Comprehensive genomic profiling (CGP) reliably detects alterations associated with a lack of benefit to anti-epidermal growth factor receptor therapy in advanced colorectal cancer, while simultaneously identifying alterations that are potentially important in guiding treatment. This study found that the use of CGP during the course of clinical care allows for the refined selection of appropriate targeted therapies and clinical trials, increasing the chance of clinical benefit and avoiding therapeutic futility.
Keywords: KRAS, Colon cancer, Biomarker, Genomic profiling, Epidermal growth factor receptor, Cetuximab
Abstract
Introduction.
A KRAS mutation represented the first genomic biomarker to predict lack of benefit from anti-epidermal growth factor receptor (EGFR) antibody therapy in advanced colorectal cancer (CRC). Expanded RAS testing has further refined the treatment approach, but understanding of genomic alterations underlying primary and acquired resistance is limited and further study is needed.
Materials and Methods.
We prospectively analyzed 4,422 clinical samples from patients with advanced CRC, using hybrid-capture based comprehensive genomic profiling (CGP) at the request of the individual treating physicians. Comparison with prior molecular testing results, when available, was performed to assess concordance.
Results.
We identified a RAS/RAF pathway mutation or amplification in 62% of cases, including samples harboring KRAS mutations outside of the codon 12/13 hotspot region in 6.4% of cases. Among cases with KRAS non-codon 12/13 alterations for which prior test results were available, 79 of 90 (88%) were not identified by focused testing. Of 1,644 RAS/RAF wild-type cases analyzed by CGP, 31% harbored a genomic alteration (GA) associated with resistance to anti-EGFR therapy in advanced CRC including mutations in PIK3CA, PTEN, EGFR, and ERBB2. We also identified other targetable GA, including novel kinase fusions, receptor tyrosine kinase amplification, activating point mutations, as well as microsatellite instability.
Conclusion.
Extended genomic profiling reliably detects alterations associated with lack of benefit to anti-EGFR therapy in advanced CRC, while simultaneously identifying alterations potentially important in guiding treatment. The use of CGP during the course of clinical care allows for the refined selection of appropriate targeted therapies and clinical trials, increasing the chance of clinical benefit and avoiding therapeutic futility.
Implications for Practice:
Comprehensive genomic profiling (CGP) detects diverse genomic alterations associated with lack of benefit to anti-epidermal growth factor receptor therapy in advanced colorectal cancer (CRC), as well as targetable alterations in many other genes. This includes detection of a broad spectrum of activating KRAS alterations frequently missed by focused molecular hotspot testing, as well as other RAS/RAF pathway alterations, mutations shown to disrupt antibody binding, RTK activating point mutations, amplifications, and rearrangements, and activating alterations in downstream effectors including PI3K and MEK1. The use of CGP in clinical practice is critical to guide appropriate selection of targeted therapies for patients with advanced CRC.
Introduction
Colorectal adenocarcinoma (CRC) is the third most common cancer in men and second in women, accounting for an estimated 694,000 deaths annually worldwide [1]. Despite screening efforts, nearly 20% of patients present with metastatic disease, which is associated with a poor prognosis and median overall survival of 24–30 months [2–5]. The monoclonal antibodies directed against the epidermal growth factor receptor (EGFR), cetuximab and panitumumab, have improved outcomes in patients without oncogenic alterations in RAS, and multiple studies have shown that KRAS mutations, as well as NRAS mutations and BRAF V600E, are associated with lack of clinical benefit from anti-EGFR therapies [6–11]. However, RAS and RAF mutations only account for lack of response to anti-EGFR therapies in up to 60% of CRC patients [12], suggesting that a significant fraction of RAS/RAF wild-type patients do not respond and additional predictors of response remain to be established.
Activating mutations in KRAS have been reported in approximately 40% of CRC, and point mutations at codons 12 or 13 of exon 2 account for 80%–90% of these alterations [12, 13]. However, additional mutations have been reported in exons 2, 3, and 4, and have also been shown to predict resistance to cetuximab and panitumumab [14–16]. Current guidelines now endorse extended RAS testing (exons 2–4 of KRAS and NRAS) to reflect the evidence that anti-EGFR therapy should be avoided in RAS-altered advanced CRC [17]. Smaller studies have suggested that RAS amplification and alterations in genes, including PIK3CA, EGFR, and ERBB2, are associated with resistance to cetuximab or panitumumab in CRC, but a large comprehensive study reporting the incidence and diversity of these alterations is lacking [18–21]. Furthermore, assessment of the genomic landscape of CRC has used mainly treatment-naïve, largely nonmetastatic primary tumors or focused assessments of specimens from patients with advanced disease [22, 23]. We present the genomic profiles and clinical implications of 4,422 CRC cases assayed in the course of clinical care, often in the setting of metastatic disease.
Materials and Methods
A series of 4,422 CRC cases was prospectively collected and assayed with a validated, hybrid-capture-based comprehensive genomic profiling (CGP) assay between August 2012 and June 2015. DNA was extracted from 40 microns of formalin-fixed, paraffin-embedded sections, and CGP was performed on hybridization-captured, adaptor-ligation-based libraries to a mean coverage depth of >650× for at least 236 cancer-related genes plus 47 introns from 19 genes frequently rearranged in cancer, as described previously [24]. Full exons for all included genes were sequenced. All classes of genomic alterations (GAs) were identified, including base-pair substitutions, insertions and deletions, copy number alterations, and rearrangements. Focal amplifications are called at segments with ≥6 copies (or ≥7 for triploid; ≥8 for tetraploid tumors) in samples with purity of >20%. Microsatellite instable (MSI-H) or stable (MSS) status as a measure of mismatch repair deficiency was determined using a proprietary computational algorithm. Tumors were classified as MSI-H or MSS using a principal component 1 cutoff value of <−8.5 or >−4, respectively. Values >−8.5 and <−4 were classified as MSI-ambiguous [25]. Results from prior KRAS molecular testing were obtained by review of available medical records provided by the treating physician. Ordinal relationships were examined using the Wilcoxon-Mann-Whitney test; the relationship between the presence of ERBB2 alterations and RAS/RAF status was examined using a chi-square test. Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol no. 20152817).
Results
Detection of KRAS Mutations by CGP
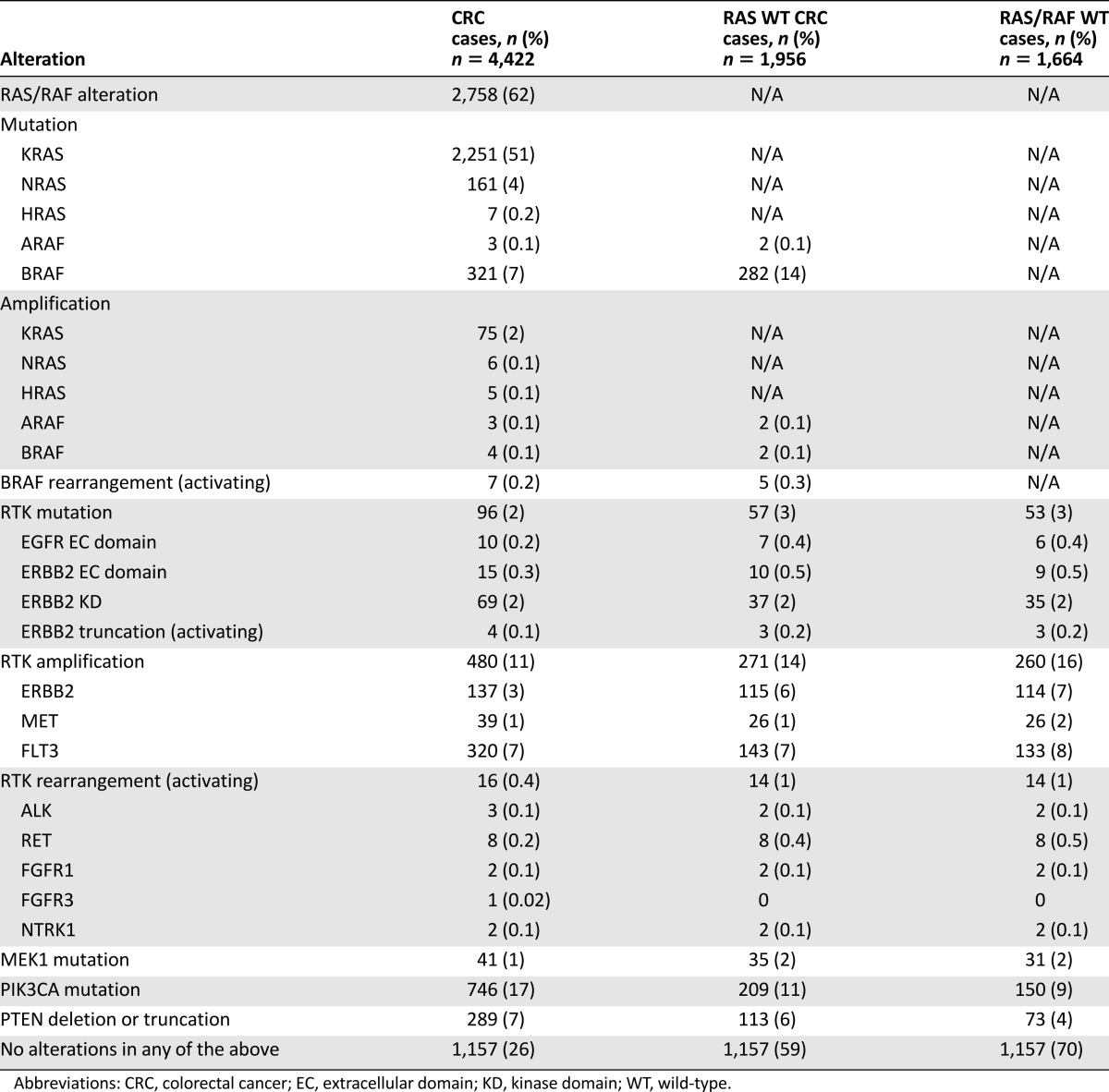
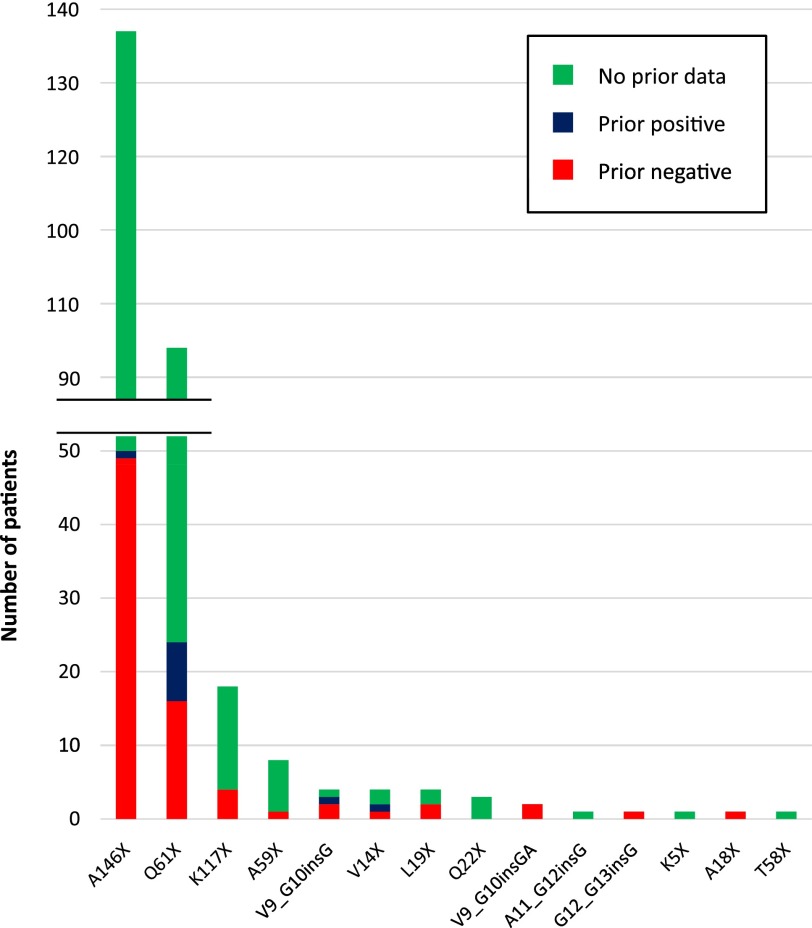
CGP identified 51% of cases (2,251 of 4,422) as positive for any KRAS mutation, including 284 cases (6.4% of total CRC cases) with activating KRAS mutations occurring outside of codons 12 and 13 (Table 1). Non-codon 12/13 KRAS alterations consisted of mutations at A146 (n = 137; 48%) and Q61 (n = 94; 33%), as well as 8 other positions at which mutations have been shown to be activating and oncogenic (Fig. 1). Eight CRC cases otherwise negative for RAS or BRAF alterations were also found to harbor KRAS insertions in the codon 12/13 hotspot region.
Table 1.
Incidence of RAS/RAF alterations and other alterations predicted to negatively affect response to anti-EGFR therapy in CRC
Figure 1.
Missed detection of KRAS non-G12/13 mutations by prior focused molecular testing. KRAS mutations detected by comprehensive genomic profiling and missed by prior focused molecular testing (red bars), detected by prior focused molecular testing (blue bars), or with no record of prior molecular testing in provided pathology report (green bars).
We examined provided pathology reports for CRC cases with KRAS non-12/13 activating mutations detected by CGP (n = 284), and prior focused KRAS molecular testing results were available for 90 cases. Of these, 79 (88%) reported a KRAS wild-type result, indicating that the KRAS non-12/13 alteration was not detected by hotspot testing in the vast majority of cases. This represents a missed clinical benefit rate of 98% (49 of 50) for patients with A146 mutations, 67% (16 of 24) for patients with Q61 mutations, and 74% (25 of 34) for activating point mutations at 9 other codons detected by CGP (Fig. 1). KRAS insertions in the 12/13 hotspot region were missed in 5 of 6 cases (83%) by prior hotspot testing (Fig. 1).
Non-KRAS Ras/Raf Family Alterations
In addition to KRAS mutations, NRAS and HRAS mutations were found in 161 (3.6%) and 7 (0.2%) of all CRC cases analyzed, respectively (Table 1). RAS amplification was observed in 86 CRC cases (1.9%), including 75 cases with KRAS amplification. BRAF mutations were observed in 321 (7.3%) CRC cases and included V600 mutations (n = 233; 72.6%), non-V600 activating mutations (n = 34; 10.6%), and inactivating mutations known to lead to feedback activating of RAF/MEK signaling (n = 54; 16.8%) [26]. All mutations at BRAF position 600 were V600E, except for one case with V600G. In addition, activating BRAF rearrangements were observed in seven cases, including three with TRIM24-BRAF fusions (Table 1).
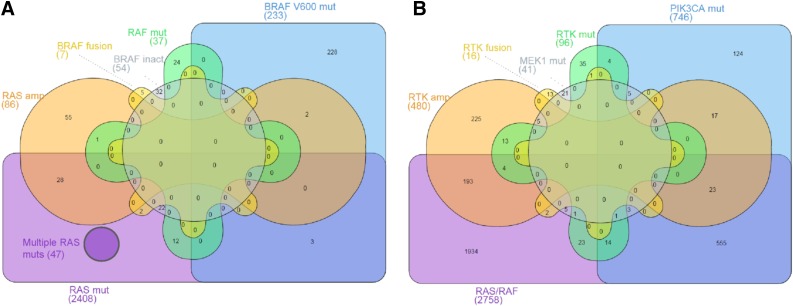
RAF and RAS alterations were not mutually exclusive in our dataset, with 47 cases harboring 2 or more RAS mutations (Fig. 2A). BRAF inactivating mutations co-occurred with RAS mutations in 22 of 54 cases (41%) and BRAF rearrangements co-occurred with RAS mutations in 2 of 7 cases (29%); however, BRAF V600 mutations were the sole RAS/RAF alteration in 228 of 233 cases (98%). RAS amplification co-occurred with other RAS/RAF alterations in 31 of 86 cases (36%) (Fig. 2A), and BRAF amplification was observed in 4 total cases, but co-occurred with a KRAS or BRAF mutation in 3 of 4 cases (data not shown).
Figure 2.
Overlap of RAS/RAF alterations and other potential drivers in colorectal cancer. Diagram represents overlap of colorectal patient cases with the indicated classes of genomic alterations detected by comprehensive genomic profiling. (A): Overlap of cases with subtypes of RAS/RAF alterations. (B): Overlap of all cases with RAF/RAS alterations with cases with subtypes of activating alterations in other potential drivers.
Abbreviations: amp, amplification; mut, mutation; RTK, receptor tyrosine kinase.
RTK Alterations
We observed RTK mutations in 96 cases (2%) including EGFR extracellular (EC) domain mutations (n = 10), and ERBB2 EC domain (n = 15) and kinase domain (KD; n = 69) mutations and activating truncations (n = 4) (Table 1). These included eight cases with EGFR G456R or S492R mutations. We also identified one case with an EGFR L858R activating kinase domain mutation, but did not observe EGFR exon 19 deletion or exon 20 insertion alterations. The most common ERBB2 mutations were S310F/Y (n = 13), R678Q (n = 18), and V842I (n = 18). Twelve cases with ERBB2 point mutations also had concurrent ERBB2 amplification. We observed amplification of FLT3, ERBB2, and MET in 7%, 3%, and 1% of cases, respectively. Activating RTK rearrangements involving RET (n = 8), ALK (n = 3), FGFR1 (n = 2), NTRK1 (n = 2), and FGFR3 (n = 1) were detected in 16 cases (0.4%), including a novel CENPF-ALK fusion. RET fusion partners included NCOA4 (n = 4), CCDC6 (n = 3), and TRIM24 (n = 1).
MEK1, PTEN, and PI3K Alterations
We identified MEK1 (MAP2K1) mutations in 41 cases (0.9%), PTEN homozygous deletion or inactivating truncation in 289 cases (7%), and mutations in the catalytic subunit of PI3K (PIK3CA) in 748 cases (17%) (Table 1). MEK1 mutations included K57E/N/T (n = 19), Q56P (n = 6), E203K (n = 6), C121S (n = 4), and other (n = 6). MEK1 mutations co-occurred with BRAF mutations in 4 of 41 cases, including 1 case with BRAF K601N, but did not co-occur specifically with BRAF V600 alterations. Of cases with PTEN-inactivating alterations, homozygous deletion and truncation were relatively equally represented (48% and 52% of cases, respectively). The majority of PIK3CA mutations were activating and occurred most commonly in exons 9 and 20, specifically at E545 (244), E542 (130), and H1047R (134).
Co-occurrence With RAS/RAF Alterations
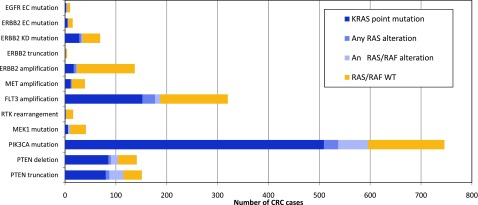
RTK alterations were not mutually exclusive with RAS/RAS alterations; most notably, FLT3 amplification co-occurred with KRAS point mutations in 48% of cases (152 of 320) and with RAS/RAF alterations in general in 58% of cases (187 of 320). MET amplification and ERBB2 amplification were less likely to co-occur with RAS/RAF alterations, and tumors harboring amplification of these receptors were extended RAS/RAF wild-type in 26 of 39 cases (67%) and 114 of 137 cases (83%), respectively (Fig. 3). Interestingly, FLT3 and MET amplification were significantly higher in RAS/RAF wild-type cases (mean copy number: FLT3, 18 copies; MET, 34 copies) compared with cases in which a RAS or RAF alteration co-occurred (mean copy number: FLT3, 12 copies; MET: 9 copies; p < .003 and p < .001, respectively); whereas ERBB2 amplification was consistently high regardless of RAS/RAF status (mean ERBB2 copy number: 60 copies in RAS/RAF wild-type cases, and 64 copies in RAS/RAF positive cases; p = .300).
Figure 3.
Quantitation of CRC cases with potential driver alterations and co-occurrence of KRAS mutations. Breakdown of specific subtypes of RTK, MEK1, PIK3CA, and PTEN alteration classes and overlap with RAS/RAF alterations. Dark blue bars specify the subset of cases that overlap specifically with KRAS point mutations.
Abbreviations: CRC, colorectal cancer; EC, extracellular domain; KD, kinase domain; RTK, receptor tyrosine kinase.
Tumors with RTK point mutations also harbored concurrent RAS/RAF alterations in a significant fraction of cases. Mutations in the EC domain of EGFR and ERBB2 were observed in the presence of co-occurring KRAS mutations in 4 of 10 cases (40%) and 6 of 15 cases (40%), respectively. Half the cases with ERBB2 KD mutations harbored concurrent RAS/RAF mutations (35 of 69 cases; 51%) (Fig. 3), and in 28 of 35 of these RAS/RAF positive cases, the concurrent alteration was an activating KRAS mutation at G12 or 13 (data not shown). Notably, cases with activating ERBB2 truncations or RTK rearrangements lacked co-occurring RAS/RAF mutations in 3 of 4 cases (75%) and 14 of 16 cases (88%) (Fig. 3).
The majority of tumors with MEK1 mutations (31 of 41; 76%) did not harbor co-occurring RAS/RAF alterations. However, PIK3CA mutations co-occurred with RAS/RAF alterations in 80% of cases (596 of 746), including 509 of 746 cases (68%) with concurrent KRAS mutations (Fig. 3), and also co-occurred with other potential driver alterations (Fig. 2B). PTEN inactivating alterations also co-occurred with RAF/RAF alterations in 75% of cases (216 of 289) (Fig. 3).
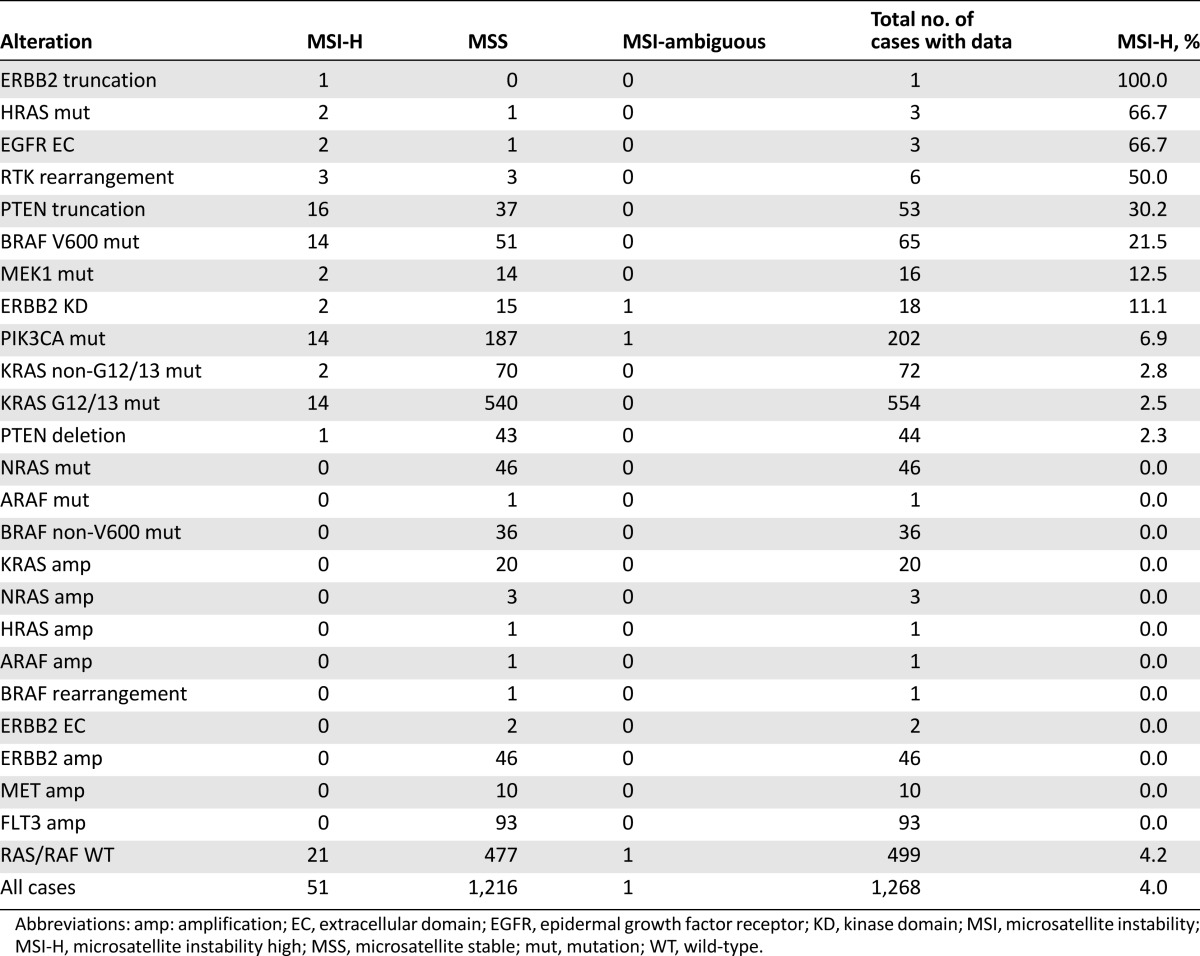
Microsatellite Instability
MSI data were available for a subset of patients with advanced CRC in this series. Overall, 4.0% of tumors (51 of 1,268) were MSI-H, and 96% of cases were MSS. Similarly, 4.2% of RAS/RAF wild-type cases were MSI-H. However, further analysis of alteration subclasses revealed that BRAF V600 mutated tumors were MSI-H in 21.5% of cases (14 of 65), and microsatellite instability was also enriched to a lesser degree in tumors with MEK1, ERBB2 KD, and PI3K mutations (Table 2). Interestingly, 30% of tumors with PTEN truncation were MSI-H, compared with only 2% of tumors with PTEN homozygous deletion. Tumors with KRAS mutations, regardless of whether the mutation affected the codon 12/13 hotspot region, were less likely overall to harbor microsatellite instability (MSI-H in 2.5% of cases). In this series, tumors with available data harboring other activating alterations, including NRAS mutations (n = 46), BRAF non-V600 mutations (n = 36), RAS amplification (n = 24), ERBB2 amplification (n = 46), MET amplification (n = 10), and FLT3 amplification (n = 93), were exclusively MSS (Table 2).
Table 2.
MSI in colorectal cases categorized by genomic alteration
Discussion
Using CGP, we set out to characterize the genomic landscape of advanced CRC in clinical practice with a focus on coexisting alterations that may underlie the poor response to EGFR-directed therapies in patients with reportedly KRAS wild-type tumors, and to identify other clinically relevant genomic alterations. Largely consistent with prior reports, we identified KRAS mutations in 51% of cases (2,251 of 4,422) and other RAS/RAF alterations in an additional 507 cases. KRAS amplification was found in 75 cases (1.7%)—slightly higher than in a previous study that showed amplification as well as mutation was associated with resistance to cetuximab and panitumumab [27]. Importantly, our series differs from prior analyses in both the size of the data set (4,422 vs. 276 in the Cancer Genome Atlas) and use of samples from advanced disease more reflective of the population considered for EGFR-directed therapies [22]. Although EGFR protein expression detected by immunohistochemistry was the original biomarker used to guide selection of anti-EGFR monoclonal antibody therapies in CRC, this test is no longer clinically relevant and was not evaluated in the current study [28]. A limitation of this study is the lack of interpretable clinical outcomes in patients with these alterations who were treated with targeted therapy. However, for patients treated with EGFR monoclonal antibodies, these therapies were given in combination with various chemotherapy backbones in the majority of cases identified, making clear assessment of the effect of EGFR inhibitor alone difficult to interpret. Furthermore, access to and treatment with targeted therapies including kinase inhibitors is still relatively uncommon in clinical practice for CRC and other gastrointestinal malignancies, making identification of these cases onerous, particularly as a number of these patients are currently enrolled in active clinical trials. Future studies focused on genomic analysis of patients treated with anti-EGFR monotherapy, as well as continued efforts to identify and assess CRC patients treated with other targeted therapies, are needed.
Using CGP, we identified nearly 1 in 3 patients with RAS/RAF wild-type tumors (507 of 1,644) that harbored at least 1 concurrent alteration that could potentially mediate resistance to an EGFR therapeutic antibody. Furthermore, in patients negative for alterations commonly detected by hotspot tests, including KRAS codons 12/13 and BRAF V600 mutations, CGP identified more than 1 in 2 patients (1,159 of 2,224) whose tumors harbored at least 1 concurrent alteration that could potentially mediate EGFR antibody resistance. In a significant portion, the additional alteration was itself a validated alternative therapeutic target in CRC. Interestingly, the response rates to combination chemotherapy and anti-EGFR therapy from phase III trials of KRAS wild-type (limited KRAS testing) only approach 60%, indirectly suggesting that our observations may partly underlie the clinical observation [12]. Potential cetuximab/panitumumab resistance alterations in RAS/RAF wild-type tumors included alterations that impact antibody binding, bypass track activation, and alteration of downstream signaling cascade nodes—all well-established resistance mechanisms in cancer. Specifically, we observed EGFR EC domain mutations, which have been shown to disrupt antibody binding, and amplification of MET and FLT3, all of which have been associated with resistance to anti-EGFR therapy in CRC [19, 29–32]. Concurrent or compensatory ERBB2 amplification is a poor prognostic indicator independent of KRAS and ERBB2 mutations, and mediates resistance in a portion of anti-EGFR-treated patients [33, 34]. We observed a 5% rate (n = 219) of ERBB2 mutation or amplification, which was enriched in RAS/RAF wild-type CRC (n = 148; 9%; p < .001).
Activating alterations downstream of RTKs are well-established resistance mechanisms to RTK-targeted therapies and we identified activating mutations in PIK3CA and MAP2K1 (MEK1) and inactivating alterations in PTEN. MEK1 mutations have been identified in CRC patients with primary resistance to panitumumab, as well as acquired resistance to BRAF-directed therapy [19, 35]. PI3K pathway alterations, including PIK3CA mutation and PTEN mutation and loss of expression, have been associated with poor overall response rate and shorter overall survival in patients with KRAS wild-type CRC treated with cetuximab and panitumumab [36–38]. However, PIK3CA mutation has been reported in both pre- and post-treatment biopsy specimens from CRC patients who have developed resistance to anti-EGFR therapies, and we and others observed high co-occurrence with other alterations, including RAS mutations, suggesting that the functional significance of PIK3CA mutation in CRC is less clear [21, 29]. The frequencies of RTK and MEK1 alterations reported here were similar to those observed in a recent smaller study, which suggested a poor response rate to anti-EGFR antibodies in patients with RAS wild-type CRC with these alterations [39]; however, we did identify some co-occurrence of MEK1 and RAS/RAF mutations.
To approximate the fraction of cases tested using CGP that were likely tested in the setting of acquired resistance to targeted therapy, we assessed whether the tumor biopsy specimen submitted for CGP was taken from a primary site (colon or rectum) or from a metastatic site (most commonly liver or lung). For cases with available data, 46% (2,009 of 4,343) were metastatic (supplemental online Table 1). Unsurprisingly, tumors with EGFR EC domain mutations known to disrupt EGFR antibody binding were found in metastatic biopsy specimens in 8 of 10 cases. Generally, RAS and RAF amplification, as well as ERBB2 EC domain mutation and MEK1 mutation, were also associated with metastatic tumors. Notably, ERBB2 KD mutations and BRAF V600 mutations were more commonly found in primary tumors of the colon or rectum (supplemental online Table 1). These observations may indicate that particular genomic alterations are more likely to occur pretreatment as primary drivers in CRC, whereas others arise post-treatment; however, further analysis is warranted.
In addition to predicting lack of sensitivity to EGFR-targeted therapies, alterations in several assayed genes have been associated with sensitivity to alternate targeted therapies. ERBB2 amplification and mutation have emerged as targetable alterations in CRC (5% in our series) and two trials targeting ERBB2 are ongoing in this molecularly defined group (i.e., NCT01960023 and NCT01862003 [http://www.clinicaltrials.gov]) [32, 40, 41]. Response to the multikinase inhibitor sorafenib was also observed in a CRC patient with FLT3 amplification [42]. Furthermore, a recent study reported a response to panitumumab in combination with trametinib in a patient with a MEK1 K57T mutation [43]. A subset of colorectal tumors with neuroendocrine features and BRAF V600E mutations is also predicted to respond to combined inhibition with BRAF and MEK inhibitors [44]. Although EGFR EC mutations can disrupt cetuximab and panitumumab binding, newer generations of therapeutic antibodies have shown promise in overcoming this resistance and may be effective in CRC patients with these alterations [45]. Although less common (0.4% overall), chromosomal rearrangements involving ALK, RET, and NTRK1 in CRC are emerging as sensitive to appropriate targeted therapies [46–48]. Interestingly, rearrangements were mutually exclusive from RAS/RAF alterations in 88% of cases (14 of 16), possibly suggesting an oncogene-driven, biologically distinct subset of CRC requiring further investigation.
Patients with mismatch-repair-deficient CRC have been shown to have significantly improved response rates to antiprogrammed death 1 (PD-1) immune checkpoint inhibitor therapy compared with patients with mismatch repair-proficient tumors [49]. Herein, we found that 4.0% of CRC cases for which data were available, including a similar fraction of RAS/RAF wild-type tumors, were mismatch-repair deficient. This is lower than the 15% MSI-H level consistently reported in the literature; however, cases submitted for CGP are often metastatic, and also are most likely biased toward those determined to be MSS by prior testing and thus have not been identified as candidates for immunotherapy [50]. Interestingly, tumors with BRAF V600 mutations or PTEN truncation appeared to be enriched for mismatch-repair deficiency, suggesting that both PD-1/PD-L1 and BRAF/MEK- or PI3K/AKT-targeted therapies, respectively, or possibly a combination of immunotherapy and kinase-targeted therapy, could be effective in these patients. In contrast, mismatch-repair deficiency was not detected in tumors with other potential driver alterations, including ERBB2, MET, and KRAS amplification. A recent report has described a similar method from MSI testing arising from next-generation sequencing [51].
Among all CRC cases analyzed, we identified non-codon 12/13 KRAS mutations in 284 (6.4%). Multiple studies have shown that alterations across KRAS exons 2, 3, and 4 similarly predict cetuximab and panitumumab resistance, and the National Comprehensive Cancer Network (NCCN) and American Society for Clinical Oncology (ASCO) guidelines have expanded to recommend extended KRAS testing, as well as testing for NRAS and BRAF mutations [6, 9, 15, 16, 52]. Additionally, KRAS insertions within or adjacent to hotspot regions have been reported; these mutations have been characterized as activating and oncogenic, and may similarly predict resistance, although this has not yet been established [53–56].
There is no uniform standard for KRAS testing and the majority of commonly used, commercially available assays assess only point mutations at codons 12 and 13, and lack sufficient coverage to detect less frequent KRAS alterations, which have been shown to predict resistance to anti-EGFR therapy [57, 58]. Molecular test results returned to treating physicians can be misleading because published validation for the majority of assays indicating performance limitations is not readily available. Unfortunately, use of standard KRAS testing without understanding the limitations can lead to incorrect reporting of false-negative results for patients harboring activating KRAS alterations, including point mutations at Q61 and A146, as well as less common activating point mutations and insertions. We report here that 88% of KRAS non-12/13 mutations detected by CGP (79 of 90) were missed by prior hotspot testing. Given the established predictive value of these alterations and the expansion of current NCCN and ASCO guidelines, addressing the limitations of KRAS testing methods currently used in clinical practice is essential.
Conclusion
Beyond standardizing expanded RAS testing methodologies, interrogating tumors for RAS amplification, EGFR EC mutations, and targetable driver alterations in other genes, including ERBB2 amplification and activating point mutations, MET amplification, and RTK rearrangements, should also be considered because these alterations occur in a significant fraction of patients with RAS/RAF wild-type CRC. CGP in the course of clinical care provides sensitive detection of all classes of RAS/RAF, RTK, PI3K/AKT, and MEK/ERK pathway alterations and should be used at a minimum for CRC patients who test negative for KRAS or BRAF mutations using limited panels, to detect alterations predicting both lack of response to EGFR antibodies as well as response to other available targeted therapies. Moreover, given the co-occurrence of canonical KRAS mutations with other oncogenic drivers, CGP may also reveal targetable alterations in KRAS mutant CRC. Thus, CGP could provide benefit when used as first-line testing for metastatic CRC. With the complexities of cancer genomics and the expansion of targeted compounds and immunotherapy, we anticipate CGP to play an increasing role in personalizing the treatment of advanced CRC.
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Material
Footnotes
For Further Reading: Roberto Moretto, Chiara Cremolini, Daniele Rossini et al. Location of Primary Tumor and Benefit From Anti-Epidermal Growth Factor Receptor Monoclonal Antibodies in Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer. The Oncologist 2016;21:988–994.
Implications for Practice: Right- and left-sided colorectal tumors have peculiar epidemiological and clinicopathological characteristics, distinct gene expression profiles and genetic alterations, and different prognoses. This study assessed the potential predictive impact of primary tumor site with regard to anti-epidermal growth factor receptor (EGFR) monoclonal antibody treatment in patients with RAS and BRAF wild-type metastatic colorectal cancer. The results demonstrated the lack of activity of anti-EGFRs in RAS and BRAF wild-type, right-sided tumors, thus suggesting a potential role for primary tumor location in driving treatment choices.
Author Contributions
Conception/Design: Andrew Rankin, Samuel J. Klempner, Philip J. Stephens, Vincent A. Miller, Siraj M. Ali, Alexa B. Schrock
Collection and/or assembly of data: Andrew Rankin, James X. Sun, Alexa B. Schrock
Data analysis and interpretation: Andrew Rankin, Samuel J. Klempner, Rachel Erlich, James X. Sun, Axel Grothey, Marwan Fakih, Thomas J. George Jr., Jeeyun Lee, Jeffrey S. Ross, Philip J. Stephens, Vincent A. Miller, Siraj M. Ali, Alexa B. Schrock
Manuscript writing: Andrew Rankin, Thomas J. George Jr., Alexa B. Schrock
Final approval of manuscript: Andrew Rankin, Samuel J. Klempner, Rachel Erlich, James X. Sun, Axel Grothey, Marwan Fakih, Thomas J. George Jr., Jeeyun Lee, Jeffrey S. Ross, Philip J. Stephens, Vincent A. Miller, Siraj M. Ali, Alexa B. Schrock
Disclosures
Andrew Rankin: Foundation Medicine (E, OI); Samuel J. Klempner: Foundation Medicine (H); Rachel Erlich: Foundation Medicine (E, OI); James X. Sun: Foundation Medicine (E, OI); Axel Grothey: Genentech, Bayer, Eisai, Boehringer Ingelheim, Boston Biomedical (RF); Marwan Fakih: Sirtex, Amgen (H), Novartis, Amgen (RF); Thomas J. George Jr.: NewLink Genetics, Bayer (C/A), Bayer, Bristol-Myers Squibb, NewLink Genetics, Eli Lilly (RF); Jeffrey S. Ross: Foundation Medicine (RF, E, OI); Philip J. Stephens: Foundation Medicine (E, OI); Vincent A. Miller: Foundation Medicine (E, OI); Siraj M. Ali: Foundation Medicine (E, OI, IP); Alexa B. Schrock: Foundation Medicine (E, OI). The other author indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Cremolini C, Loupakis F, Antoniotti C, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 4.Cremolini C, Loupakis F, Falcone A. FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2015;372:291–292. doi: 10.1056/NEJMc1413996. [DOI] [PubMed] [Google Scholar]
- 5.Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 6.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 7.Douillard JY, Siena S, Cassidy J, et al. Final results from PRIME: Randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:1346–1355. doi: 10.1093/annonc/mdu141. [DOI] [PubMed] [Google Scholar]
- 8.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 9.Van Cutsem E, Köhne C-H, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 10.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 11.Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: The OPUS study. Ann Oncol. 2011;22:1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 12.Misale S, Di Nicolantonio F, Sartore-Bianchi A, et al. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 13.De Roock W, De Vriendt V, Normanno N, et al. KRAS, BRAF, PIK3CA, and PTEN mutations: Implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 14.Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris VK, Lucas FAS, Overman MJ, et al. Clinicopathologic characteristics and gene expression analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal cancer. Ann Oncol. 2014;25:2008–2014. doi: 10.1093/annonc/mdu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loupakis F, Ruzzo A, Cremolini C, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCCN Guidelines v.3.2015. Available at https://www.nccn.org/professionals/physician_gls/f_guidelines.asp. Accessed June 8, 2016.
- 18.Bertotti A, Sassi F. Molecular pathways: Sensitivity and resistance to anti-EGFR antibodies. Clin Cancer Res. 2015;21:3377–3383. doi: 10.1158/1078-0432.CCR-14-0848. [DOI] [PubMed] [Google Scholar]
- 19.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 21.Perrone F, Lampis A, Orsenigo M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall MJ, Gowen K, Sanford EM, et al. Evaluation of microsatellite instability (MSI) status in gastrointestinal (GI) tumor samples tested with comprehensive genomic profiling (CGP) J Clin Oncol. 2016;34(suppl 4S):528a. [Google Scholar]
- 26.Wan PTC, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 27.Valtorta E, Misale S, Sartore-Bianchi A, et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int J Cancer. 2013;133:1259–1265. doi: 10.1002/ijc.28106. [DOI] [PubMed] [Google Scholar]
- 28.Ross JS. Biomarker-based selection of therapy for colorectal cancer. Biomarkers Med. 2011;5:319–332. doi: 10.2217/bmm.11.38. [DOI] [PubMed] [Google Scholar]
- 29.Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21:2157–2166. doi: 10.1158/1078-0432.CCR-14-2821. [DOI] [PubMed] [Google Scholar]
- 30.Braig F, März M, Schieferdecker A, et al. Epidermal growth factor receptor mutation mediates cross-resistance to panitumumab and cetuximab in gastrointestinal cancer. Oncotarget. 2015;6:12035–12047. doi: 10.18632/oncotarget.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montagut C, Dalmases A, Bellosillo B, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer [published correction appears in Nat Med 2012;18:1445] Nat Med. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 32.Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–673. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5:832–841. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahronian LG, Sennott EM, Van Allen EM, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 37.Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 38.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 39.Hechtman JF, Zehir A, Yaeger R, et al. Identification of targetable kinase alterations in patients with colorectal carcinoma that are preferentially associated with wild-type RAS/RAF. Mol Cancer Res. 2016;14:296–301. doi: 10.1158/1541-7786.MCR-15-0392-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Disel U, Germain A, Yilmazel B, et al. Durable clinical benefit to trastuzumab and chemotherapy in a patient with metastatic colon adenocarcinoma harboring ERBB2 amplification. Oncoscience. 2015;2:581–584. doi: 10.18632/oncoscience.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurwitz H, Hainsworth JD, Swanton C, et al. Targeted therapy for gastrointestinaI (GI) tumors based on molecular profiles: Early results from MyPathway, an open-label phase IIa basket study in patients with advanced solid tumors. J Clin Oncol. 2016;34(suppl 4S):653a. doi: 10.1200/JCO.2017.75.3780. [DOI] [PubMed] [Google Scholar]
- 42.Moreira RB, Peixoto RD, de Sousa Cruz MR. Clinical response to sorafenib in a patient with metastatic colorectal cancer and FLT3 amplification. Case Rep Oncol. 2015;8:83–87. doi: 10.1159/000375483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 2016;6:147–153. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klempner SJ, Gershenhorn B, Tran P, et al. BRAFV600E mutations in high-grade colorectal neuroendocrine tumors may predict responsiveness to BRAF-MEK combination therapy. Cancer Discov. 2016;6:594–600. doi: 10.1158/2159-8290.CD-15-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arena S, Siravegna G, Mussolin B, et al. MM-151 overcomes acquired resistance to cetuximab and panitumumab in colorectal cancers harboring EGFR extracellular domain mutations. Sci Transl Med. 2016;8:324ra14. doi: 10.1126/scitranslmed.aad5640. [DOI] [PubMed] [Google Scholar]
- 46.Yakirevich E, Resnick MB, Mangray S, et al. Oncogenic ALK Fusion in rare and aggressive subtype of colorectal adenocarcinoma as a potential therapeutic target. Clin Cancer Res. 2016;22:3831–3840. doi: 10.1158/1078-0432.CCR-15-3000. [DOI] [PubMed] [Google Scholar]
- 47.Russo M, Misale S, Wei G, et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6:36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 48.Le Rolle A-F, Klempner SJ, Garrett CR, et al. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget. 2015;6:28929–28937. doi: 10.18632/oncotarget.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34:2141–2147. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allegra CJ, Rumble RB, Hamilton SR, et al. Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology provisional clinical opinion update 2015. J Clin Oncol. 2016;34:179–185. doi: 10.1200/JCO.2015.63.9674. [DOI] [PubMed] [Google Scholar]
- 53.Bollag G, Adler F, elMasry N, et al. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem. 1996;271:32491–32494. doi: 10.1074/jbc.271.51.32491. [DOI] [PubMed] [Google Scholar]
- 54.Tong JHM, Lung RWM, Sin FMC, et al. Characterization of rare transforming KRAS mutations in sporadic colorectal cancer. Cancer Biol Ther. 2014;15:768–776. doi: 10.4161/cbt.28550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Macedo MP, de Lima LGCA, Begnami MDFS, et al. KRAS insertions in colorectal cancer: what do we know about unusual KRAS mutations? Exp Mol Pathol. 2014;96:257–260. doi: 10.1016/j.yexmp.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Wójcik P, Kulig J, Okoń K, et al. KRAS mutation profile in colorectal carcinoma and novel mutation--internal tandem duplication in KRAS. Pol J Pathol. 2008;59:93–96. [PubMed] [Google Scholar]
- 57.Tougeron D, Lecomte T, Pagès JC, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2013;24:1267–1273. doi: 10.1093/annonc/mds620. [DOI] [PubMed] [Google Scholar]
- 58.Malapelle U, Carlomagno C, de Luca C, et al. KRAS testing in metastatic colorectal carcinoma: Challenges, controversies, breakthroughs and beyond. J Clin Pathol. 2014;67:1–9. doi: 10.1136/jclinpath-2013-201835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.