Abstract
The facultative intracellular bacterium Listeria monocytogenes is being developed as a cancer vaccine platform because of its ability to induce potent innate and adaptive immunity. For successful clinical application, it is essential to develop a Listeria platform strain that is safe yet retains the potency of vaccines based on wild-type bacteria. Here, we report the development of a recombinant live-attenuated vaccine platform strain that retains the potency of the fully virulent pathogen, combined with a >1,000-fold reduction in toxicity, as compared with wild-type Listeria. By selectively deleting two virulence factors, ActA (ΔactA) and Internalin B (ΔinlB), the immunopotency of Listeria was maintained and its toxicity was diminished in vivo, largely by blocking the direct internalin B-mediated infection of nonphagocytic cells, such as hepatocytes, and the indirect ActA-mediated infection by cell-to-cell spread from adjacent phagocytic cells. In contrast, infection of phagocytic cells was not affected, leaving intact the ability of Listeria to stimulate innate immunity and to induce antigenspecific cellular responses. Listeria ΔactA/ΔinlB-based vaccines were rapidly cleared from mice after immunization and induced potent and durable effector and memory T-cell responses with no measurable liver toxicity. Therapeutic vaccination of BALB/c mice bearing murine CT26 colon tumor lung metastases or palpable s.c. tumors (>100 mm3) with recombinant Listeria ΔactA/ΔinlB expressing an endogenous tumor antigen resulted in breaking of self-tolerance and long-term survival. We propose that recombinant Listeria ΔactA/ΔinlB expressing human tumor-associated antigens represents an attractive therapeutic strategy for further development and testing in human clinical trials.
Cancer immunotherapy represents a promising treatment strategy that has produced some tantalizing clinical responses for a variety of malignant diseases. Although promising, there continues to be a strong need for a practical immunization strategy that can be routinely adopted to specific malignancies and that consistently yields durable and robust therapeutic antitumor responses.
Progress in molecular and cellular immunology, combined with increasing understanding of pathogen physiology and host–pathogen interaction has facilitated the design and use of attenuated bacteria as conventional vaccine vectors. However, the practical utility of live attenuated vaccines relies on achieving a proper balance between the virulence/toxicity and immunogenicity of the vaccine. The potency of a pathogen to elicit adaptive immunity is related in part to its ability to stimulate significant innate immunity through recognition of microbial pathogen-associated molecular patterns by Toll-like receptors. Microbial encounter with professional antigen-presenting cells (APC), such as dendritic cells, results in activation and maturation (1) as well as secretion of high levels of T helper-1-type cytokines (2). This interaction initiates adaptive immune responses and therefore links innate and adaptive immunity.
Recombinant microbial-based vectors expressing model tumor antigens, including Listeria monocytogenes, have been shown to elicit robust innate and antigen-specific cellular responses in a number of models, including tumor-bearing animals (3, 4). Despite the potential safety concerns for using wild-type Listeria as a vaccine vector, there are a number of desirable features of the natural biology of this bacterium that make it an attractive platform for continued development toward clinical evaluation. The central rationale is that the intracellular lifecycle of Listeria enables effective stimulation of CD4+/CD8+ T cell immunity. There are also numerous practical features of Listeria-based vaccines, including its anticipated ease of production in defined media, combined with relatively simple vaccine construction by using well developed techniques for bacterial engineering (5). Listeria is not hindered by constraints on the size of the heterologous sequence, a limitation common to viral-based vector systems. However, Listeria is a food-borne pathogen, and unique approaches to retain the immunogenicity of this platform while attenuating its pathogenicity are essential for eventual clinical efficacy trials in humans.
Here, we describe our work constructing a live-attenuated vaccine strain, Listeria ΔactA/ΔinlB (InlB, Internalin B). We hypothesized that by combining specific attenuating genetic mutations affecting Listeria cell-to-cell spread and tropism for nonphagocytic cells, it should be possible to segregate vaccine immunogenicity because of uptake of bacteria by phagocytic cells, including professional APC, from overt Listeria toxicity due in part to infection of nonphagocytic cells, such as hepatocytes. By deletion of two distinct virulence determinants, we believe that the ΔactA/ΔinlB-based vaccine strain appropriately addresses toxicity concerns related to Listeria while preserving its immunogenicity and is thus an attractive vaccine platform strain for continued development.
Materials and Methods
Bacterial Host Strains and Recombinant Vectors. All Listeria strains used in this study were derived from the L. monocytogenes wild-type strain 10403S (6) (Supporting Methods and Table 1, which are published as supporting information on the PNAS web site). Listeria strains with in-frame deletions of the indicated genes were generated by splicing by overlapping extension PCR and allelic exchange with established methods (7). The pPL2 genetic integration vector (5) was used to derive ovalbumin (OVA) and AH1-A5 recombinant Listeria strains containing a single copy of the antigen expression cassette within the Listeria genome adjacent to the gene encoding tRNA arginine. Expression of secreted heterologous antigens was driven from the hly promoter as a fusion protein with the N-terminal region of listeriolysin O (LLO) as described in ref. 8. The AH1-A5 epitope was inserted in-frame within OVA by using a unique AvaII site.
In Vitro Infectivity. Infectivity of human cell lines and primary cells, such as monocytes and hepatocytes, was assessed in vitro as described in ref. 9. Human primary hepatocytes were obtained from In Vitro Technologies (Baltimore). Human primary monocytes were purified from a buffy-coat from healthy volunteers (Sacramento Blood Bank Center, Sacramento, CA) by using a Ficoll–Hypaque gradient and CD14 microbeads (Miltenyi Biotec, Auburn, CA). HepG2 cells (human hepatocellular carcinoma cell line, American Type Culture Collection) and primary human hepatocytes were infected at a multiplicity of infection of 10. THP-1 cells (human monocyte cell line, American Type Culture Collection) and primary human monoctyes were infected at an multiplicity of infection of 1 and 100, respectively. After the incubation with gentamycin (50 μg/ml), cells were lysed with sterile water, and serial dilutions were plated onto brain heart infusion agar to determine the level of infectivity.
Listeria Pathogenicity Studies. To determine liver enzyme levels, mice were infected i.v. with Listeria strains at a dose equal to 0.1 LD50. Serum liver enzyme levels for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined at the indicated time after vaccination. The in vivo growth of attenuated Listeria mutants was determined as described in ref. 10. Briefly, C57BL/6 mice were infected with the indicated Listeria strains at a dose equal to 0.1 LD50. The level of infection in each mouse was determined at the indicated time after Listeria challenge by enumerating bacteria in liver and spleen organ homogenates. All animals were treated according to National Institutes of Health guidelines, and protocols requiring animal experimentation received prior approval from the Cerus Animal Care and Use Committee.
Antigen-Specific T Cell Response. The frequency of IFN-γ and tumor necrosis factor type α (TNF-α) secreting CD8+ T lymphocytes specific for OVA, glycoprotein gp70 (AH1 or AH1-A5), and LLO (LLO190-201 and LLO296-304) were determined by intracellular cytokine staining as described in refs. 11 and 12. For the detection of OVA-specific CD8+ T cells the H-2Kb-restricted epitope SL8 (SIINFEKL) was used (13). gp70-specific immunity was assessed by using the endogenous H-2Ld-restricted epitope AH1 (SPSYVYHQF) and the altered peptide ligand AH1-A5 (SPSYAYHQF) (14). Cells were stained for cell-surface markers with anti-CD8α-peridinin chlorophyll protein (clone 53-6.7, Pharmingen). Cells were fixed in 2% paraformaldehyde, permeabilized with Perm/Wash buffer (Pharmingen), and incubated with anti-IFN-γ-allophycocyanin (clone XMG1.2, eBioscience, San Diego) and anti-TNF-α-phycoerythrin (clone MP6-XT22, eBioscience). Samples were acquired on a FACS-Calibur flow cytometer, and the data were analyzed with cellquest software (Becton Dickinson Immunocytometry Systems).
In Vivo Cytotoxicity Assay. In vivo cytotoxic activity of antigen-specific T cells was determined in female BALB/c mice vaccinated with Listeria ΔactA/ΔinlB-AH1-A5 by using the in vivo cytotoxicity assay as described in ref. 15. The carboxyfluorescein diacetate-succinimidyl ester (CFSE)hi-labeled cells were pulsed with the AH1 or AH1-A5 peptide, whereas the CFSElow-labeled cells were pulsed with the β-galactosidase control peptide. A 1:1 ratio of CFSElow- to CFSEhi-labeled cells was injected i.v. into BALB/c mice that were vaccinated 7 days prior. At 18 h after injection of the targets cells, spleens were removed and the ratio of CFSElow to CFSEhi cells was determined by flow cytometry.
In Vivo Tumor Studies. Female BALB/c mice were implanted i.v. with 2 × 105 CT26 cells on day 0. Four days later, mice were randomized and vaccinated i.v. with the indicated Listeria strains or Hanks' balanced salt solution (HBSS) as negative control. Lungs were harvested 19 days after tumor challenge, fixed in Bouin's solution and the number of metastases was assessed. To assess survival, mice were killed when any signs of stress or labored breathing were observed.
Statistical Analyses. All experiments were repeated at least twice to confirm reproducibility. Tumor survival results were analyzed by one-way ANOVA and two-tailed Student's t test for statistical significance.
Results
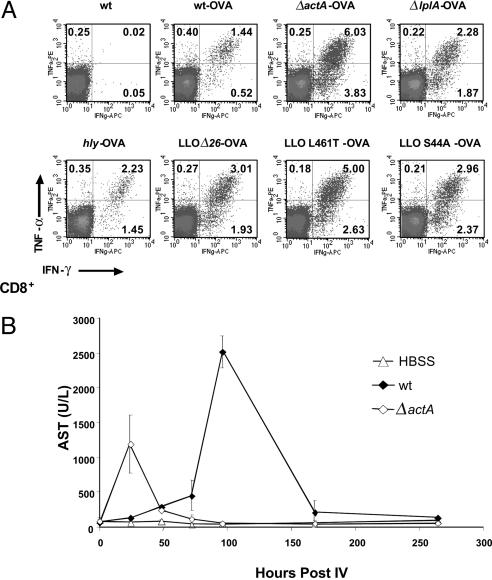
Attenuated Listeria Strains Deleted in actA Induce OVA-Specific CD8+ T Cell Responses with the Highest Magnitude. For clinical application as a tumor vaccine, two independent deletions in virulence genes are preferred to minimize the possibility for reversion to wild type. Identification of the vaccine platform strain was performed in a two-step process. First, we screened Listeria mutants deleted for a single virulence factor based on the relative potency to induce primary (Fig. 1) and memory T cell responses (Table 1). Second, we introduced deletions into the leading single mutant strains with the goal of maintaining potency but improving safety. Initially, we selected a wide range of attenuated mutants deficient or altered in a single virulence gene (e.g., hly, actA, or lplA) as well as gene products that might contribute to infection of nonphagocytic cells, such as InlB (16, 17). The loss of certain virulence determinants was demonstrated by an up to 10,000-fold attenuation in pathogenicity in mice, as determined by median lethality (LD50). To determine the relative potency of these Listeria vaccine strains to induce antigen-specific effector and memory T cell-mediated immunity, OVA fused with a truncated form of LLO to facilitate antigen secretion was used as a model antigen. Because of the defined integration of a single copy of the LLO–OVA fusion gene into the genome, all recombinant strains had similar growth characteristics in liquid broth, expressed and secreted OVA at similar levels (within a 2-fold variation) based on Western Blot analysis, and the LD50 of the resulting strains was within a factor of five of the unmodified parental strains (data not shown). A single vaccination of mice induced potent OVA-specific effector CD8+ T cell (Fig. 1 A) as well as LLO190-201-specific CD4+ T cell responses (data not shown). Depending on the strain used for vaccination, differences in magnitude (percentage) of the response to OVA were observed, with the ΔactA and LLO L461T mutants routinely eliciting the strongest responses. However, no significant difference in spleen size or number of splenocytes in mice vaccinated with various Listeria mutants was observed. The potency to induce OVA-specific primary responses did not correlate with the number of bacteria administered or with the strain's ability to induce protective memory T cell responses. Certain defined genetic mutations, such as ΔLLO, ΔPEST, and ΔlplA, resulted in a significant reduction or complete loss of the ability to induce protective immunity. Vaccination of mice with the ΔactA or LLO L461T strain resulted in almost complete protection, comparable with mice vaccinated with wild-type Listeria. Based on the ability to induce potent effector and memory T cell responses as well as a 1,000-fold attenuation compared with wild type, we selected the Listeria ΔactA for further strain development.
Fig. 1.
Recombinant Listeria strains induce a potent OVA-specific effector T cell response that is associated with transient liver toxicity. (A) Female C57BL/6 mice were vaccinated i.v. (IV) with 0.1 LD50 of indicated attenuated Listeria-OVA strain. OVA-specific immunity was assessed by intracellular cytokine staining on day 7 after a single vaccination. Representative dot blots of SL8-stimulated spleen cells gated on CD8+ T cells are shown. The percentage of IFN-γ+CD8+ T cells in the absence of SL8 stimulation was <0.03% (data not shown). (B) Vaccination of mice with wild type (wt) or the attenuated Listeria ΔactA mutant induces transient liver toxicity. Three C57BL/6 mice per group were vaccinated i.v. with 5 × 104 colony-forming units (CFU) of wild type or 1 × 107 colony-forming units of ΔactA. Serum levels of the liver enzyme AST (in units per liter) were determined at different times after infection.
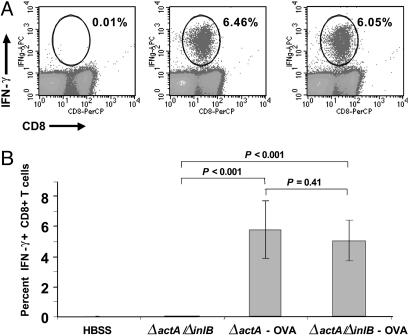
Attenuated Listeria Strain Deleted in actA and inlB Maintains Its Immunogenicity. Next, we constructed double-deletion mutants by in-frame deletion of actA by allelic exchange in ΔinlB, LLO L461T, and ΔlplA strains. The deletion of inlB was chosen to test whether the potency of ΔactA to induce cellular immunity could be segregated from its toxicity (Fig. 1B) by altering its tropism for nonphagocytic cells, such as hepatocytes. The deletion of inlB on the background of ΔactA did not result in a significant reduction in lethality (LD50) as compared with Listeria ΔactA (Table 1). Vaccination of mice with double mutants expressing OVA clearly demonstrated that ΔactA/ΔinlB maintained its ability to induce potent OVA-specific primary responses (Fig. 2) and memory responses (Table 1) that were comparable with the ΔactA single mutant. Listeria ΔactA/ΔinlB was significantly more potent compared with other double mutants tested. Listeria ΔactA/ΔinlB provided an increased therapeutic window, as shown by induction of OVA-specific CD8+ T cell responses that were equivalent to that observed with wild-type Listeria, but at a 3-log lower dose (Fig. 5A, which is published as supporting information on the PNAS web site). We also determined whether Listeria ΔactA/ΔinlB induced potent OVA-specific T cell responses when administered by means of routes that are preferable for clinical applications. To date, most studies in mice have investigated the interaction of immune cells with Listeria when either given i.v., i.p., or by means of oral delivery. We tested these immunization routes and additional routes of administration, including s.c., intradermal, and i.m. Oral delivery was not assessed in this study based on previous findings by Cossart and coworkers (18), demonstrating that the mouse model is inappropriate to study oral delivery because of poor interaction between invasins, such as internalin A, and host cell receptors that are required for mucosal invasion by Listeria. i.m. but not s.c. or intradermal administration of attenuated ΔactA/ΔinlB resulted in potent OVA-specific CD8+ T cell immunity, comparable with i.v. vaccinations (Fig. 5B).
Fig. 2.
The introduction of the inlB deletion on the ΔactA mutant does not abrogate the ability of strains to induce potent antigen-specific T cell responses. C57BL/6 mice were vaccinated i.v. with 1 × 107 colony-forming units (CFU) of the indicated strain. OVA-specific CD8+ T cell immunity was assessed on day 7 after vaccination by using intracellular cytokine staining. (A) Representative dot blots of SL8-stimulated spleen cells are shown. (Left) ΔactA/ΔinlB. (Center) ΔactA-OVA. (Right) ΔactA/ΔinlB-OVA. PerCP, peridinin chlorophyll protein. (B) The average percentage of OVA-specific CD8+ T cells of eight mice is shown.
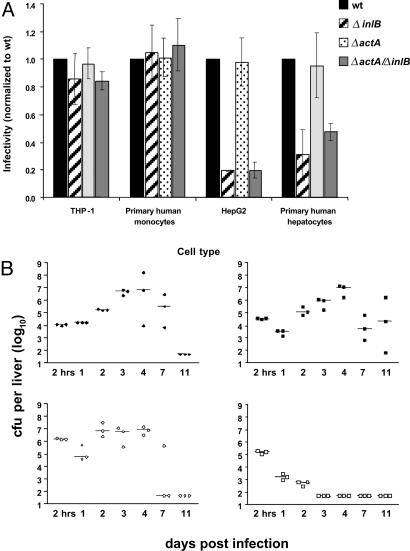
Deletion of ActA and InlB Renders Bacteria Unable to Infect Nonphagocytic Parenchymal Cells in Vitro. Internalin proteins, such as InlB, have been shown to confer the unique ability of Listeria to infect nonphagocytic cells, such as hepatocytes. To assess the ability of the double mutant ΔactA/ΔinlB to infect phagocytic as well as nonphagocytic cells in vitro, we compared the infection by wild type, ΔactA, ΔinlB, and ΔactA/ΔinlB strains of Listeria in human nonphagocytic and phagocytic cell lines and primary cells. As shown in Fig. 3A, all strains were able to infect THP-1 cells and human monocytes to a similar extent, indicating that the absence of ActA or InlB does not affect infection of phagocytic cells. However, infection of hepatocytes was significantly decreased for Listeria strains lacking InlB. Infectivity was reduced by ≈60% in primary human hepatocytes and by ≈80% in HepG2 cells when infecting with either of the InlB-null mutant strains (Fig. 3A).
Fig. 3.
The inability of Listeria ΔactA/ΔinlB to infect nonphagocytic cells in vitro results in accelerated clearance in vivo. (A) In vitro infectivity of the human hepatocyte cell line (HepG2), fresh human hepatocytes (two donors), a human monocyte cell line (THP-1), and primary human monocytes (three donors) is shown. For all strains, the rate of infection is normalized to the rate of infection by wild-type (wt) Listeria. The averages of three (or two for primary human hepatocytes) independent experiments are shown. (B) The in vivo growth kinetic in liver was assessed for wild type (Upper Left), ΔactA (Lower Left), ΔinlB (Upper Right), and ΔactA/ΔinlB (Lower Right). C57BL/6 mice were injected i.v. with 0.1 LD50 of the indicated strain and bacteria per organ were determined from three mice at each time point. A representative of two experiments is shown.
Accelerated Clearance of ΔactA/ΔinlB Infection Without the Induction of Liver Damage. Although attenuated strains of Listeria can be administered at higher doses compared with wild type, for the development of a safe vaccine it is important that the infection can be cleared rapidly, without damaging the primary organs of infection, i.e., liver or spleen. In that regard, we compared the in vivo growth kinetics of wild type, ΔinlB, ΔactA, and ΔactA/ΔinlB in female C57BL/6 mice (Fig. 3B and Fig. 6A, which is published as supporting information on the PNAS web site). Infection of mice with wild-type Listeria resolved within 7–11 days after administration as described in ref. 19. The number of wild-type Listeria steadily increased over the time period of 4 days by 10,000-fold and decreased to undetectable levels in spleen and liver by day 11. Interestingly, the ΔinlB mutant demonstrated similar kinetics and magnitude of expansion in spleen as well as liver. This result stands in contrast with previous studies by Lingnau et al. (17) in which retarded growth of the ΔinlB mutant in liver and spleen had been observed. The number of ΔactA Listeria increased 10-fold over the first 48 h in the liver but not in the spleen, and the bacteria count was maintained for up to 4 days before it eventually decreased; the infection was cleared by day 7 after infection. In contrast, the ΔactA/ΔinlB double mutant was cleared significantly more rapidly from the liver and spleen by days 3–4. To assess the level of tissue damage and liver pathology, serum samples were collected at different time points during the course of an infection and liver enzyme levels and the proinflammatory cytokine profile (data not shown) were assessed. ΔactA but not ΔactA/ΔinlB induced liver enzymes, such as AST or ALT, 24 h after infection (Fig. 6B). Moreover, histopathological changes in the liver throughout the course of infection were significantly milder in mice receiving the ΔactA/ΔinlB double mutant when compared with wild type-, ΔactA-, or ΔinlB-infected mice (data not shown). Interestingly, wild type and ΔactA induced higher serum levels of IFN-γ, MCP-1, and IL-6 at 24 h after infection compared with the ΔinlB and ΔactA/ΔinlB mutants (data not shown). However, the induction of proinflammatory cytokines did not seem to correlate with the ability of a strain to induce potent antigen-specific T cell responses.
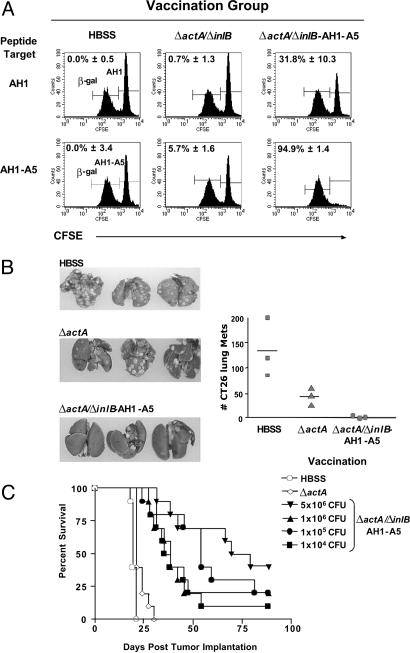
Vaccination with Recombinant ΔactA/ΔinlB Expressing AH1-A5 Breaks Self-Tolerance and Results in Tumor Regression and Prolongation of Life. The previous experiments demonstrated that a single vaccination with attenuated Listeria strains expressing a foreign antigen, such as OVA, elicits antigen-specific CD8+ T cell immunity of high magnitude. However, the induction of immunity to a truly native tumor antigen requires breaking of self-tolerance. To test whether Listeria-based vaccinations result in breaking of self-tolerance, we constructed the ΔactA/ΔinlB that expresses the altered T cell epitope, AH1-A5, of the endogenous tumor antigen gp70 of the murine colon tumor CT26 (14, 20). A single vaccination with ΔactA/ΔinlB-AH1-A5 administered either i.v. or i.m. (data not shown) resulted in the induction of 2.2% AH1-A5-specific CD8+ T cells in the spleen (Fig. 7A, which is published as supporting information on the PNAS web site). Half of the response (1.1%) was specific to the endogenous T cell epitope AH1. The AH1-specific CD8+ T cell response was significantly increased by administering a boost vaccination 4 weeks after the primary vaccination (Fig. 7B). The boost vaccination could be administered as early as 2 weeks after the primary immunization with similar results (data not shown). Antigen-specific cytolytic activity in vaccinated mice was determined by using the in vivo cytotoxicity assay. Vaccination of mice with ΔactA/ΔinlB expressing AH1-A5 induced a potent cytotoxic T cell response demonstrated by the disappearance of AH1-A5 as well as AH1 peptide-loaded splenocytes in vaccinated mice but not HBSS- or ΔactA/ΔinlB parental control-immunized mice (Fig. 4A). We next tested whether a vaccination with ΔactA/ΔinlB-AH1-A5 induces therapeutic immunity in an experimental lung metastases tumor model by using the colon tumor-derived CT26 cell line (Fig. 4B). The therapeutic vaccination of mice with the ΔactA/ΔinlB double mutant expressing AH1-A5 resulted in a significant reduction of lung metastases compared with vehicle control group (P < 0.05). More importantly, therapeutic vaccination of CT26 tumor-bearing mice with ΔactA/ΔinlB-AH1-A5, but not with the ΔactA control strain, resulted in a significant prolongation of life with 40% long-term survivors (P < 0.0001) (Fig. 4C). Even at vaccination doses 10,000-fold below the experimental LD50, a potent antitumor response was induced that resulted in a median survival of >35 days compared with 21 days for mice treated with buffer or the Listeria vector only (P < 0.001). The robustness of the therapeutic anti-CT26 tumor response after Listeria-based vaccination was furthermore demonstrated in that vaccination could be delayed as much as 10 days after tumor implantation when tumors reached a size of >100 mm3 resulting in comparable tumor growth delay (Fig. 7C). The therapeutic anti-CT26 response was completely mediated by CD8+ T (depletion data not shown).
Fig. 4.
Recombinant attenuated Listeria ΔactA/ΔinlB AH1-A5 breaks tolerance to self that results in potent therapeutic antitumor activity and prolongation of life. (A) Female BALB/c mice were vaccinated once with 0.1 LD50 of the indicated strain. Cytotoxic activity was determined by in vivo cytotoxicity assay. CFSEhi AH1 (Upper) or AH1-A5 (Lower) peptide-pulsed BALB/c splenocytes (3 × 106) and CFSElow nonpulsed splenocytes (3 × 106) were coinjected i.v. into BALB/c mice that were naïve or 7 days prior to vaccination with the indicated Listeria strain. In vivo killing of CFSE-labeled target cells was assessed 18 h later. Histograms are gated on CFSE+ cells in host mice. The number represents the average percentage of specific killing of three individual mice normalized to HBSS-injected mice. (B) Female BALB/c mice were implanted i.v. with 2 × 105 CT26 cells on day 0. Four days later, mice received a single vaccination with a dose equal to 0.1 LD50 of the indicated strain. Lungs were harvested on day 19, fixed in Bouin's solution, and the number of surface lung metastases were counted. Representative lungs are shown. (C) Female BALB/c mice were implanted i.v. with 2 × 105 CT26 cells on day 0. Four days later, mice were vaccinated with 0.1 LD50 of the indicated strain or vehicle control (n = 10). Survival was monitored over the course of the experiment. Therapeutic vaccination of mice with ΔactA/ΔinlB AH1-A5 both resulted in a significant survival benefit (one-way ANOVA, P < 0.0001) compared with HBSS or ΔactA controls.
Discussion
There exists a relatively abundant literature demonstrating the potency of Listeria-based vaccines targeting selected infectious disease or model tumor antigens, in both prophylactic and therapeutic settings in animal models of disease. However, to our knowledge, there has not been an effort to develop a genetically defined mutant vaccine platform that combines potency similar to wild-type Listeria with reduced pathogenicity. The results of this study demonstrate that by selectively deleting two determinants of Listeria pathogenesis, we can generate a vaccine strain that is more immunogenic yet considerably less toxic than wild-type bacteria. Immunization of tumor-bearing mice with this strain encoding a tumor antigen resulted in long-term survival as a result of breaking tolerance against an endogenous antigen.
The rationale for the combined deletions of actA and inlB was to separate the immunogenicity of Listeria-based vaccines from toxicity because of infection of nonphagocytic cells, such as hepatocytes. Because ΔinlB single mutants can spread from cell to cell, infection of hepatocytes in vivo can occur indirectly by means of Kupffer cells. We hypothesized that the ΔinlB in vivo phenotype would be revealed only on the background of Listeria ΔactA, having defective cell-to-cell spread. This notion is supported by Gregory et al. (21), who demonstrated that Listeria ΔinlB infected and propagated within mouse hepatocytes in vivo despite its dramatically reduced capacity to infect nonphagocytic cells in vitro. As demonstrated here, the deletion of both actA and inlB resulted in an attenuated strain that is both unable to spread effectively from cell to cell and has a reduced capacity to infect hepatocytes directly or indirectly. Consequently, Listeria ΔactA/ΔinlB was cleared rapidly from the liver and spleen as compared to either single mutant alone. In contrast to the ΔactA or ΔinlB single mutants, vaccination of mice with Listeria ΔactA/ΔinlB did not result in any measurable induction of liver toxicity, as determined by release of liver enzymes. Furthermore, it has been suggested that the infection of the CNS by Listeria is mediated through routes that require cell-to-cell spread as well as InlB-dependent infection (22, 23). Preliminary results in an acute Listeria infection model in guinea pigs supports the hypothesis that the ΔactA/ΔinlB double mutant has lost its ability to infect the CNS.
The accelerated clearance of Listeria ΔactA/ΔinlB infection in mice after i.v. administration suggests strongly that in vivo bacterial proliferation and expansion in liver and spleen after 48 h is not required for the induction of a productive T cell response. This finding is supported by Mercado et al. (24), who demonstrated that the kinetics or magnitude of a T cell response after priming with Listeria is independent of the duration of infection in vivo. The idea that the innate immune response to Listeria during the first 24 h sets in motion an immunological program that determines the extent and duration of the T cell response has been also supported by Badovinac et al. (25), who demonstrated that the kinetics of the T cell response is independent of duration of infection or amount of antigen displayed. In contrast to the conclusions of Mercado et al., our dose–response data from mice given different doses of Listeria ΔactA/ΔinlB-OVA vaccines support the idea proposed by Badovinac et al. that the magnitude of the response depends on the amount of antigen displayed at the initiation of the immune response. More importantly, the attenuation of the double mutant (>1,000-fold) means that more bacteria can be administered for each vaccination, resulting in a much larger amount of antigen presented during the crucial time of T cell priming. This result means that because of the attenuated phenotype and the associated accelerated clearance, antigen-specific T cell responses are initiated without the potentially harmful in vivo expansion of Listeria that is associated with wild-type Listeria infection.
Listeria has evolved mechanisms for intracellular growth and spread while minimizing cytotoxicity, largely through confining the activity of the pore-forming cytolysin LLO to the low-pH environment of the phagosome. Particular Listeria hly mutants have a cytotoxic phenotype because of the retention of activity of LLO in the cytoplasm, which results in premature host cell lysis. Somewhat paradoxically, cytotoxic mutants have diminished virulence. We evaluated a number of Listeria mutants with various degrees of cytotoxicity (related to the cytoplasmic enzymatic activity of LLO) for their capacity to induce an effector immune response. Interestingly, although the LLO L461T mutant strain was highly immunogenic, the combination of that mutation with ΔactA resulted in a strain that was poorly immunogenic. Other single mutants with phenotypes of increased cytotoxicity, including LLO Δ26, LLO S44A, and LLO S44A/LLO L461T were considerably impaired in their capacity to induce a primary immune response. We speculate that the appropriate level of Listeria infection-induced cell death may be a critical factor for direct priming or crosspriming of CD8+ T cells, to induce an effector and memory T cell immunity (26). Early events during infection may cause qualitatively and quantitatively differences in activation of APC at the site of infection and may result in the induction of distinct T cell responses, i.e., effector versus memory T cell immunity (27–30). By using a range of cytotoxic mutants, we are currently in the process of characterizing these early events during infection and the role of CD4+ T helper cells for the induction of effector and memory CD8+ T cell responses.
The ΔactA/ΔinlB double mutant vaccine platform was systematically selected from a large panel of genetically defined attenuated Listeria strains. A single immunization of mice with Listeria ΔactA/ΔinlB induced protective immunity against lethal challenge with wild-type Listeria, which requires secondary CD8+ T cell expansion and formation of memory of CD8+ T cells (31). A single immunization with the vaccine strain given through a variety of routes also induced potent CD8+ T cell immunity to a strong foreign antigen (OVA) and to the native tumor-associated antigen gp70. Furthermore, Listeria ΔactA/ΔinlB-based vaccination broke self-tolerance after a single immunization that translated into a therapeutic antitumor response and long-term survival. In stark contrast, delays in tumor growth but not complete tumor regressions have been observed in a therapeutic setting with other broadly used vaccine strategies targeting gp70, including antigen-pulsed dendritic cells, recombinant vaccinia virus, or adenovirus (14, 32–34). Furthermore, immunization of B16 melanoma-bearing mice with a Listeria vaccine containing a single attenuating mutation expressing tyrosinase-related protein 2 resulted in a significant decrease in pulmonary tumor nodules, demonstrating further that recombinant Listeria can overcome tolerance to an endogenous tumor-associated antigen (19). Listeria ΔactA/ΔinlB AH1-A5 vaccination resulted in complete tumor regression, even though CT26 tumors have been shown to evade anti-tumor immunity by down-regulating gp70 expression upon exposure to IFN-γ (35). Listeria ΔactA/ΔinlB maintained its immunogenicity, even when administered at doses that were several logs below its LD50. Thus, deleting actA and inlB increased the therapeutic window for the safe use of Listeria-based vaccines.
In summary, we have selected a genetically defined live-attenuated Listeria platform strain with potent immunogenicity. Because its capacity to directly or indirectly infect particular nonphagocytic cells is largely abrogated and as a consequence of an accelerated clearance in vivo, we believe that this strain is an ideal candidate for further clinical development of potent Listeria-based vectors for active cancer immunotherapy. An evaluation of Listeria ΔactA/ΔinlB vaccines encoding antigens related to human cancer in preclinical animal studies would prove useful for the clinical development of this candidate.
Supplementary Material
Acknowledgments
We thank Gary Bolton and Anthony Garcia for help with the mouse tumor experiments, Aaron Reames for performing the in vivo cytotoxicity assay, and Dr. Anne North for critical review of the manuscript. D.A.P. was supported by U.S. Public Health Service Grants AI29619 and AI27655.
Abbreviations: OVA, ovalbumin; LLO, listeriolysin O; APC, antigen-presenting cells; CFSE, carboxyfluorescein diacetate-succinimidyl ester; AST, aspartate aminotransferase; ALT, alanine aminotransferase; TNF-α, tumor necrosis factor type α.
References
- 1.Kolb-Maurer, A., Gentschev, I., Fries, H. W., Fiedler, F., Brocker, E. B., Kampgen, E. & Goebel, W. (2000) Infect. Immun. 68, 3680–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolb-Maurer, A., Kammerer, U., Maurer, M., Gentschev, I., Brocker, E. B., Rieckmann, P. & Kampgen, E. (2003) FEMS Immunol. Med. Microbiol. 35, 255–262. [DOI] [PubMed] [Google Scholar]
- 3.Paterson, Y. & Ikonomidis, G. (1996) Curr. Opin. Immunol. 8, 664–669. [DOI] [PubMed] [Google Scholar]
- 4.Jensen, E. R., Selvakumar, R., Shen, H., Ahmed, R., Wettstein, F. O. & Miller, J. F. (1997) J. Virol. 71, 8467–8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauer, P., Chow, M. Y., Loessner, M. J., Portnoy, D. A. & Calendar, R. (2002) J. Bacteriol. 184, 4177–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, D. K. & Hinrichs, D. J. (1987) J. Immunol. 139, 2005–2009. [PubMed] [Google Scholar]
- 7.Camilli, A., Tilney, L. G. & Portnoy, D. A. (1993) Mol. Microbiol. 8, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikonomidis, G., Paterson, Y., Kos, F. J. & Portnoy, D. A. (1994) J. Exp. Med. 180, 2209–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Riordan, M., Yi, C. H., Gonzales, R., Lee, K. D. & Portnoy, D. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13861–13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auerbuch, V., Brockstedt, D. G., Meyer-Morse, N., O'Riordan, M. & Portnoy, D. A. (2004) J. Exp. Med. 200, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prussin, C. & Metcalfe, D. D. (1995) J. Immunol. Methods 188, 117–128. [DOI] [PubMed] [Google Scholar]
- 12.Geginat, G., Schenk, S., Skoberne, M., Goebel, W. & Hof, H. (2001) J. Immunol. 166, 1877–1884. [DOI] [PubMed] [Google Scholar]
- 13.Brockstedt, D. G., Podsakoff, G. M., Fong, L., Kurtzman, G., MuellerRuchholtz, W. & Engleman, E. G. (1999) Clin. Immunol. 92, 67–75. [DOI] [PubMed] [Google Scholar]
- 14.Slansky, J. E., Rattis, F. M., Boyd, L. F., Fahmy, T., Jaffee, E. M., Schneck, J. P., Margulies, D. H. & Pardoll, D. M. (2000) Immunity 13, 529–538. [DOI] [PubMed] [Google Scholar]
- 15.Mueller, S. N., Jones, C. M., Smith, C. M., Heath, W. R. & Carbone, F. R. (2002) J. Exp. Med. 195, 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cossart, P., Pizarro-Cerda, J. & Lecuit, M. (2003) Trends Cell Biol. 13, 23–31. [DOI] [PubMed] [Google Scholar]
- 17.Lingnau, A., Domann, E., Hudel, M., Bock, M., Nichterlein, T., Wehland, J. & Chakraborty, T. (1995) Infect. Immun. 63, 3896–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B. & Cossart, P. (1999) EMBO J. 18, 3956–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starks, H., Bruhn, K. W., Shen, H., Barry, R. A., Dubensky, T. W., Brockstedt, D., Hinrichs, D. J., Higgins, D. E., Miller, J. F., Giedlin, M., Bouwer, A. G. (2004) J. Immunol. 173, 420–427. [DOI] [PubMed] [Google Scholar]
- 20.Huang, A. Y., Gulden, P. H., Woods, A. S., Thomas, M. C., Tong, C. D., Wang, W., Engelhard, V. H., Pasternack, G., Cotter, R., Hunt, D., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9730–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory, S. H., Sagnimeni, A. J. & Wing, E. J. (1997) Infect. Immun. 65, 5137–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dramsi, S., Levi, S., Triller, A. & Cossart, P. (1998) Infect. Immun. 66, 4461–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drevets, D. A. (1999) Infect. Immun. 67, 3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mercado, R., Vijh, S., Allen, S. E., Kerksiek, K., Pilip, I. M. & Pamer, E. G. (2000) J. Immunol. 165, 6833–6839. [DOI] [PubMed] [Google Scholar]
- 25.Badovinac, V. P., Porter, B. B. & Harty, J. T. (2002) Nat. Immunol. 3, 619–626. [DOI] [PubMed] [Google Scholar]
- 26.Schaible, U. E., Winau, F., Sieling, P. A., Fischer, K., Collins, H. L., Hagens, K., Modlin, R. L., Brinkmann, V. & Kaufmann, S. H. (2003) Nat. Med. 9, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 27.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., Von Herrath, M. G. & Schoenberger, S. P. (2003) Nature 421, 852–856. [DOI] [PubMed] [Google Scholar]
- 28.Harrington, L. E., Galvan, M., Baum, L. G., Altman, J. D. & Ahmed, R. (2000) J. Exp. Med. 191, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaech, S. M., Wherry, E. J. & Ahmed, R. (2002) Nat. Rev. Immunol. 2, 251–262. [DOI] [PubMed] [Google Scholar]
- 30.Shen, H., Miller, J. F., Fan, X., Kolwyck, D., Ahmed, R. & Harty, J. T. (1998) Cell 92, 535–545. [DOI] [PubMed] [Google Scholar]
- 31.Harty, J. T., Lenz, L. L. & Bevan, M. J. (1996) Curr. Opin. Immunol. 8, 526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kershaw, M. H., Hsu, C., Mondesire, W., Parker, L. L., Wang, G., Overwijk, W. W., Lapointe, R., Yang, J. C., Wang, R. F., Restifo, N. P. & Hwu, P. (2001) Cancer Res. 61, 7920–7924. [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura, M., Iwahashi, M., Nakamori, M., Ueda, K., Matsuura, I., Noguchi, K. & Yamaue, H. (2002) Clin. Cancer Res. 8, 2742–2749. [PubMed] [Google Scholar]
- 34.Casares, N., Lasarte, J. J., de Cerio, A. L., Sarobe, P., Ruiz, M., Melero, I., Prieto, J. & Borras-Cuesta, F. (2001) Eur. J. Immunol. 31, 1780–1789. [DOI] [PubMed] [Google Scholar]
- 35.Beatty, G. L. & Paterson, Y. (2000) J. Immunol. 165, 5502–5508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.