Abstract
Tuberous sclerosis complex (TSC) is a multisystem disorder that results from heterozygous mutations in either TSC1 or TSC2. The primary organ systems that are affected include the brain, skin, lung, kidney, and heart, all with variable frequency, penetrance, and severity. Neurological features include epilepsy, autism, and intellectual disability. There are more than 1,500 known pathogenic variants for TSC1 and TSC2, including deletion, nonsense, and missense mutations, and all pathogenic mutations are inactivating, leading to loss of function effects on the encoded proteins TSC1 and TSC2. These proteins form a complex to constitutively inhibit mechanistic target of rapamycin (mTOR) signaling cascade, and as a consequence, mTOR signaling is constitutively active within all TSC-associated lesions. The mTOR inhibitors rapamycin (sirolimus) and everolimus have been shown to reduce the size of renal and brain lesions and improve pulmonary function in TSC, and these compounds may also decrease seizure frequency. The clinical application of mTOR inhibitors in TSC has provided one of the first examples of precision medicine in a neurodevelopmental disorder.
Keywords: TSC, epilepsy, genetics, mTOR, rapamycin
Introduction
Tuberous sclerosis complex (TSC) was first fully clinically detailed by Bourneville.1 TSC is a multisystem disorder associated with hamartomas or benign tumor growths in the brain, heart, lung, eye, or kidney.2 Over the past 25 years, knowledge about genetic and cell signaling abnormalities in TSC has rapidly advanced, based on both animal model and clinical research, culminating in 2012 in the US Food and Drug Administration approval of a targeted therapy for renal and central nervous system involvement in TSC. Currently, TSC is estimated to affect one in 50,000 individuals worldwide and to occur in one in 6,000 to one in 10,000 live births across all ethnic demographies.3 This review summarizes some of the recent developments in TSC genetics, diagnostic criteria, evaluation, and surveillance, as well as evaluates some of the salient cell biology of the TSC-mechanistic target of rapamycin (mTOR) pathway.
Clinical presentation
The symptoms and severity of TSC vary widely between individuals, and there is great phenotypic variability. Even across family pedigrees with identical genotypes, there can be great clinical variability (a phenomenon known as pleiotropy) where, for example, some family members develop severe neurological diseases (seizures and autism), whereas others have TSC features confined to the skin or kidneys with minimal neurological involvement.4 The most common organ systems affected in TSC are the brain, skin, kidney, lung (in females), and heart. Hamartomas lead to impaired organ function, typically by compression of surrounding structures; in female TSC patients, lymphangioleiomyomatosis (LAM) results in cystic changes and parenchymal lung destruction. The major causes of neurological disability in TSC, including seizures, intellectual disabilities, and autism, are linked to structural developmental abnormalities in the brain known as cortical tubers, a type of focal cortical dysplasia.5 Tubers form during fetal brain development and remain throughout life. Seizures remain the most common presenting sign for TSC (>80% of TSC patients have seizures) and can develop at any point in life from infancy (infantile spasms) to adulthood. The incidence of autism and intellectual disability in TSC is ~40% and follows a bimodal distribution, with some individuals exhibiting severe intellectual disability, whereas others having only mild impairments.6 Indeed, normal intellectual function occurs commonly in TSC. Benign glial brain tumors known as subependymal giant cell astrocytomas (SEGAs) may grow within the lateral, third, or fourth ventricles in ~10%–20% of TSC patients, leading to hydrocephalus, increased intracranial pressure, and even death. Progressive LAM in some individuals leads to irrevocable loss of pulmonary function. LAM cells express estrogen and progesterone receptors, which might in part explain the predominance in females. Renal angiomyolipomas (AMLs) are hamartomas deriving from the renal parenchyma and may be multiple and bilateral. AMLs often spare renal function until they are bilateral and extensive in size and number. AMLs cause destruction of renal tissue by direct compression in the kidney. In addition, the vascular supply of AMLs may contain aneurysmal dilations, leading to a risk of rupture and hemorrhage, especially when AMLs exceed 4 cm in size. Additionally, there is a link to polycystic kidney disease (PKD) in TSC, as the PKD1 locus is adjacent to the TSC2 gene on chromosome 16, and the appearance of PKD may be associated with hypertension and renal failure.7 Indeed, lung and kidney disease in TSC patients can lead to a shortened life span compared with the unaffected individuals, with renal disease being the most common cause of death. Interestingly, cardiac rhabdomyomas spontaneously resolve in >50% of TSC patients and cause no clinical difficulties. In babies with TSC, cardiac rhabdomyomas may cause congestive heart failure or dysrhythmias.
TSC is diagnosed by clinical criteria, which were recently updated in 2012.3 The current clinical criteria dictate two types of TSC diagnosis – definite and possible – predicated on the detection of “major” and “minor” diagnostic criteria (Table 1). The major criteria include cortical tubers, subependymal nodules, SEGAs, retinal astrocytic hamartomas, pulmonary LAM, renal AMLs, cardiac rhabdomyomas, facial angiofibromas (previously called adenoma sebaceum), ungual fibromas, hypomelanotic macules (previously referred to as ash leaf spots), and the Shagreen patch (collagenous fibroadenoma). Individuals with two major features or one major feature with two minor features or positive genetic testing meet criteria for “definite” TSC.3 Those with either one major feature, one major and one minor feature, or two or more minor features meet criteria for “possible” TSC. Once a definite diagnosis is made, determination of a familial or sporadic occurrence is necessary for appropriate genetic counseling, and then, long-term clinical surveillance can be implemented. As an autosomal dominant disorder, inheritance is 50% in children of an affected proband. A new feature of the updated diagnostic criteria includes the use of genetic testing as a major diagnostic criterion such that a TSC diagnosis can be made solely with the identification of a clearly pathogenic genetic mutation, eg, nonsense variant, even in the absence of clinical signs and symptoms. However, genetic testing is not an absolute requirement for diagnosis, and most cases are diagnosed based on clinical findings alone in the absence of genetic testing. The reasoning behind this is that a mutation in TSC1 or TSC2 will not be identified (no mutation identified or the so-called NMI) in 10%–15% of patients with clinically defined TSC.8 Therefore, negative genetic testing does not exclude the diagnosis of TSC.
Table 1.
TSC diagnostic criteria3
| Definite diagnosis: two major features or one major feature with two minor features or positive genetic testing | |
| Possible diagnosis: either one major feature, one major and one minor feature, or two or more minor features | |
| Major features | Minor features |
| Hypomelanotic macules (three or more, at least 5 mm diameter) | Confetti skin lesions |
| Angiofibromas (two or more) or fibrous cephalic plaque | Dental enamel pits (three or more) |
| Ungual fibromas (two or more) | Intraoral fibromas (two or more) |
| Shagreen patch | Retinal achromatic patch |
| Multiple renal hamartomas | Multiple renal cysts |
| Cortical dysplasias (three or more, includes tubers and white matter radial migration lines) | Nonrenal hamartomas |
| SENs (two or more) | |
| SEGA | |
| Cardiac rhabdomyoma | |
| LAM | |
| AMLs (two or more) |
Note: Adapted from Pediatr Neurol 2013;49(4), Northrup H, Krueger DA, Tuberous sclerosis complex diagnostic update: recommendations of the 2012 international tuberous sclerosis complex consensus conference. Copyright (2012), with permission from Elsevier.3
Abbreviations: TSC, tuberous sclerosis complex; SENs, subependymal nodules; SEGA, subependymal giant cell astrocytoma; LAM, lymphangioleiomyomatosis; AMLs, angiomyolipomas.
TSC genetics
TSC1 is located on chromosome 9q34, and TSC2 is located on chromosome 16p13 (Table 2).9,10 TSC1 is a 23 exon gene encoding an 8.6 kb transcript and a 30 kDa protein, known as TSC1 or hamartin. TSC2 encodes a 5.5 kb transcript and a 180 kDa protein, known as TSC2 or tuberin.11 TSC1 and TSC2 are widely expressed across cell types and organ systems, and there is high interspecies sequence conservation of these genes and proteins, from Drosophila to humans. TSC1 and TSC2 bind with a third protein, TBC1D7, to function as part of a heteromeric protein complex to regulate cell growth, cell size, cell cycle, and proliferation12,13 through the mTOR pathway (see “Cell Biology and mTOR signaling in TSC”).
Table 2.
Genes associated with TSC
| Gene name | TSC1 | TSC2 |
| OMIM entry | 605284 | 191092 |
| Protein name | Hamartin | Tuberin |
| Cytogenetic location | 9q34.13 | 16p13.3 |
| Most common types of mutations | Frameshift/protein truncation > nonsense | Frameshift/protein truncation = deletion/insertion/duplication = nonsense mutation |
| Number of unique variants reporteda | 847 | 2,395 |
Note:
As of April 2016 in the Leiden Open Variation Database.
Abbreviations: TSC, tuberous sclerosis complex; OMIM, Online Mendelian Inheritance in Man.
Patients with TSC have mutations in either TSC1 or TSC2, not both genes. In addition, mutations in the genes that encode other regulatory components of the mTOR pathway, eg, TBC1D7 and Rheb, have not been associated with TSC, ie, there is no known TSC3 gene. Mutations in TSC1 are often small insertions or deletions (indels) that result in shortened (truncated) protein, whereas TSC2 mutations include large deletions, indels, nonsense, and missense mutations.14,15 Splicing errors account for a small number of TSC1 and TSC2 mutations. Each mutation type is believed to either functionally inactivate TSC1 or TSC2 or prevent TSC2 from forming the complex with TSC1, culminating in loss of tonic inhibition of the mTOR pathway. Indeed, deleterious variants are functionally validated in vitro based on their effect (enhancement) on mTOR signaling. A potential disease causing mutation may be identified in ~75%–90% of patients with TSC. All commercially available testing is performed on DNA extracted from peripheral blood lymphocytes, and more recently, analysis of buccal swab or saliva has been shown to be successful for genotyping. Germline mutations found in blood are also detected in tissue samples, eg, brain, lung, kidney, from the same individuals. In ~10%–15% of TSC individuals, a pathogenic TSC1 or TSC2 mutation is not detected (NMI) using conventional testing methods.
Estimates are that two out of three TSC cases are sporadic mutations not linked to a family pedigree and, thus, arise de novo at the initial time of zygote formation. In contrast, approximately one out of three TSC cases can be linked to a pedigree and are inherited in an autosomal dominant manner. Currently, a comprehensive listing of TSC1 and TSC2 variants with results of functional assays and predicted pathogenicity can be found at the Leiden Open Variation Database managed by the Leiden University Medical Center (www.lovd.nl/TSC1 and www.lovd.nl/TSC2). As of 2016, there are 847 unique TSC1 variants and 2,395 unique TSC2 variants accounted for in this database.16
Multiple studies have suggested that TSC2 mutations account for the majority (~51%–82%) of all patients in whom a pathogenic mutation was identified.15,17 Indeed, sporadic TSC cases more often result from TSC2 than TSC1 mutations, with a range of 3:1–7:1. In contrast, inherited TSC cases result nearly equally from TSC1 and TSC2 mutations. In a small number of patients with TSC, large genomic mutations affecting TSC2 and the adjacent PKD1 gene can lead to a contiguous gene syndrome with clinical features of both TSC and PKD (autosomal dominant PKD). These patients are distinguished from those with typical TSC-associated renal disease by the severity and early onset of the cystic kidney changes.18
Different mutations in the TSC1 and TSC2 genes suggest that missense and small in-frame shift mutations may be pathogenic. For example, in one series, of the 152 amino acid substitutions and in-frame insertions/deletions, 86 (56%) were found to be pathogenic in vitro systems, and they increased the phosphorylation of p70 S6 kinase (p70S6K), reflecting activation of mTOR. Variations in TSC2 resulted in a wider variety of protein changes in the TSC1/2 complex.19 Fifty-five variations resulted in changes to the N-terminal, whereas 54 variations caused changes to the C-terminal amino acid region. Changes in these amino acids decreased the binding stability of the TSC1/TSC2 complex as well as decreased production of TSC1. Changes in the N-terminal amino acids seemed to have a higher effect on the stability to the TSC1/2 complex than the changes in the C-terminal ones did. Overall, variations to either TSC1 or TSC2 that inhibited or had no effect on phosphor activation of the downstream p70S6 kinase were deemed not pathogenic.9
Genotype–phenotype relationships
Genetic testing is not required for the diagnosis of TSC. However, in cases when the diagnosis is not clear by clinical criteria, genetic testing can help establish a diagnosis. Additionally, with the current criteria, the diagnosis of TSC can be made with genetic testing even in the absence of any signs or symptoms of the disease. In these cases, individuals for whom a diagnosis has been made with genetic testing can be referred for appropriate clinical screening and management as some features of TSC will appear over the course of a lifetime, ie, renal AML or LAM, and thus, clinical evaluation can be updated.
A number of series with long-term follow-up have provided insights into disease spectrum in TSC. Broadly speaking, patients with TSC2 mutations tend to have more severe disease than those with TSC1 mutations. For example, patients with TSC2 mutations exhibit an earlier onset of seizures and epilepsy, lower scores on tests of cognition, more skin findings, higher incidence of SEGAs, more numerous renal and hepatic AML, and greater tuber burden than those with TSC1 mutations.20,21 However, there may be significant overlap in disease severity between TSC1 and TSC2 patients, and many individuals with TSC1 mutations have severe multiorgan system involvement. Patients within a pedigree, ie, same mutation, can have different disease manifestations, eg, pleiotropy or can exhibit variable penetrance of any one phenotypic feature. Therefore, making predictions about disease severity in any individual patient based on their genotype alone is not yet feasible in clinical practice.
The “two-hit” hypothesis is an important concept in understanding lesion formation in TSC as well as the variable clinical expression in TSC. For example, all patients with either germline or sporadic TSC are heterozygotes (exceptions are somatic mosaic and germline mosaic individuals), with each cell in the body containing a single heterozygous TSC1 or TSC2 mutation. It is widely believed among investigators that lesion formation in TSC likely results from a somatic “second-hit” mutation in the other unaffected TSC1 or TSC2 allele, resulting in loss of heterozygosity (LOH) or biallelic inactivation. When both alleles contain mutations, the result is biallelic inactivation, thus reducing the function of the TSC1–TSC2 complex. From a functional perspective, this means that the mTOR signaling cascade no longer has inhibitory modulation by the TSC1–TSC2 complex and can signal unchecked. Thus, AML in the kidney or SEGAs in the brain have been shown to exhibit LOH, while LOH has been difficult to confirm in cardiac rhabdomyomas and cortical tubers.22,23 Single-cell DNA sequencing or deep sequencing of tuber DNA has identified somatic mutations in subpopulations of tubers, suggesting that other factors or modifier genes may be involved in the development of hamartomas in TSC.24 Conceptually, phenotypic severity in each patient with TSC is largely, though not exclusively, dictated by hamartoma burden, and in most cases, hamartomas in TSC result from a combination of germline and somatic second-hit mutations. More recently, epigenetic changes may contribute to lesion formation in TSC, including microRNAs that regulate TSC1 and TSC2 protein expression.25,26
Recommendations for genetic testing in TSC
Genetic testing may be important for counseling and management of TSC patients. Although TSC is usually diagnosed on clinical criteria alone, results of genetic testing are now considered as major diagnostic criteria if a clearly deleterious mutation is identified and certain circumstances dictate that a clinician might want to pursue genetic testing. For example, genetic testing should be offered to any TSC individual of reproductive age for family planning purposes in the case where a couple with an affected child wishes to have additional children. The risk of having a second child with TSC when the parents have no signs or symptoms of TSC is very low (~2%), though not zero, and near the risk within the general population. If parents wish to undergo genetic testing, the affected child should be tested first to detect the pathogenic mutation. Then, parental testing can be performed targeting that specific mutation. If the same mutation is identified in either parent, the risk of having a subsequent affected child is ~50%. If the parent is found to be negative, then the recurrence risk is ~2%. For parents with positive testing who wish to have further children, preimplantation genetic testing is available. Because positive genetic testing is considered as an independent diagnostic criterion for TSC, the condition of asymptomatic carrier does not exist. Individuals with positive genetic testing should undergo thorough evaluation using the consensus guidelines for newly diagnosed individuals with TSC even in the absence of any obvious signs or symptoms.
Genetic testing might also be considered in individuals who have a possible diagnosis of TSC based on the diagnostic criteria listed in Table 1 (either one major feature or two or more minor features). Detection of a mutation would impact future surveillance of the patient as well as prognosis. A unique situation arises when, for example, an individual has a single manifestation of TSC, eg, a single cortical tuber or AML, without any other signs of TSC. While these individuals may have a very mild phenotype, an alternative possibility is somatic mosaicism in which either TSC1 or TSC2 mutations are confined to single or a few organ systems. Commercial genetic testing in this case will likely not detect a mutation, although high coverage deep sequencing in a research laboratory might, which is highly relevant for family planning counseling for prospective parents.
When evaluating a possible family pedigree, the closest three generations of living patient relatives should be screened for signs using the diagnostic criteria, being vigilant because some family members may have a very mild disease phenotype. Family members (parents, siblings, children) should have a thorough evaluation using the consensus guidelines for newly diagnosed individuals with TSC. Parents who wish to find out whether a seemingly unaffected sibling of the proband is affected should first undergo genetic testing themselves. If the parents of a child with a known TSC mutation do not have the same mutation, it is exceedingly unlikely that a clinically unaffected sibling has TSC.
Cell biology and mTOR signaling in TSC
The mammalian target of rapamycin or mTOR is a serine/threonine protein that belongs to the phosphatidylinositol 3-kinase-related kinase family.27 It acts through two distinct protein complexes, mTORC1 and mTORC2. mTORC1 is characterized by the presence of regulatory-associated protein of mTOR (Raptor) and is effectively inhibited by rapamycin,28 a macrolide produced by Streptomyces hygroscopicus bacteria. mTORC1 responds to amino acids leucine and arginine, stress, oxygen, energy, and growth factors to control and promote anabolic processes such as protein synthesis, lipid synthesis, and limiting of catabolic reactions such as autophagy.29 mTORC2 is characterized by the presence of rapamycin-insensitive companion of mTOR (Rictor) and nonresponsiveness to rapamycin.30,31 This complex responds to growth factors and regulates cell survival, metabolism, and cytoskeletal integrity.28 mTORC1 has five components in its catalytic subunit: Raptor, mammalian lethal with SEC13 protein 8 (mLST8), proline-rich AKT substrate 40 kDa (PRAS40), and DEP domain-containing mTOR-interacting protein (Deptor). mTORC2 has six different components: Rictor, mammalian stress-activated protein kinase interacting protein 1 (mSIN1), protein observed with Rictor-1 (Protor-1), mLST8, and Deptor.30
The mTOR pathway provides rheostatic control of cell growth and autophagy and serves as a primary regulator of cell growth and proliferation in response to cellular nutrition, energy levels, and growth factor stimulation.32 The mTOR signaling pathway responds to different environmental factors and then integrates the diverse environmental cues into appropriate cellular responses.31,33,34 Homologous pathways have been found in yeast, Drosophila, and other eukaryotes, suggesting that they are highly conserved among species.27,35 TSC1 and TSC2 are genes that encode proteins hamartin and tuberin, the proteins that form the TSC1 and TSC2 complex (TSC1:TSC2). The complex functions as the main negative regulator of mTOR through GTPase-activating protein (GAP), acting on Ras-homolog enriched in the brain (Rheb). If GTP is bound to Rheb, it becomes a potent activator of mTOR. Therefore, TSC1:TSC2 inhibits this activation by stimulating the GTPase activity of Rheb, which in turn cleaves a phosphate group from GTP, resulting in the inactive GDP-bound form.28,35 In the absence of TSC1:TSC2, the levels of Rheb-GTP increase leading to the activation of the mTOR–Raptor pathway and constitutive deregulation of protein synthesis and cell growth.35
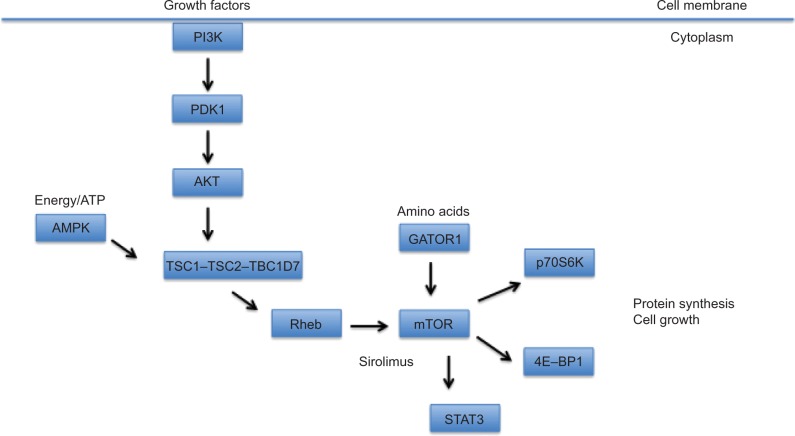
Deregulation of mTOR signaling pathway has been linked to a wide variety of neurodevelopmental disorders, including TSC as well as focal cortical dysplasia, hemimegalencephaly, and megalencephaly.31 The mTOR pathway has also been directly linked to a number of cancer subtypes. mTOR is regulated upstream by several protein kinases, such as PI3K, PDK1, PTEN, AKT, LKB1, and AMPK (Figure 1). LKB1 is an important inhibitor of the mTOR pathway since it responds to energy starvation and is necessary to protect the cell from death triggered by stress. LKB1 also enhances phosphorylation of AMPK-dependent phosphorylation sites on TSC2, suggesting its importance in activating TSC2:TSC1 under conditions of energy starvation.36 However, the identification of TSC1 and TSC2 genes, as causal to TSC, has served as a pivot point for understanding mTOR regulation. TSC1 and TSC2 proteins serve as a meeting point for the complex intracellular signaling pathways that include insulin signaling (PI3K–AKT), mitogenic signaling (MAPK), and energy sensing (AMPK). The main output pathway of TSC2:TSC1 is the mTOR protein complex, and under normal homeostatic conditions, TSC2:TSC1 stops mTOR activation.34,36 The activity of both TSC1 and TSC2 is mediated by phosphorylation at multiple sites. TSC1 is phosphorylated by cyclin-dependent kinase 1 (CDK1)/cyclin B and by IKKβ.TSC2 is phosphorylated by AKT and AMPK as well as extracellular signaling-regulated kinase (Erk) and glycogen synthase kinase 3 (GSK3).
Figure 1.
A schematic diagram depicting the TSC-mTOR signaling pathway.
Notes: mTOR is modulated by the TSC1–TSC2–TBC1D7 complex, integrating inputs from growth factors via PI3K, amino acids via the GATOR1 complex, and cellular energy levels (ATP) via AMPK. Enhanced mTOR signaling in response to these cues or as a consequence of TSC1 or TSC2 mutations leads to increased protein synthesis and cell growth.
Abbreviations: TSC, tuberous sclerosis complex; mTOR, mechanistic target of rapamycin; PI3K, PI3 kinase; p70S6K, p70 S6 kinase.
In eukaryotic cells under normal physiological conditions, TSC1 and TSC2, along with TBC1D7, form a heteromeric complex that serves as a crucial upstream negative regulator of mTOR activity. The complex acts as a guanosine triphosphatase activator (GAP) protein that prevents the phosphorylation of the G-protein Rheb. When Rheb is inactive, it is unable to activate mTORC1, thus inhibiting cell growth.4,31,37 Under growth-promoting conditions, growth factors activate the intracellular PI3K–AKT–mTOR pathway and TSC2 is then phosphorylated by Akt, decreasing GAP activity and increasing mTOR activity. When hypoxic or low energy (ATP) conditions are signaled to the pathway, TSC2 is phosphorylated and GAP activity is increased.28,37 A mutation in either TSC1 or TSC2 genes interferes with inhibitory control of mTOR, leading to hyperactivation, continued cell proliferation, and in some cases, oncogenic transformation.36 Activation of mTOR pathway leads to enhanced protein synthesis via the downstream effectors of mTOR, p70S6K, ribosomal protein S6, and translation initiation factors, such as 4E-binding proteins (4E-BP1). Indeed, deleterious putatively pathogenic variants are functionally validated in vitro based on their effect (enhancement) on mTOR signaling, eg, increased phosphorylation of p70S6K. Loss of TSC2:TSC1 can also lead to endoplasmic reticulum stress, which leads to protein unfolding. This stress response impacts mTOR-mediated negative feedback inhibition and raises the cell’s vulnerability to apoptosis.36 TBC1D7 is the stably associated and ubiquitous third core subunit of the TSC2:TSC1 complex. A knockdown of the TBC1D7 subunit has been shown to decrease the association of TSC1 and TSC2, reducing GAP activity toward Rheb, along with increased mTOR activity, delayed induction of autophagy, and enhanced cell growth under low energy conditions.38
In many organ systems, TSC lesions, including renal AMLs, cortical tubers, and SEGAs, exhibit constitutively activated mTOR signaling evidenced by enhanced phosphorylation of p70S6K, ribosomal S6, and STAT3 proteins. It is believed that TSC1 or TSC2 heterozygosity will not confer mTOR activation and that a somatic second-hit mutation in the unaffected allele is needed.4,39 Biallelic inactivation in the form of LOH or small mutations has been identified in renal and brain lesions.
Death-associated protein kinase has also been identified as a negative regulator of TSC2:TSC1. Its subsequent phosphorylation of TSC2 leads to the dissociation of the heterodimer. TSC2:TSC1 is also inhibited by the binding of forkhead transcription factor (FoxO1) to TSC2, ultimately activating the mTOR signaling cascade.2 Abnormal β-catenin signaling has also been argued as a component of TSC pathogenesis.40 Another inhibitor of the mTOR signaling pathway is p53, through the regulation of AMPK, PTEN, and TSC2.2 TSC2:TSC1 activation is mediated by AMPK and GSK3. Depletion of ATP results in enhanced phosphorylation of TSC2 and decreased phosphorylation of mTOR effectors, S6K and 4E-BP1. Energy depletion also induces LKB1-dependent activation of AMPK, which then activates the TSC2:TSC1 complex.
The mTOR complex phosphorylates S6K1 and 4E-BP1. Catalytic activity of S6K1 is regulated by multiple phosphorylation events, and it is a kinase that activates ribosomal subunit protein S6, which leads to the recruitment of ribosomes and protein translation.37 Additionally, S6K1 establishes a negative feedback loop on mTOR signaling by negative regulation of insulin receptor substrate-1. Taking a step back, it is evident that enhanced activation of the PI3K–AKT pathway leads to activation of the mTOR cascade, which results in enhanced phosphorylation of S6K1, and later, attenuation of PI3K–AKT signaling. 4E-BP1 has been implicated in the regulation of metabolism and adipogenesis41 and inhibits translation initiation factor (eIF4E). When phosphorylated by mTOR, 4E-BP1 loses control of eIF4E.37
VEGF has also been suggested as a biomarker for abnormal TSC2:TSC1 function due to its high presence in TSC1 heterozygous animals with hepatic hemangiomas. Furthermore, treatment of TSC1 heterozygous animals with repeated doses of rapamycin decreases serum VEGF levels and results in histological changes in the tumors. Phosphorylation of TSC1 by IKKβ results in dissociation of the TSC2:TSC1 complex, enhanced levels of GTP-bound Rheb, and subsequent increase in the activation of mTOR, causing an increase in VEGF and accelerated angiogenesis.2
Clinical utility of mTOR inhibitors in TSC
Inhibition of mTOR can be achieved pharmacologically by rapamycin (sirolimus), which causes dissociation of mTORC1 from its binding partner Raptor, thus successfully inactivating it. Rapalogs (rapamycin derivatives) have also been proven to be valuable candidates to inhibit mTORC1 and are being used to treat different cancer malignancies in addition to TSC.4 Everolimus, a derivative of sirolimus, has been successfully used to treat SEGA.42 Everolimus treatment led to a reduction in SEGA volume within 3 months, an effect that persisted for 5 years. Additional studies suggest that everolimus may be useful for the treatment of epilepsy in TSC.
Summary and future directions
The mTOR cascade has now been linked to a host of neurological disorders at both ends of its signaling spectrum. In disorders characterized by exuberant mTOR signaling, inhibition of mTOR itself or targeting downstream substrates provides hopeful avenues for therapy. Indeed, advances over the next 5–10 years will hopefully identify specific substrates, eg, precision targets, for distinct neurological disorders. In contrast, enhancing mTOR signaling in disorders where mTOR activation is diminished will be a challenge since few if any pharmacological compounds can target mTOR for activation. For both types of neurological disorders, establishment of appropriate timing and duration of therapy will be of paramount importance.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bourneville DM. Sclerose tubereuse des circonvolutions cerebrales. Arch Neurol. 1880;1:81–91. French. [Google Scholar]
- 2.Orlova KA, Crino PB. The tuberous sclerosis complex. Ann N Y Acad Sci. 2010;1184:87–105. doi: 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Northrup H, Krueger DA. Tuberous sclerosis complex diagnostic update: recommendations of the 2012 international tuberous sclerosis complex consensus conference. Pediatr Neurol. 2013;49(4):243–254. doi: 10.1016/j.pediatrneurol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohrman MH. Emerging treatments in the management of tuberous sclerosis complex. Pediatr Neurol. 2012;46(5):267–275. doi: 10.1016/j.pediatrneurol.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Jansen FE, Vincken KL, Algra A, et al. Cognitive impairment in tuberous sclerosis complex is a multifactorial condition. Neurology. 2008;70(12):916–923. doi: 10.1212/01.wnl.0000280579.04974.c0. [DOI] [PubMed] [Google Scholar]
- 6.Curatolo P. Mechanistic target of rapamycin (mTOR) in tuberous sclerosis complex-associated epilepsy. Pediatr Neurol. 2015;52(3):281–289. doi: 10.1016/j.pediatrneurol.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Henske EP, Neumann HP, Scheithauer BW, Herbst EW, Short MP, Kwiatkowski DJ. Loss of heterozygosity in the tuberous sclerosis (TSC2) region of chromosome band 16p13 occurs in sporadic as well as TSC-associated renal angiomyolipomas. Genes Chromosomes Cancer. 1995;13(4):295–298. doi: 10.1002/gcc.2870130411. [DOI] [PubMed] [Google Scholar]
- 8.Tsai V, Crino P. Tuberous sclerosis complex: genetic basis and management strategies. Adv Genomics Genet. 2012;2012:19–31. [Google Scholar]
- 9.European Chromosome 16 TSC Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 10.van Slegtenhorst M, de Hoogt R, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277(5327):805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 11.Xu L, Sterner C, Maheshwar MM, et al. Alternative splicing of the tuberous sclerosis 2 (TSC2) gene in human and mouse tissues. Genomics. 1995;27(3):475–480. doi: 10.1006/geno.1995.1079. [DOI] [PubMed] [Google Scholar]
- 12.Santiago Lima AJ, Hoogeveen-Westerveld M, Nakashima A, et al. Identification of regions critical for the integrity of the TSC1-TSC2-TBC1D7 complex. PLoS One. 2014;9(4):e93940. doi: 10.1371/journal.pone.0093940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412(2):179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rendtorff ND, Bjerregaard B, Frödin M, et al. Analysis of 65 tuberous sclerosis complex (TSC) patients by TSC2 DGGE, TSC1/TSC2 MLPA, and TSC1 long-range PCR sequencing, and report of 28 novel mutations. Hum Mutat. 2005;26(4):374. doi: 10.1002/humu.20227. [DOI] [PubMed] [Google Scholar]
- 15.Dabora SL, Jozwiak S, Franz DN, et al. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am J Hum Genet. 2001;68(1):64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povey S, Ekong R. Tuberous Sclerosis Database. Leiden Open Variation Database; 2016. [Accessed May 14, 2016]. webpage on the Internet. Available from: http://chromium.lovd.nl/LOVD2/TSC. [Google Scholar]
- 17.Sancak O, Nellist M, Goedbloed M, et al. Mutational analysis of the TSC1 and TSC2 genes in a diagnostic setting: genotype – phenotype correlations and comparison of diagnostic DNA techniques in Tuberous Sclerosis Complex. Eur J Hum Genet. 2005;13(6):731–741. doi: 10.1038/sj.ejhg.5201402. [DOI] [PubMed] [Google Scholar]
- 18.Boehm D, Bacher J, Neumann HP. Gross genomic rearrangement involving the TSC2-PKD1 contiguous deletion syndrome: characterization of the deletion event by quantitative polymerase chain reaction deletion assay. Am J Kidney Dis. 2007;49(1):e11–e21. doi: 10.1053/j.ajkd.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 19.Hoogeveen Westerveld M, Wentink M, van den Heuvel D, et al. Functional assessment of variants in the TSC1 and TSC2 genes identified in individuals with tuberous sclerosis complex. Hum Mutat. 2011;32(4):424–435. doi: 10.1002/humu.21451. [DOI] [PubMed] [Google Scholar]
- 20.Kothare SV, Singh K, Chalifoux JR, et al. Severity of manifestations in tuberous sclerosis complex in relation to genotype. Epilepsia. 2014;55(7):1025–1029. doi: 10.1111/epi.12680. [DOI] [PubMed] [Google Scholar]
- 21.Rakowski SK, Winterkorn EB, Paul E, Steele DJ, Halpern EF, Thiele EA. Renal manifestations of tuberous sclerosis complex: incidence, prognosis, and predictive factors. Kidney Int. 2006;70(10):1777–1782. doi: 10.1038/sj.ki.5001853. [DOI] [PubMed] [Google Scholar]
- 22.Crino P, Aronica E, Baltuch G, Nathanson KL. Biallelic TSC gene inactivation in tuberous sclerosis complex. Neurology. 2010;74(21):1716–1723. doi: 10.1212/WNL.0b013e3181e04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niida Y, Stemmer-Rachamimov AO, Logrip M, et al. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am J Hum Genet. 2001;69(3):493–503. doi: 10.1086/321972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin W, Chan JA, Vinters HV, et al. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol. 2010;20(6):1096–1105. doi: 10.1111/j.1750-3639.2010.00416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patursky-Polischuk I, Kasir J, Miloslavski R, et al. Reassessment of the role of TSC, mTORC1 and microRNAs in amino acids-meditated translational control of TOP mRNAs. PLoS One. 2014;9(10):e109410. doi: 10.1371/journal.pone.0109410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dombkowski AA, Batista CE, Cukovic D, et al. Cortical tubers: windows into dysregulation of epilepsy risk and synaptic signaling genes by microRNAs. Cereb Cortex. 2016;26(3):1059–1071. doi: 10.1093/cercor/bhu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17(8):666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 28.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(S2):S43–S51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laplante M, Sabatini DM. MTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crino PB. mTOR: a pathogenic signaling pathway in developmental brain malformations. Trends Mol Med. 2011;17(12):734–742. doi: 10.1016/j.molmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37(1):19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 33.Xu K, Liu P, Wei W. MTOR signaling in tumorigenesis. Biochim Biophys Acta. 2014;1846(2):638–654. doi: 10.1016/j.bbcan.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang K, Fingar DC. Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Napolioni V, Curatolo P. Genetics and molecular biology of tuberous sclerosis complex. Curr Genomics. 2008;9(7):475–487. doi: 10.2174/138920208786241243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingswood JC, de Vries PJ. Tuberous sclerosis complex. Paediatr Child Health. 2015;25(10):467–473. [Google Scholar]
- 37.Curatolo P, Bombardieri R, Jozwiak S. Tuberous sclerosis. Lancet. 2008;372(9639):657–668. doi: 10.1016/S0140-6736(08)61279-9. [DOI] [PubMed] [Google Scholar]
- 38.Dibble CC, Elis W, Menon S, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47(4):535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 40.Mak BC, Kenerson HL, Aicher LD, Barnes EA, Yeung RS. Aberrant β-catenin signaling in tuberous sclerosis. Am J Pathol. 2005;167(1):107–116. doi: 10.1016/s0002-9440(10)62958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai S, Sitzmann JM, Dastidar SG, et al. Muscle-specific 4E-BP1 signaling activation improves metabolic parameters during aging and obesity. J Clin Invest. 2015;125(8):2952–2964. doi: 10.1172/JCI77361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krueger DA, Care MM, Holland K, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363(19):1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]