Abstract
Proteomics is a systems physiology discipline to address the large-scale characterization of protein species within a biological system, be it a cell, a tissue, a body biofluid, an organism, or a cohort population. Building on advances from chemical analytical platforms (e.g., mass spectrometry and other technologies), proteomics approaches have contributed powerful applications in cardiovascular biomedicine, most notably in: 1) the discovery of circulating protein biomarkers of heart diseases from plasma samples; and 2) the identification of disease mechanisms and potential therapeutic targets in cardiovascular tissues, in both preclinical models and translational studies. Contemporary proteomics investigations offer powerful means to simultaneously examine tens of thousands of proteins in various samples, and understand their molecular phenotypes in health and disease. This concise review introduces study design considerations, example applications and use cases, as well as interpretation and analysis of proteomics data in cardiovascular biomedicine.
Keywords: mass spectrometry, molecular phenotyping, protein signatures, post-translational modifications, protein arrays, protein dynamics
Overview
Over the past 20 years, there has been an accelerating growth in the application of proteomics in cardiovascular biomedicine, building upon pioneering studies with translational significance (1,2) and mechanistic insights (3–8). The majority of human genes functions to create proteins, which are the molecular workhorses that carry out virtually all metabolic, signaling, and physiological functions in life. Many human diseases may be characterized by the elicited changes in proteome configurations. In complex late-onset diseases in particular, considerable environmental components exist, such that individual genetic variants are often poorly predictive of disease states. Telltale molecular signatures of acquired cardiovascular diseases may instead manifest through intermediate phenotypes, including transcript and protein abundance. Beyond what may be learned from gene and transcript information alone, molecular changes at the protein layer, in particular, can provide independent insights into disease mechanisms in the following 3 areas:
First, it has gradually emerged from large-scale studies that transcript and protein changes in a system may, at times, correspond poorly—with transcript levels explaining as little as 10% to 30% of variations in protein abundance. Although the exact contribution of transcripts to protein-level abundance is debated, it is evident that in the heart and other organs, a large number of post-transcriptional modulators can alter disease-driver protein expression and function during pathogenesis, without changes in transcript abundance (9,10). These modulators include components of the nonsense-mediated decay pathway, long noncoding RNAs, and microRNAs (11–14).
Secondly, plasma proteins provide an accessible readout of the status of potentially all tissues, and are the sources of many current biomarkers in use (Table 1). Clinically useful biomarkers can include circulating proteins that have been secreted or leaked directly into the plasma from resident cells (e.g., myocytes) following diseases or injuries (15), and are thus spatially uncoupled from the transcript change at the tissue of origin. The tissues of origin of circulating proteins may be unknown, inaccessible, or invasive to procure, making it impractical to hunt for these potential biomarkers via transcript measurements, instead of protein measurements.
Table 1.
Selected Protein Biomarkers for Cardiovascular Diseases
| Protein Biomarker | Abbreviation | Disease Relevance | Required Assay Sensitivity (Estimate) |
*Discovery Period |
Reference |
|---|---|---|---|---|---|
| Apolipoprotein A-I | APOA | Cardiovascular event risk |
~1 mg/ml | Mid-1980s | (96) |
| Apolipoprotein B | APOB | Cardiovascular event risk |
~1 mg/ml | Mid-1980s | (97) |
| B-type natriuretic peptide |
BNP | Heart failure, acute coronary syndrome |
~100 pg/ml | Early 2000s | (98) |
| C-reactive protein | CRP | Cardiovascular event risk |
~10 µg/ml | Late 1990s | (99) |
| Creatine kinase- myocardial band |
CK-MB | Acute myocardial infarct, myocardial necrosis |
~1 ng/ml | 1960s–1970s | (100) |
| Cystatin-C | CST3 | Cardiovascular event risk |
~1 µg/ml | 2000s | (101) |
| Fibrinogen | FBN | Cardiovascular event risk |
~1 mg/ml | 1980s | (99) |
| Lipoprotein- associated phospholipase A2 |
Lp-PLA2 | Coronary heart disease risk |
~100 ng/ml | 2000s | (102) |
| Myeloperoxidase | MPO | Ischemic heart disease; acute coronary syndrome |
~10 ng/ml | 1980s | (103) |
| Myoglobin | MYO | Myocardial infarction, necrosis |
~ 10 ng/ml | Late 1970s | (104) |
| Serum amyloid A | SAA | Coronary artery disease |
~10 µg/ml | 1990s | (105) |
| Troponin I | cTnI | Myocardial injury, myocardial infarction |
~10 pg/ml | 1970s–1990s | (106) |
| Troponin T | cTnT | Myocardial injury, myocardial infarction |
~10 pg/ml | 1970s–1990s | (107) |
Thirdly, disease processes may be mediated not by the altered abundance of gene products, but by other functional parameters of the proteome, including protein post-translational modifications (PTMs), protein-protein interactions, and protein degradation, which take place after the protein molecules are synthesized and cannot be predicted from genetic information a priori.
Proteomics technologies allow researchers to measure protein function on a large scale, and hence interrogate the molecular layer that closely abuts physiological phenotypes (16). However, the complexity of the human proteome also presents a daunting analytical challenge for large-scale characterization. For example, circulating proteins in human plasma, found at a concentration of ~70 mg/ml, are estimated to comprise at least 10,000 distinct protein species (17) over a concentration range of >10 orders of magnitude 10-billion-fold differences) (18). Approximately 3,000 proteins in plasma may be viewed as classical resident plasma proteins, which tend to occupy the higher end of the abundance spectrum. These resident proteins include extremely abundant species, such as albumin, immunoglobulins, transferrins, and fibrinogens, which are found in the mg/ml concentration range and account for >90% of plasma protein content. Other plasma proteins come from secretion or leakage from various organs before being diluted into the 5 liters of peripheral blood, and are found in the µg/ml to ng/ml range. Very low-abundance proteins, including interleukins 6 and 12, tumor necrosis factor (TNF)-alpha, and other cytokines, circulate in the pg/ml range, and are typically masked by higher-abundance proteins in untargeted proteomics analyses (Figure 1). Adding to this complexity, plasma proteins may be proteolytically processed, creating numerous degradation product species, and repetitive sequences in proteins can give a spurious appearance of higher abundance. These challenges are compounded by the absence of a protein equivalent of the polymerase chain reaction, such that amplification of minute samples is not possible.
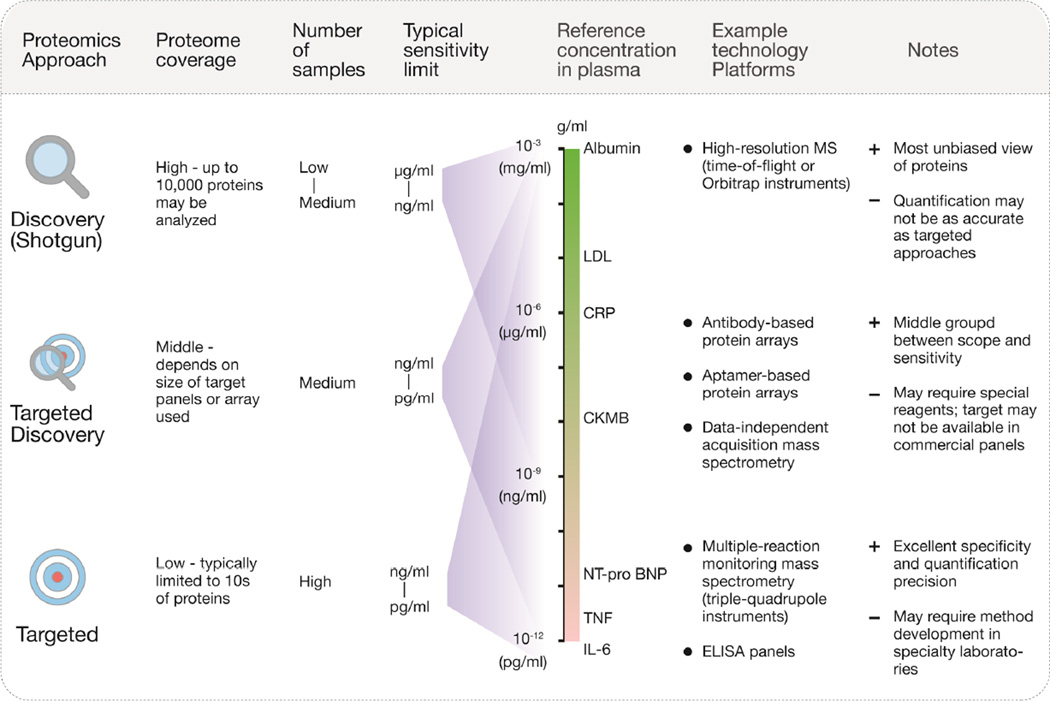
Figure 1. Sensitivity and Coverage of Various Proteomics Study Designs.
Properties of 3 common proteomics study designs are listed. Discovery (shotgun), targeted, and targeted discovery proteomics approaches take different strategies to address proteome coverage, number of samples, and sensitivity limits. Discovery proteomics experiments have high coverage (up to 10,000 proteins in a single sample), but have limitations in throughput; fewer samples/subjects can be analyzed. Targeted discovery approaches focus on analyzing a panel of high-potential targets in sufficient numbers of samples. Targeted proteomics is able to achieve the highest sensitivity, which allows the detection of low-abundance plasma markers, such as TNF and IL-6, but at the expense of scope and throughput. Graph (center) shows the typical sensitivity range of discovery, targeted discovery, and targeted approaches, juxtaposed with the concentrations of selected plasma proteins and disease markers. Example technological platforms and additional notes are shown on the right. CK-MB = creatine kinase-myocardial band; CRP = C-reactive protein; ELISA = enzyme-linked immunosorbent assay; IL-6 = interleukin 6; LDL = low-density lipoprotein; MS = mass spectrometry; NT-proBNP = N-terminal B-type natriuretic peptide; TNF = tumor necrosis factor.
In the following 3 segments of this review, we will discuss 3 aspects of proteomics applications in cardiovascular biomedical research: 1) study design for biomarker research and discovery, with discussions on approaches to circumvent analytical challenges; 2) characterization of multidimensional protein parameters in disease mechanism research; and 3) interpretation and validation of proteomics data/methodologies.
Study Designs for Biomarker Research and Discovery
With precision medicine initiatives demanding a broader pool of measurable proxies of individual characteristics, the call for investigations to discover new candidate biomarkers will likely continue to increase. Successful screens of protein biomarker candidates require obtaining data: 1) with good coverage of all the proteins present in a sample; and 2) in a sufficient number of subjects to ensure adequate power for discovery. Unlike microarrays or RNA sequencing (RNA-seq), a single proteomics experiment often cannot simultaneously obtain high coverage and throughput, necessitating tradeoffs in experimental designs. The advantages and disadvantages of technological platforms should therefore be evaluated comprehensively on the basis of the scope and depth of analysis required, and determined prior to the commencement of chemical analysis. Three common designs, all of which can provide both qualitative and quantitative data on proteins, are: 1) global discovery; 2) targeted; and 3) targeted discovery. Each has distinct strengths and may be better suited to particular experimental goals. Each may be performed in suitable technological platforms that offer different throughputs, analytical depth, and sensitivity. We provide here a brief summary on the strengths and considerations for each type of approach. The Central Illustration presents an overview of data acquisition and analysis; see Table 2 for a glossary of selected technological terms commonly in use.
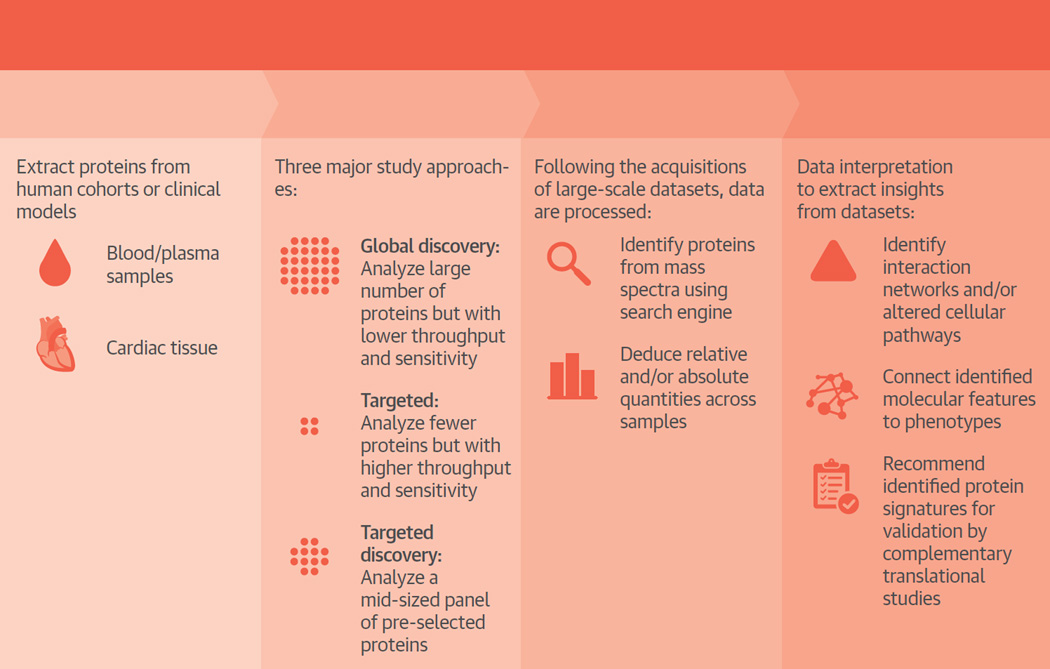
Central Illustration. Overview of Proteomics Analyses for Cardiovascular Diseases.
This figure lists major components in common proteomics workflows, as well as their associated experimental considerations in biomarker discovery and disease studies. From top to bottom, protein samples are collected from the plasma of human cohorts or cardiac tissues in animal models according to study goals (either biomarker discovery or mechanistic studies). Three major study approaches (discovery, targeted, and targeted discovery) take different strategies between proteome coverage and analytical throughput, and utilize different technological platforms, including mass spectrometry and protein arrays. Following the acquisition of large-scale datasets, data are processed to identify the protein species present and to deduce their relative quantities across samples. Subsequently, a number of statistical and bioinformatics workflows are used in data interpretation to extract insights from datasets. Network analysis casts proteins in the context of interaction networks and/or altered cellular pathways. Statistical learning and modeling methods connect the identified molecular features to orthogonal phenotypes, identify signatures, and offer information on subject classification or predictive analysis. The identified protein signatures will require validation, which can be achieved by complementary translational studies, including in vitro biochemical analysis and large cohorts. PTM = post-translational modifications.
Table 2.
Glossary of Selected Technologies Used in Cardiovascular Proteomics
| Terms | Description |
|---|---|
| Amino acid sequence |
A segment of the primary structure of the protein where individual amino acids are linked by peptide bonds. |
| Aptamer array | A non-MS proteomics technology where single-stranded DNA oligonucleotides with affinity for specific protein targets are used as a means to detect and quantify up to ~1,000 different protein targets. |
| Liquid Chromatography |
A common front-end separation technology in proteomics experiments to fractionate complex sample mixture. It is used to resolve and enrich peptides by their chemical properties such as size, hydrophobicity, or charge, and the like. |
| Mass spectrometry | An analytical technology to measure the mass of an analyte (e.g., peptide, protein, amino acid, and so on) in the format of mass-to- charge ratio. |
| Mass spectrum | The data output of MS. A mass spectrum is a plot displaying the relative intensity or abundance of the analyte ions (y-axis) and their mass-to-charge ratio (x-axis). |
| Mass/charge (m/z) | The principal measurement of a mass spectrometer: the mass of the ionized analytes divided by the charge they carry. Also commonly referred to as mass-to-charge ratio. |
| Multiple-reaction monitoring |
A targeted proteomics approach in which a particular type of mass spectrometer is programmed to only measure a few peptides and their fragment ions of a target protein of interest, while filtering out all other peptides and proteins in the sample, thus enhancing specificity and sensitivity of the detection/quantification of the targets. Also known as selected reaction monitoring (SRM). |
| Search engine | A software tool used in sequence database search to identify proteins from MS data (e.g., SEQUEST (110)). |
| Sequence database search |
A method to identify proteins from MS data by matching the collected experimental mass spectra to theoretical mass spectra generated from a protein sequence database, such as UniProt (19) or RefSeq (111). |
| Spectral library search |
A method to identify peptides by matching MS data to previously known experimental spectra in a library, such as COPaKB (21) or PeptideAtlas (20). |
| Shotgun proteomics | A method to identify and quantify proteins on a large scale, in which proteins are first digested into short segments (peptides) that are amenable to sequence determination via tandem MS analysis. The identity of each present protein is then inferred from the sequenced peptides. Also known as bottom-up proteomics. |
| Tandem mass spectrometry |
An analytical method involving the fragmentation of peptides (or other analytes) via multiple stages of MS analyses to obtain sequence/structural information of the analytes. Also known as MS/MS or MS2. |
| Tandem mass tags (TMT)/iTRAQ |
Commercially available chemical labels containing stable isotopes, which can be used to label protein samples for multiplexed analysis and comparison. |
iTRAQ = isobaric tags for relative and absolute quantitation; MS = mass spectrometry
Global Discovery Proteomics
Global discovery approaches offer an unbiased view of the protein species that may be found in a sample. Shotgun proteomics is one of the most widely used and standardized global discovery strategies. In shotgun analysis, proteins are first enzymatically digested (e.g., using trypsin) into peptides in vitro, and then subjected to tandem mass spectrometry (MS) analysis. The resulting mass spectra are then searched against a protein sequence database (19) or spectral library (20,21) for protein identification. Various downstream data analyses, such as spectral counting and network analysis may be performed to extract other qualitative and quantitative information from the data. A major strength of shotgun proteomics is that it casts the widest net towards potential biomarkers (covering thousands of proteins) (22) and does not require the development of individual assays for each target (23,24). Two weaknesses are its lower overall sensitivity, which may bias it against biologically significant proteins at low abundance (see later discussion on Improving Detection of Low-Abundance Proteins and Throughput), and less accurate quantification compared with targeted proteomics.
Targeted Proteomics
Targeted proteomics offers the highest sensitivity and specificity among MS-based proteomics approaches. MS-based targeted approaches require the programming of a specific instrument (e.g., a triple quadrupole mass spectrometer), and development of methods (e.g., multiple reaction monitoring [MRM]) to monitor only a limited number of analytes (via predetermined peptide mass/charge and fragment information) to achieve its high sensitivity. It was demonstrated that targeted MS, in conjunction with stable isotope dilution, was able to verify a panel of cardiovascular biomarker candidates at a limit of quantification (LOQ) ranging from 2 to 15 ng/ml (25). Targeted MS is also applicable to post-translationally modified proteins with PTM site specificity (26), with the absolute quantity of 14 phosphorylation sites on cardiac Troponin I as a notable example (27). Ongoing effort is being devoted to the development of targeted protein assays aimed at cardiac biomarker analysis (28–30). A disadvantage of targeted proteomics is that the practice of only utilizing 1 or a few peptides in the experiments may not represent the behavior of the full-length protein (e.g., due to proteolysis). Specific assays must be developed for each protein being targeted, which requires substantial technical expertise from specialized laboratories. However, once developed, other investigators and facilities in clinical and research settings may readily adapt the assays. Due to the labor and costs required to validate assays, the number of proteins that can be realistically analyzed in a targeted biomarker search may be limited (~100 or fewer). Accordingly, targeted proteomics is most suitable for characterizing subsets of likely candidate biomarkers with prior disease implications. A limited number of non-MS-based antibody assays are also available for targeted quantification of low-abundance proteins in the plasma (e.g., enzyme-linked immunosorbent assays [ELISA]).
Targeted Discovery Approaches
A middle ground between a targeted and an untargeted approach is sometimes sought, which may be called targeted discovery. The goal is to attain the best of both worlds: focusing on a finite panel of targets, while maintaining sufficient throughput to analyze large number of samples. Oftentimes, this comes at the expense of scope (number of proteins analyzed) and flexibility, but, on the upside, panels of “likely bets” can be preselected by manufacturers or by the investigators to increase the probability of discovery (31). A number of MS-based (e.g., sequential window acquisition of all theoretical mass spectra [SWATH] (32)) and non-MS-based affinity proteomics (e.g., antibody- and aptamer-based arrays (31,33,34)), are being used to discover candidate biomarkers. Antibody-based arrays have been utilized to discover markers for ischemic stroke, using a proteomics chip of 92 high-potential cardiovascular proteins (35), including B-type natriuretic peptide (BNP), growth differentiation factor (GDF)-15, matrix metalloproteinase (MMP)-12, and others. Aptamer arrays that use nucleotides in place of antibodies for target binding have been utilized to discover protein myocardial infarct markers and to evaluate a panel of blood proteins as risk factors for prediction of coronary heart diseases (33,34,36,37).
Improving Detection Of Low-Abundance Proteins And Throughput
To improve detection of low-abundance proteins in plasma for MS-based analysis, commercial immunodepletion columns, including the Human IgY14 and SuperMix columns (Sigma-Aldrich, St. Louis, Missouri), are used to remove top- to medium-abundance proteins. Simplifying the sample mixture helps unmask low-abundance proteins for detection, but many proteins or fragments of interest may be inadvertently codepleted by the process. Typically, a sample containing 100 µl of plasma may only yield ~140 µg of proteins after 98% of high-abundance species are depleted, an amount sufficient for a limited number of replicate analyses. To target challenging proteins in plasma and other samples, additional preprocessing steps, including size-exclusion chromatography, multidimensional liquid chromatography, and immunocapture may be used, but necessitate greater starting amounts of samples (e.g., 0.01 to 1 ml of plasma) and additional processing time.
To achieve multiplexing, minimize experimental variations, and increase throughput, multiple protein samples can be barcoded with stable isotopes and analyzed in a single experiment at the expense of costs. Commercially available isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tag (TMT) labels, small amine-reactive molecules covalently linked to differential mass reporters and mass balancers, can be used to tag peptides such that up to 10 sets of barcoded samples may be coanalyzed in 1 MS experiment. During tandem MS, the mass reporters separate from the balancers and act as surrogates for comparison of protein quantity of each sample. This multiplexing strategy has been profitably used to identify potential infarct markers in a study that analyzed the plasma of 16 patients at multiple time points following septal ablation treatment. With the aid of 4-plex iTRAQ labels, the proteomics result quantified >3,000 human plasma proteins in 16 patients and discovered 333 regulated proteins (38), including known markers of spontaneous myocardial infarction, such as creatine kinase-myocardial band (CK-MB) and troponin. Using 6-plex TMT labels, the proteomic analysis of the plasma samples of ~1,000 human subjects has been recently demonstrated (39). The popularization of new 10-plex TMT labels will further increase throughput.
Multidimensional Protein Parameters in Disease Mechanism Research
Contemporary proteomics applications in basic disease mechanism research often incorporate additional experimental techniques that target increasing numbers of “dimensions” of protein properties. Whereas previous proteomics studies focused largely on gene product abundance (up- and down-regulation in quantity as primary drivers of changes in function), there has been a growing appreciation of other, equally significant proteome parameters that modulate cardiovascular biology. These parameters include post-translational regulation, protein-protein interactions, and protein turnover, each of which may be altered in disease and present potential therapeutic targets (9). These parameters are aiding in efforts to fully comprehend the disease proteome as a dynamic, multidimensional entity. Ongoing developments in these areas may expand the parameter space from which future biomarkers may be found.
Protein Post-Translational Regulation
Post-translational regulation through covalent modification of proteins is a major mechanism of cardiac signaling. Over 200 protein PTMs that can alter the chemistry and interaction surface of a protein, and thereby its structure, activity, binding partners, or subcellular localization, are known (40). Although over 500,000 PTM sites are known to exist (41), available site-specific antibodies target relatively few of them. MS-based proteomics techniques do not require separate reagents (i.e., antibodies) for each site, and hence can be effectively used to examine new sites or sites for which antibodies are not available.
For a number of reasons, PTM studies often require additional sample preparation and enrichment considerations (e.g., with titanium dioxide [TiO2] (42)) before data acquisition,. First, modified proteins frequently exist in lower abundance than the total protein population (although in some instances, such as in the case of secreted glycoproteins, almost every protein molecule is modified). For example, if only 10% of a protein species is modified, then the modified proteins are effectively 10 times more difficult to detect than the total (modified plus unmodified) population. Secondly, whereas identifying a protein only requires detection of any of its multiple proteolytic peptides along its primary structure, PTM analysis requires the precise detection of the peptide containing the PTM site, which sometimes can lie in a region of the protein that is not amenable to MS detection (e.g., sequences without trypsin digestion sites). Thirdly, many PTMs are chemically labile and biologically transient, making their capture and enrichment before signal measurement essential.
Post-translational modifications under intense investigation for their roles in cardiovascular diseases include phosphorylation (43,44), acetylation (45,46), SUMOylation (47), glycosylation (48,49), S-nitrosylation (50,51), ubiquitination (52), and others (53,54). The remainder of this section highlights proteomics approaches to understand protein phosphorylation, acetylation, and oxidative modifications.
Multiple major kinase pathways operate in the heart, including protein kinase A (PKA), protein kinase B (PKB/Akt), mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), protein kinase C (PKC), glycogen synthase kinase 3b (GSK3b), calcium/calmodulin-dependent protein kinase II (CamKII), and others; hence, it is no surprise that phosphoproteomics is commonly applied in the study of heart diseases. A well-known example of a phosphorylated protein essential for cardiac energetics is pyruvate dehydrogenase (PDH) (55), which converts pyruvate to acetyl coenzyme A for respiration, and whose activity is dictated by its phosphorylation by PDH kinases (PDKs). PDH phosphorylation thus serves as a major regulatory checkpoint of cardiac energy metabolism. Classically, phosphorylation sites were mapped by phosphorus 32 (32P)-labeling and Edman degradation. Up to ~50,000 phosphorylation sites may now be identified in a single study using shotgun proteomics (56) combined with enrichment strategies. Mapping in the heart has unearthed a large number of phosphorylation sites, including those common to multiple tissues and many that are unique to the cardiovascular system (57). Targeted mapping efforts may also be directed towards selected cellular compartments (e.g., mitochondria (58)), pathways (e.g., those targeted by phosphodiesterase 9A [PDE9A]/phosphodiesterase 5 [PDE5] (59) and CamKII (60)), specific proteins (e.g., the myosin-binding protein C [MyBP-C] phosphoproteome (61)), or multiple specific sites on the same proteins (26).
Acetylation of histones plays a key role in gene expression by modulating the accessibility of transcriptional elements to DNA wound around the histones. A family of enzymes known as histone acetyltransferases (HATs) catalyzes the addition of the acetyl group to proteins, whereas histone deacetylases (HDACs) revert the modification. Although protein acetylation was classically described on histones, other proteins are also modified. Protein acetylation participates in nonchromatin processes, including metabolism and protein localization, as well as protein stability (45,46), and can be mapped with proteomics techniques following enrichment by anti-acetyl-lysine antibodies (62,63). Over one-third of mitochondrial proteins are subjected to acetylation, including a large number of metabolic enzymes. Acetylation in mitochondria may function in cardioprotective contexts (46), whereas hyperacetylation of succinate dehydrogenase in mitochondria may drive energy imbalance in the development of heart failure (63).
Reactive oxygen and nitrogen species can confer covalent modifications (e.g., S-nitrosylation, S-sulfhydration, and S-glutathionylation) on cysteine residues in redox-sensitive proteins. These modifications can change the activity and binding surfaces of target proteins, and thus directly participate in cardiovascular biology in a number of disease contexts. For example, it is known that nitric oxide (NO) serves as a critical signaling messenger in ischemic preconditioning. NO signals partly by causatively increasing protein S-nitrosylation, which has been proposed to mediate cardioprotection by reducing other, pernicious forms of cysteine oxidation on the target proteins during reperfusion injury (64,65). To detect oxidative modification en masse, biotin switch proteomics are used that first chemically block free thiols, then uses a specific reducing agent to reduce different reductive states of oxidative PTMs (e.g., ascorbic acids may be used to reduce only S-nitrosylated thiols, but not other modifications), and finally labels the freed thiols with biotin or isotope-labeled tags (cys-TMT or iodo-TMT) (51,66).
Protein Temporal Dynamics
Protein homeostasis and proteolysis are broadly implicated in heart diseases and injuries. Classical studies of protein turnover required pulse-chase of radioisotopes and isolation of target proteins. With recent advances, proteomics studies can measure turnover dynamics of thousands of proteins in multiple tissues and organisms (67,68). Combining stable isotope labeling, mathematical modeling, and shotgun proteomics, recent investigations into the turnover of ~5,000 cardiac proteins in mouse cardiac hypertrophy (69,70) revealed novel correlations between proteome profiles and phenotypic functional limitations. For example, although the hypertrophic heart is known to switch in fuel preference from fatty acid to glucose, the expression of glycolytic proteins does not change accordingly (71). Instead, the turnover of most glycolytic proteins was found to accelerate drastically in hypertrophy without concomitant expression changes. Thus static expression profiling experiments alone would not have discovered this aspect of cardiac pathophysiology had there been no a priori knowledge of increased glycolysis, whereas a large-scale experiment of protein turnover might have led to the generation of relevant hypotheses. A possible connection between turnover and function is that old proteins may have reduced activity compared with young, functional proteins. For example, the glycolysis enzyme triose-phosphate isomerase (TPI) can become chemically deamidated upon catalytic cycling. In another example, the stem cell factor/KIT ligand (KITLG) has been found to lose biological potency by 50-fold after chemical deamidation (72).
Protein-Protein Interactions
Physical liaison between proteins is a primary means whereby information is transduced along signaling pathways. The target specificity and activity of PKA are markedly regulated by which of the ~40 A-kinase anchoring proteins (AKAPs) it primarily interacts with (73), to mediate kinase signaling, and which can change during heart disease (74). Protein-protein interaction analysis is pursued to characterize signaling networks, discover the components of protein complexes, and identify the contexts in which a protein may function in health and disease (75). A powerful approach to analyze protein-protein interaction is affinity purification-mass spectrometry (AP-MS) (76,77), which precipitates bait proteins, along with their interacting partners, using antibodies or another purification method. Using unbiased AP-MS, a recent study discovered the association of the cardiac transcription factor TBX5 with the nucleosome remodeling deacetylase (NuRD) repressor complex, revealing a novel mechanism for TBX5 in cardiac development and congenital heart diseases (78). Although it is the current mainstay of interactome research, AP-MS specificity can vary. Even with careful washing, nonspecific binding occurs and contaminants can remain. Thus, identified interactors should be verified by complementary methods. New methods are currently being developed to ameliorate this challenge, including in silico filtering of interactome data (79,80) and cross-linking mass spectromety (XL-MS) (81). The latter uses reactive chemicals to physically link together proteins in apposition for confident MS identification (reviewed in (82)).
Interpretation and Validation of Proteomics Data
Several considerations are provided here regarding how to interpret proteomic results, as well as how to utilize new workflows and technologies.
Scope and sensitivity
With incomplete coverage, the old adage that the absence of evidence is not evidence of absence is especially true for proteomics experiments. The number of proteins and peptides in a shotgun experiment is an important indicator of the depth of coverage in data acquisition. Hence, if a known biomarker is not reported or reproduced in a discovery-based experiment in the cohort, a pertinent question is whether the technological platform used has sufficient sensitivity and precision to support reproducible measurements at endogenous concentration.
The predictive value of a putative biomarker depends not only on the magnitude of its difference in normal versus diseased/at-risk individuals in the population, but also on the effect size, defined as the number of standard deviations between 2 measured means. Hence, for the purpose of protein biomarker discovery, the coefficient of variation (CV) of measurement is essential for determining whether there is sufficient power to distinguish significant differences between healthy and disease samples. Beyond the precision characteristics of the technological platforms, sample processing, as well as biological variations in human cohorts (both intrasubject and intersubject), may also impart significant variance in protein measurements. Thus, individual putative protein markers often require lengthy verification and validation pipelines that involve large cohorts (83). Proteomics assays have reported typical CVs in the range of <5% for intralaboratory measurements (34,84) to <20% for interlaboratory measurements (85). Other measurements of analytical performance that can have a drastic impact on the likelihood of payoff in downstream verification and validation processes include the limit of detection (LOD), limit of quantification (LOQ), and linearity of measurement, which are covered at length elsewhere (83).
False Discovery Rate
As in other large-scale approaches, both the number of identified proteins and the significance of quantitative comparisons in a quantitative proteomics experiment may become inflated if controls for false discovery rates (FDRs) are not adequate. In protein identification, the FDR of a database search provides an estimate for the proportion of incorrect protein identification in the result list. An FDR of 1% is commonly deemed acceptable, meaning that in a profiling experiment claiming to identify 5,000 proteins, up to 50 may be misidentifications. FDR is sometimes calculated with the aid of decoy databases, which contain scrambled or reversed protein sequences that are compared to experimental data, along with real protein sequences. The number of identified decoy sequences provides an estimate of the number of false discoveries among regular database sequence matches. For quantitative proteomics comparisons between samples, as many hypotheses as the number of proteins compared are being tested per sample pair. Multiple-testing corrections (e.g., Bonferroni corrections) are used to control for type I errors (incorrectly rejecting the null hypothesis) that arise from making multiple comparisons (1 per protein per sample pair). However, it must be noted that even infinitesimal p values are no indication that results are biomedically important (86), and do not obviate the need to validate with follow-up studies, including in vitro experimentation or in cohorts.
Comparison with Existing Techniques
In addition to technical considerations, criteria to nominate a differentially expressed protein as a candidate biomarker can vary. Currently, no gold standard exists for what constitutes a good candidate (87). Decision justifications in published reports range from formal statistical inferences of individual proteins (88), panels of proteins after feature selection (89), to additional criteria, such as cutoffs in the raw magnitude of fold-change (e.g., >5-fold changes) in at least n number of patients (90), to more custom modeling that adjust for covariates in risk assessments (31,35). In determining whether the presented candidates are worthy of prioritized verification studies, it stands to reason that the results of discovery experiments should be carefully scrutinized for both nominal significance (e.g., p values), and for the magnitude and variance of biological changes (effect size and confidence interval).
Ultimately, a candidate biomarker or protein with potential biomedical significance needs to be validated by orthogonal approaches in independent cohorts or models. In silico methods may be used to triage a list of differentially expressed proteins and prioritize candidates for validation, for example, by determining their tissue expression specificity and half-lives. Promising biomarkers may be further credentialed with targeted discovery approaches that quantify the status of the candidates in larger numbers of subjects and conditions. For instance, protein profiles may be compared in paired controls before and after unrelated procedures, or in peripheral blood versus proximal tissue fluids, such as coronary sinus effluents, to filter out spurious associations. An in-depth discourse on biomarker verification and validation can be found elsewhere (83).
Outlook
With continued technological progress, new interactions of proteomics with genetics and genomics are now becoming possible. Proteomics studies applied to large cohorts can be integrated with genetic polymorphisms to identify protein quantitative trait loci (pQTL) that control protein abundance, and perhaps thereby control phenotypic traits. Proteogenomics studies, combining RNA-seq and proteomics data from common datasets, have been undertaken in cancer research to identify the protein coding consequences of cancer variants and mutations (91), whereas a recent multiomics study in 80 mouse genetic backgrounds demonstrated that multiple types of omics data can complement each other to characterize mitochondrial functional regulation (92). Further integration of proteomics and genomics methods will allow examination of nonstandard gene products, including short open reading frames, unannotated transcripts, and alternative splicing isoform proteins. By greatly expanding the depth of the proteome that may be experimentally accessed, the development of isoform proteomics, in particular, will have important implications for cardiovascular research. These isoforms may present with differential expression levels or patterns, localizations, interactions, and PTMs in different cell types and during disease progression, including in ischemic cardiomyopathy (93) and hypertrophy (94). Although RNA-seq has discovered many alternative isoforms at the transcript level, the majority of isoform transcripts have not been validated at the protein level. Many alternatively spliced transcripts may lead to frame-shifts and be targeted by nonsense-mediated decay. The functional significance of many transcript isoforms may soon be addressed by proteomics using custom sequence databases translated from RNA-seq transcripts.
Broader applications will also be facilitated by continued improvements in throughput and coverage. For example, newly-developed neutron encoding (NeuCode) techniques will allow over 30 samples to be combined for MS analysis (95), leading to a 5-fold increase in throughput for investigating large cohorts. With new affinity proteomics platforms, such as protein and aptamer arrays, we have already seen further development of preselected cardiovascular-specific protein panels (33), which could allow the disease status of the likeliest candidates to be screened across large sample pools. In summary, with core proteomics technologies having now matured to provide high protein and sample coverage, new applications are increasingly available to interrogate the dynamic parameters that connect gene expression to physiological functions. We envision that proteomics-based discovery-driven platforms will continue to support molecular phenotyping of individuals in the era of precision medicine, a9nd will provide ample opportunities for innovative breakthroughs in cardiovascular medicine for years to come.
Acknowledgments
This work was supported by NIH K99-HL127302 to Dr. Lam; NIH R37-HL063901 and NIH U54-GM114833 to Dr. Ping; and NIH ZIA-HL006059 and NIH ZIA-HL002066 to Dr Murphy.
ABBREVIATIONS AND ACRONYMS
- FDR
false discovery rate
- MS
mass spectrometry
- PTM
post-translational modification
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
References
- 1.McDonough JL, Labugger R, Pickett W, et al. Cardiac troponin I is modified in the myocardium of bypass patients. Circulation. 2001;103:58–64. doi: 10.1161/01.cir.103.1.58. [DOI] [PubMed] [Google Scholar]
- 2.Labugger R, Organ L, Collier C, et al. Extensive troponin I and T modification detected in serum from patients with acute myocardial infarction. Circulation. 2000;102:1221–1226. doi: 10.1161/01.cir.102.11.1221. [DOI] [PubMed] [Google Scholar]
- 3.Weekes J, Wheeler CH, Yan JX, et al. Bovine dilated cardiomyopathy: proteomic analysis of an animal model of human dilated cardiomyopathy. Electrophoresis. 1999;20:898–906. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<898::AID-ELPS898>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Ping P, Zhang J, Pierce WM, et al. Functional proteomic analysis of protein kinase C ε signaling complexes in the normal heart and during cardioprotection. Circ Res. 2001;88:59–62. doi: 10.1161/01.res.88.1.59. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson RD, Vondriska TM, Biederman KJ, et al. Protein kinase C ε signaling complexes include metabolism- and transcription/translation-related proteins: complimentary separation techniques with LC/MS/MS. Mol Cell Proteomics. 2002;1:421–433. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Lindsey ML, Goshorn DK, Comte-Walters S, et al. A multidimensional proteomic approach to identify hypertrophy-associated proteins. Proteomics. 2006;6:2225–2235. doi: 10.1002/pmic.200500013. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Picht E, Ginsburg KS, et al. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel α1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 8.Ge Y, Rybakova IN, Xu Q, et al. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc Natl Acad Sci U S A. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larance M, Lamond AI. Multidimensional proteomics for cell biology. Nat Rev Mol Cell Biol. 2015;16:269–280. doi: 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Vogel C, Abreu Rde S, Ko D, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Q, Stepaniants SB, Mao M, et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 14.Lundberg E, Fagerberg L, Klevebring D, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall KD, Edwards MA, Krenz M, et al. Proteomic mapping of proteins released during necrosis and apoptosis from cultured neonatal cardiac myocytes. Am J Physiol Cell Physiol. 2014;306:C639–C647. doi: 10.1152/ajpcell.00167.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Eyk JE. Proteomics: unraveling the complexity of heart disease and striving to change cardiology. Curr Opin Mol Ther. 2001;3:546–553. [PubMed] [Google Scholar]
- 17.Nanjappa V, Thomas JK, Marimuthu A, et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014;42:D959–D965. doi: 10.1093/nar/gkt1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Pundir S, Magrane M, Martin MJ, et al. Searching and navigating UniProt databases. Curr Protoc Bioinformatics. 2015;50:1.27.1–1.27.10. doi: 10.1002/0471250953.bi0127s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusebauch U, Deutsch EW, Campbell DS, et al. Using PeptideAtlas, SRMAtlas, and PASSEL: comprehensive resources for discovery and targeted proteomics. Curr Protoc Bioinformatics. 2014;46:13.25.1–13.25.28. doi: 10.1002/0471250953.bi1325s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zong N, Li H, Li H, et al. Integration of cardiac proteome biology and medicine by a specialized knowledgebase. Circ Res. 2013;113:1043–1053. doi: 10.1161/CIRCRESAHA.113.301151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riley NM, Hebert AS, Coon JJ. Proteomics moves into the fast lane. Cell Syst. 2016;2:142–143. doi: 10.1016/j.cels.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Fonslow BR, Shan B, et al. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshishian H, Addona T, Burgess M, et al. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam MP, Lau E, Scruggs SB, et al. Site-specific quantitative analysis of cardiac mitochondrial protein phosphorylation. J Proteomics. 2013;81:15–23. doi: 10.1016/j.jprot.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Kirk JA, Ji W, et al. Multiple reaction monitoring to identify site-specific troponin I phosphorylated residues in the failing human heart. Circulation. 2012;126:1828–1837. doi: 10.1161/CIRCULATIONAHA.112.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Percy AJ, Yang J, Hardie DB, et al. Precise quantitation of 136 urinary proteins by LC/MRM-MS using stable isotope labeled peptides as internal standards for biomarker discovery and/or verification studies. Methods. 2015;81:24–33. doi: 10.1016/j.ymeth.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Fu Q, Chen Z, Zhang S, et al. Multiple and selective reaction monitoring using triple quadrupole mass spectrometer: preclinical large cohort analysis. Methods Mol Biol. 2016;1410:249–264. doi: 10.1007/978-1-4939-3524-6_15. [DOI] [PubMed] [Google Scholar]
- 30.Keshishian H, Addona T, Burgess M, et al. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerstein HC, Paré G, McQueen MJ, et al. Outcome Reduction With Initial Glargine Intervention Trial Investigators. Identifying novel biomarkers for cardiovascular events or death in people with dysglycemia. Circulation. 2015;132:2297–2304. doi: 10.1161/CIRCULATIONAHA.115.015744. [DOI] [PubMed] [Google Scholar]
- 32.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin Appl. 2015;9:307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 33.Ngo D, Sinha S, Shen D, et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation. 2016;134:270–285. doi: 10.1161/CIRCULATIONAHA.116.021803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951. [DOI] [PubMed] [Google Scholar]
- 35.Lind L, Siegbahn A, Lindahl B, et al. Discovery of new risk markers for ischemic stroke using a novel targeted proteomics chip. Stroke. 2015;46:3340–3347. doi: 10.1161/STROKEAHA.115.010829. [DOI] [PubMed] [Google Scholar]
- 36.Sabatine MS. Using aptamer-based technology to probe the plasma proteome for cardiovascular disease prediction. JAMA. 2016;315:2525–2526. doi: 10.1001/jama.2016.6110. [DOI] [PubMed] [Google Scholar]
- 37.Gramolini A, Lau E, Liu PP. Identifying low-abundance biomarkers: aptamer-based proteomics potentially enables more sensitive detection in cardiovascular diseases. Circulation. 2016;134:286–289. doi: 10.1161/CIRCULATIONAHA.116.022940. [DOI] [PubMed] [Google Scholar]
- 38.Keshishian H, Burgess MW, Gillette MA, et al. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol Cell Proteomics. 2015;14:2375–2393. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cominetti O, Nuñez Galindo A, Corthésy J, et al. Proteomic biomarker discovery in 1000 human plasma samples with mass spectrometry. J Proteome Res. 2016;15:389–399. doi: 10.1021/acs.jproteome.5b00901. [DOI] [PubMed] [Google Scholar]
- 40.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang KY, Su MG, Kao HJ, et al. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016;44:D435–D446. doi: 10.1093/nar/gkv1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thingholm TE, Jørgensen TJ, Jensen ON, et al. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 43.Lundby A, Andersen MN, Steffensen AB, et al. In vivo phosphoproteomics analysis reveals the cardiac targets of β-adrenergic receptor signaling. Sci Signal. 2013;6:rs11. doi: 10.1126/scisignal.2003506. [DOI] [PubMed] [Google Scholar]
- 44.Schechter MA, Hsieh MK, Njoroge LW, et al. Phosphoproteomic profiling of human myocardial tissues distinguishes ischemic from non-ischemic end stage heart failure. PLoS ONE. 2014;9:e104157. doi: 10.1371/journal.pone.0104157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karamanlidis G, Lee CF, Garcia-Menendez L, et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen TT, Wong R, Menazza S, et al. Cyclophilin D modulates mitochondrial acetylome. Circ Res. 2013;113:1308–1319. doi: 10.1161/CIRCRESAHA.113.301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang W, Paschen W. SUMO proteomics to decipher the SUMO-modified proteome regulated by various diseases. Proteomics. 2015;15:1181–1191. doi: 10.1002/pmic.201400298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barallobre-Barreiro J, Gupta SK, Zoccarato A, et al. Glycoproteomics reveals decorin peptides with anti-myostatin activity in human atrial fibrillation. Circulation. 2016;134:817–832. doi: 10.1161/CIRCULATIONAHA.115.016423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker BL, Palmisano G, Edwards AV, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol Cell Proteomics. 2011;10:M110.006833. doi: 10.1074/mcp.M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang SB, Foster DB, Rucker J, et al. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ Res. 2011;109:750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohr MJ, Aponte A, Sun J, et al. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circ Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zong N, Ping P, Lau E, et al. Lysine ubiquitination and acetylation of human cardiac 20S proteasomes. Proteomics Clin Appl. 2014;8:590–594. doi: 10.1002/prca.201400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fert-Bober J, Giles JT, Holewinski RJ, et al. Citrullination of myofilament proteins in heart failure. Cardiovasc Res. 2015;108:232–242. doi: 10.1093/cvr/cvv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindsey ML, Iyer RP, Zamilpa R, et al. A novel collagen matricryptin reduces left ventricular dilation post-myocardial infarction by promoting scar formation and angiogenesis. J Am Coll Cardiol. 2015;66:1364–1374. doi: 10.1016/j.jacc.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerbey AL, Randle PJ, Cooper RH, et al. Regulation of pyruvate dehydrogenase in rat heart. Mechanism of regulation of proportions of dephosphorylated and phosphorylated enzyme by oxidation of fatty acids and ketone bodies and of effects of diabetes: role of coenzyme A, acetyl-coenzyme A and reduced and oxidized nicotinamideadenine dinucleotide. Biochem J. 1976;154:327–348. doi: 10.1042/bj1540327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma K, D'Souza RC, Tyanova S, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 57.Lundby A, Secher A, Lage K, et al. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat Commun. 2012;3:876. doi: 10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng N, Zhang J, Zong C, et al. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10:M110.000117. doi: 10.1074/mcp.M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee DI, Zhu G, Sasaki T, et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519:472–476. doi: 10.1038/nature14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scholten A, Preisinger C, Corradini E, et al. Phosphoproteomics study based on in vivo inhibition reveals sites of calmodulin-dependent protein kinase II regulation in the heart. J Am Heart Assoc. 2013;2:e000318. doi: 10.1161/JAHA.113.000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kooij V, Holewinski RJ, Murphy AM, et al. Characterization of the cardiac myosin binding protein-C phosphoproteome in healthy and failing human hearts. J Mol Cell Cardiol. 2013;60:116–120. doi: 10.1016/j.yjmcc.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svinkina T, Gu H, Silva JC, et al. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol Cell Proteomics. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horton JL, Martin OJ, Lai L, et al. Mitochondrial protein hyperacetylation in the failing heart. JCI Insight. 2016;2:e84897. doi: 10.1172/jci.insight.84897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Aponte AM, Menazza S, et al. Additive cardioprotection by pharmacological postconditioning with hydrogen sulfide and nitric oxide donors in mouse heart: S-sulfhydration vs. S-nitrosylation. Cardiovasc Res. 2016;110:96–106. doi: 10.1093/cvr/cvw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J, Nguyen T, Aponte AM, et al. Ischaemic preconditioning preferentially increases protein S-nitrosylation in subsarcolemmal mitochondria. Cardiovasc Res. 2015;106:227–236. doi: 10.1093/cvr/cvv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murray CI, Chung HS, Uhrigshardt H, et al. Quantification of mitochondrial S-nitrosylation by CysTMT6 switch assay. Methods Mol Biol. 2013;1005:169–179. doi: 10.1007/978-1-62703-386-2_14. [DOI] [PubMed] [Google Scholar]
- 67.Kim TY, Wang D, Kim AK, et al. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics. 2012;11:1586–1594. doi: 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price JC, Holmes WE, Li KW, et al. Measurement of human plasma proteome dynamics with 2H2O and liquid chromatography tandem mass spectrometry. Anal Biochem. 2012;420:73–83. doi: 10.1016/j.ab.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 69.Lam MP, Wang D, Lau E, et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J Clin Invest. 2014;124:1734–1744. doi: 10.1172/JCI73787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lau E, Cao Q, Ng DC, et al. A large dataset of protein dynamics in the mammalian heart proteome. Sci Data. 2016;3:160015. doi: 10.1038/sdata.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kolwicz SC, Jr, Tian R. Glucose metabolism and cardiac hypertrophy. Cardiovasc Res. 2011;90:194–201. doi: 10.1093/cvr/cvr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu YR, Chang WC, Mendiaz EA, et al. Selective deamidation of recombinant human stem cell factor during in vitro aging: isolation and characterization of the aspartyl and isoaspartyl homodimers and heterodimers. Biochemistry. 1998;37:2251–2262. doi: 10.1021/bi972372z. [DOI] [PubMed] [Google Scholar]
- 73.Burgers PP, van der Heyden MA, Kok B, et al. A systematic evaluation of protein kinase A-A-kinase anchoring protein interaction motifs. Biochemistry. 2015;54:11–21. doi: 10.1021/bi500721a. [DOI] [PubMed] [Google Scholar]
- 74.Aye TT, Soni S, van Veen TA, et al. Reorganized PKA-AKAP associations in the failing human heart. J Mol Cell Cardiol. 2012;52:511–518. doi: 10.1016/j.yjmcc.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 75.Pankow S, Bamberger C, Calzolari D, et al. ΔF508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris JH, Knudsen GM, Verschueren E, et al. Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat Protoc. 2014;9:2539–2554. doi: 10.1038/nprot.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huttlin EL, Ting L, Bruckner RJ, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waldron L, Steimle JD, Greco TM, et al. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rizzetto S, Priami C, Csikász-Nagy A. Qualitative and quantitative protein complex prediction through proteome-wide simulations. PLoS Comput Biol. 2015;11:e1004424. doi: 10.1371/journal.pcbi.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goldfarb D, Hast BE, Wang W, et al. Spotlite: web application and augmented algorithms for predicting co-complexed proteins from affinity purification--mass spectrometry data. J Proteome Res. 2014;13:5944–5955. doi: 10.1021/pr5008416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gingras AC, Gstaiger M, Raught B, et al. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 82.Leitner A, Faini M, Stengel F, et al. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem Sci. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 84.Percy AJ, Chambers AG, Yang J, et al. Multiplexed MRM-based quantitation of candidate cancer biomarker proteins in undepleted and non-enriched human plasma. Proteomics. 2013;13:2202–2215. doi: 10.1002/pmic.201200316. [DOI] [PubMed] [Google Scholar]
- 85.Abbatiello SE, Schilling B, Mani DR, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. 2015;14:2357–2374. doi: 10.1074/mcp.M114.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goodman SN. Aligning statistical and scientific reasoning. Science. 2016;352:1180–1181. doi: 10.1126/science.aaf5406. [DOI] [PubMed] [Google Scholar]
- 87.de Lemos JA, Rohatgi A, Ayers CR. Applying a big data approach to biomarker discovery: running before we walk? Circulation. 2015;132:2289–2292. doi: 10.1161/CIRCULATIONAHA.115.019648. [DOI] [PubMed] [Google Scholar]
- 88.Keshishian H, Burgess MW, Gillette MA, et al. Multiplexed, quantitative workflow for sensitive biomarker discovery in plasma yields novel candidates for early myocardial injury. Mol Cell Proteomics. 2015;14:1–45. doi: 10.1074/mcp.M114.046813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin X, Subramanian S, Hwang SJ, et al. Protein biomarkers of new-onset cardiovascular disease: prospective study from the systems approach to biomarker research in cardiovascular disease initiative. Arterioscler Thromb Vasc Biol. 2014;34:939–945. doi: 10.1161/ATVBAHA.113.302918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Addona TA, Shi X, Keshishian H, et al. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat Biotechnol. 2011;29:635–643. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams EG, Wu Y, Jha P, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kong SW, Hu YW, Ho JW, et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ Cardiovasc Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song HK, Hong SE, Kim T, et al. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS ONE. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hebert AS, Merrill AE, Bailey DJ, et al. Neutron-encoded mass signatures for multiplexed proteome quantification. Nat Methods. 2013;10:332–334. doi: 10.1038/nmeth.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stubbs P, Seed M, Lane D, et al. Lipoprotein(a) as a risk predictor for cardiac mortality in patients with acute coronary syndromes. Eur Heart J. 1998;19:1355–1364. doi: 10.1053/euhj.1998.1043. [DOI] [PubMed] [Google Scholar]
- 97.McQueen MJ, Hawken S, Wang X, et al. INTERHEART study investigators. Lipids, lipoproteins, and apolipoproteins as risk markers of myocardial infarction in 52 countries (the INTERHEART study): a case-control study. Lancet. 2008;372:224–233. doi: 10.1016/S0140-6736(08)61076-4. [DOI] [PubMed] [Google Scholar]
- 98.de Lemos JA, McGuire DK, Drazner MH. B-type natriuretic peptide in cardiovascular disease. Lancet. 2003;362:316–322. doi: 10.1016/S0140-6736(03)13976-1. [DOI] [PubMed] [Google Scholar]
- 99.Emerging Risk Factors Collaboration. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med. 2012;367:1310–1320. doi: 10.1056/NEJMoa1107477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puleo PR, Guadagno PA, Roberts R, et al. Early diagnosis of acute myocardial infarction based on assay for subforms of creatine kinase-MB. Circulation. 1990;82:759–764. doi: 10.1161/01.cir.82.3.759. [DOI] [PubMed] [Google Scholar]
- 101.Shlipak MG, Sarnak MJ, Katz R, et al. Cystatin C and the risk of death and cardiovascular events among elderly persons. N Engl J Med. 2005;352:2049–2060. doi: 10.1056/NEJMoa043161. [DOI] [PubMed] [Google Scholar]
- 102.Daniels LB, Laughlin GA, Sarno MJ, et al. Lipoprotein-associated phospholipase A2 is an independent predictor of incident coronary heart disease in an apparently healthy older population: the Rancho Bernardo Study. J Am Coll Cardiol. 2008;51:913–919. doi: 10.1016/j.jacc.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brennan ML, Penn MS, Van Lente F, et al. Prognostic value of myeloperoxidase in patients with chest pain. N Engl J Med. 2003;349:1595–1604. doi: 10.1056/NEJMoa035003. [DOI] [PubMed] [Google Scholar]
- 104.Kavsak PA, MacRae AR, Newman AM, et al. Effects of contemporary troponin assay sensitivity on the utility of the early markers myoglobin and CKMB isoforms in evaluating patients with possible acute myocardial infarction. Clin Chim Acta. 2007;380:213–216. doi: 10.1016/j.cca.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 105.Johnson BD, Kip KE, Marroquin OC, et al. Serum amyloid A as a predictor of coronary artery disease and cardiovascular outcome in women: the National Heart, Lung, and Blood Institute-Sponsored Women's Ischemia Syndrome Evaluation (WISE) Circulation. 2004;109:726–732. doi: 10.1161/01.CIR.0000115516.54550.B1. [DOI] [PubMed] [Google Scholar]
- 106.Adams JE, III, Bodor GS, Dávila-Román VG, et al. Cardiac troponin I. A marker with high specificity for cardiac injury. Circulation. 1993;88:101–106. doi: 10.1161/01.cir.88.1.101. [DOI] [PubMed] [Google Scholar]
- 107.Reichlin T, Hochholzer W, Bassetti S, et al. Early diagnosis of myocardial infarction with sensitive cardiac troponin assays. N Engl J Med. 2009;361:858–867. doi: 10.1056/NEJMoa0900428. [DOI] [PubMed] [Google Scholar]
- 108.Danese E, Montagnana M. An historical approach to the diagnostic biomarkers of acute coronary syndrome. Ann Transl Med. 2016;4:194. doi: 10.21037/atm.2016.05.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ladenson JH. A personal history of markers of myocyte injury [myocardial infarction] Clin Chim Acta. 2007;381:3–8. doi: 10.1016/j.cca.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 110.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 111.O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]