodontid distribution and that of Rhyacotriton, imply a North American origin of the clade with expansion into Europe and Central and South America; however, the current species distributions do not retain any signal that reflects the early biogeographic history of this ancient group within geologically and climatically dynamic North America. Fossil evidence dates back only to the Miocene, when Aneides, Plethodon, and Batrachoseps are found in Western North America, and Hydromantes is found in Slovakia (45–47).
Conclusion
We present the phylogenetic relationships among 27 plethodontid and outgroup mitochondrial genomes. Our results are surprising; we do not recover three of the four major groups currently recognized. Analyses of complete mitochondrial genomes have been used to resolve the relationships among lineages in other ancient groups, yielding both expected and unexpected phylogenies [e.g., ratites (48), insects (49), teleosts (50), and hexapods (51)]. Both methodological limitations and biological processes can cause gene trees to differ from species trees (52–54). We advocate the use of multiple independent genetic markers for taxonomic revision and look forward to forthcoming nuclear data; if these mitochondrial genome results are corroborated, retention of some taxonomic group names will be possible, although the lineages contained within the new clades will change. Consideration of multiple markers will lead to a more complete understanding of the history of Plethodontidae. In addition, it will teach us about the different processes that have shaped the evolutionary histories of the genetic markers themselves. Finally, it will instruct future researchers on the strengths and weaknesses of different datasets and analytical approaches in reconstructing relationships among ancient lineages.
Supplementary Material
Acknowledgments
We thank M. Fourcade, J. Keuhl, and D. Engle for technical assistance; D. Hillis, T. Reeder, A. Leaché, M. Mahoney, E. Rosenblum, V. Vredenburg, and M. Wake for comments on the manuscript; M. Brandley for discussion; J. Huelsenbeck for modifications to mrbayes; and A. Seago (University of California, Berkeley) for the larva in Fig. 3. Part of this work was performed by the Lawrence Berkeley National Laboratory, University of California, under the auspices of the U.S. Department of Energy, Office of Biological and Environmental Research, contract no. DE-AC03-76SF00098. R.L.M. was supported by a National Science Foundation predoctoral fellowship and a National Institutes of Health training grant. Additional funds came from a National Science Foundation doctoral dissertation improvement grant 0105824 (to R.L.M. and D.B.W.) and the AmphibiaTree Project (National Science Foundation Grant EF-0334939).
Abbreviations: AIC, Akaike Information Criterion; BP, nonparametric bootstrap proportion; PP, Bayesian posterior probability; ML, maximum likelihood; MP, maximum parsimony; MLBP, ML bootstrap proportion; PC, Clade Plethodon + C; np, n partition.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY728212–AY728235).
References
- 1.Wake, D. B. (1966) Mem. Soc. Calif. Acad. Sci. 4, 1–111. [Google Scholar]
- 2.Wake, D. B. & Larson, A. (1987) Science 238, 42–48. [DOI] [PubMed] [Google Scholar]
- 3.Wake, D. B. (1991) Am. Nat. 138, 543–567. [Google Scholar]
- 4.Wake, D. B. (1993) Herpetologica 49, 229–237. [Google Scholar]
- 5.Hillis, D. M. (1998) Syst. Biol. 47, 3–8. [DOI] [PubMed] [Google Scholar]
- 6.Zhang, P., Chen, Y.-Q., Zhou, H., Wang, X.-L. & Qu, L.-H. (2003) Mol. Phylogenet. Evol. 28, 620–626. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, P., Chen, Y.-Q., Liu, Y.-F., Zhou, H. & Qu, L.-H. (2003) Gene 311, 93–98. [DOI] [PubMed] [Google Scholar]
- 8.Zardoya, R., Malaga-Trillo, E., Veith, M. & Meyer, A. (2003) Gene 317, 17–27. [DOI] [PubMed] [Google Scholar]
- 9.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 10.Wyman, S. K., Jansen, R. K. & Boore, J. L. (2004) Bioinformatics, in press.. [DOI] [PubMed]
- 11.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 12.Nylander, J. A. A., Ronquist, F., Huelsenbeck, J. P. & Nieves-Aldrey, J. L. (2004) Syst. Biol. 53, 47–67. [DOI] [PubMed] [Google Scholar]
- 13.Brandley, M. B., Schmitz, A. & Reeder, T. W. (2005) Syst. Biol., in press. [DOI] [PubMed]
- 14.Lavine, M. & Schervish, M. J. (1999) Am. Stat. 53, 119–122. [Google Scholar]
- 15.Huelsenbeck, J. P. & Imennov, N. S. (2002) Syst. Biol. 51, 155–165. [DOI] [PubMed] [Google Scholar]
- 16.Kass, R. E. & Raftery, A. E. (1995) J. Am. Stat. Assoc. 90, 773–795. [Google Scholar]
- 17.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 18.Swofford, D. L. (1998) PAUP*4.0B10 (Sinauer, Sunderland, MA).
- 19.Reeder, T. W. (2003) Mol. Phylogenet. Evol. 27, 384–397. [DOI] [PubMed] [Google Scholar]
- 20.Buckley, T. R. (2002) Syst. Biol. 51, 509–523. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan, J. & Swofford, D. L. (1997) J. Mamm. Evol. 4, 77–86. [Google Scholar]
- 22.Felsenstein, J. (2004) Inferring Phylogenies (Sinauer, Sunderland, MA).
- 23.Larson, A. & Dimmick, W. W. (1993) Herp. Monogr. 7, 77–93. [Google Scholar]
- 24.Suzuki, Y., Glazko, G. V. & Nei, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16138–16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erixon, P., Svennblad, B., Britton, T. & Oxelman, B. (2003) Syst. Biol. 52, 665–673. [DOI] [PubMed] [Google Scholar]
- 26.Lombard, R. E. & Wake, D. B. (1986) Syst. Zool. 35, 532–551. [Google Scholar]
- 27.Titus, T. A. & Larson, A. (1996) Syst. Biol. 45, 451–472. [Google Scholar]
- 28.Duellman, W. E. & Hillis, D. M. (1987) Herpetologica 43, 141–173. [Google Scholar]
- 29.Wilder, I. W. & Dunn, E. R. (1920) Copeia 1920, 63–68. [Google Scholar]
- 30.Noble, G. K. (1925) J. Morphol. Physiol. 40, 341–416. [Google Scholar]
- 31.Beachy, C. K. & Bruce, R. C. (1992) Am. Nat. 139, 839–847. [Google Scholar]
- 32.Dunn, E. R. (1928) Am. Nat. 62, 236–248. [Google Scholar]
- 33.Dunn, E. R. (1926) Salamanders of the Family Plethodontidae (Smith College, Northampton, MA).
- 34.Ruben, J. A., Reagan, N. L., Verrell, P. A. & Boucot, A. J. (1993) Am. Nat. 142, 1038–1051. [Google Scholar]
- 35.Ruben, J. A. & Boucot, A. J. (1989) Am. Nat. 134, 161–169. [Google Scholar]
- 36.Deban, S. M. & Marks, S. B. (2002) Zool. J. Linn. Soc. 134, 375–400. [Google Scholar]
- 37.Wake, D. B. (1982) in Environmental Adaptation and Evolution, eds. Mossakowski, D. & Roth, G. (Fischer, Stuttgart), pp. 51–66.
- 38.Larsen, J. H. J., Beneski, J. T. J. & Wake, D. B. (1989) J. Exp. Zool. 252, 25–33. [Google Scholar]
- 39.Lombard, R. E. & Wake, D. B. (1977) J. Morphol. 153, 39–80. [DOI] [PubMed] [Google Scholar]
- 40.Wake, D. B. & Deban, S. M. (2000) in Feeding: Form, Function, Phylogeny, ed. Schwenk, K. (Academic, San Diego), pp. 95–116.
- 41.Deban, S. M., Wake, D. B. & Roth, G. (1997) Nature 389, 27–28.9288962 [Google Scholar]
- 42.Deban, S. M., O'Reilly, J. C. & Nishikawa, K. C. (2001) Am. Zool. 41, 1280–1298. [Google Scholar]
- 43.Wake, D. B. & Dresner, I. G. (1967) J. Morphol. 122, 265–306. [DOI] [PubMed] [Google Scholar]
- 44.Wake, D. B. (1987) Ann. Mo. Bot. Gard. 74, 242–264. [Google Scholar]
- 45.Tihen, J. A. & Wake, D. B. (1981) J. Herp. 15, 35–40. [Google Scholar]
- 46.Clark, J. M. (1985) J. Herp. 19, 41–47. [Google Scholar]
- 47.Venczel, M. & Sanchiz, B. (2004) Amphibia–Reptilia, in press.
- 48.Cooper, A., Lalueza-Fox, C., Anderson, S., Rambaut, A., Austin, J. & Ward, R. (2001) Nature 409, 704–707. [DOI] [PubMed] [Google Scholar]
- 49.Stewart, J. B. & Beckenbach, A. T. (2003) Mol. Phylogenet. Evol. 26, 513–526. [DOI] [PubMed] [Google Scholar]
- 50.Miya, M., Takeshima, H., Endo, H., Ishiguro, N. B., Inoue, J. G., Mukai, T., Satoh, T. P., Yamaguchi, M., Kawaguchi, A., Mabuchi, K., et al. (2003) Mol. Phylogenet. Evol. 26, 121–138. [DOI] [PubMed] [Google Scholar]
- 51.Nardi, F., Spinsanti, G., Boore, J. L., Carapelli, A., Dallai, R. & Frati, F. (2003) Science 299, 1887–1889. [DOI] [PubMed] [Google Scholar]
- 52.Pamilo, P. & Nei, M. (1988) Mol. Biol. Evol. 5, 568–583. [DOI] [PubMed] [Google Scholar]
- 53.Maddison, W. P. (1997) Syst. Biol. 46, 523–536. [DOI] [PubMed] [Google Scholar]
- 54.Ballard, J. W. O. & Whitlock, M. C. (2004) Mol. Ecol. 13, 729–744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.