Abstract
Extraordinary progress in the structure and immunobiology of the human respiratory syncytial virus glycoproteins has been accomplished during the last few years. Determination of the fusion (F) glycoprotein structure folded in either the prefusion or the postfusion conformation was an inspiring breakthrough not only to understand the structural changes associated with the membrane fusion process but additionally to appreciate the antigenic intricacies of the F molecule. Furthermore, these developments have opened new avenues for structure-based designs of promising hRSV vaccine candidates. Finally, recent advances in our knowledge of the attachment (G) glycoprotein and its interaction with cell-surface receptors have revitalized interest in this molecule as a vaccine, as well as its role in hRSV immunobiology.
Human respiratory syncytial virus (hRSV) was recently classified in the genus Orthopneumovirus of the newly created Pneumoviridae family within the order Mononegavirales, detached from the original Paramyxoviridae family [1]. hRSV is an enveloped virus with a genome made of a single-stranded RNA molecule of negative polarity and about 15.2 kb in length. Molecules of nucleoprotein (N) wrap around the entire length of this RNA to form a stable ribonucleoprotein (RNP) complex, which encodes 11 proteins, three of which are membrane-bound glycoproteins (G, F and SH) (for a review, [2]). The G glycoprotein was originally described as the receptor-binding or attachment protein [3]. F was identified by Walsh and Hruska [4] as the fusion protein that fuses the viral and cell membranes enabling the virus RNP to reach the cell cytoplasm. Finally, SH was initially described as a viroporin—a class of small viral proteins that modify membrane permeability [5]—and was later found to form pentameric pore-like structures in the membrane that confer cation-selective channel-like activity, compatible with its initial designation as a viroporin [6].
It is widely accepted that protection against hRSV is conferred mainly by neutralizing antibodies. For instance, high levels of neutralizing antibodies correlate with protection of human adult volunteers to hRSV challenge [7], as well as a lower risk of hRSV infection in children [8] and in the elderly [9]. Therefore, the surface glycoproteins, particularly F, have recently received much attention as targets of neutralizing and protective antibodies and as potential antigens to be included in a hRSV vaccine [10]. These aspects of hRSV vaccinology are the topic of this review.
Structure and function of the hRSV G glycoprotein
The G protein is synthesized as a polypeptide precursor of about 300 amino acids (depending on the viral strain) with a single hydrophobic domain (residues 38–63) near the N-terminus that acts as a combined signal and membrane anchor domain [11] (Fig. 1). This hydrophobic region targets the nascent chain, as it emerges from the ribosome, to the endoplasmic reticulum and ensures translocation of the polypeptide chain across the membrane while anchoring the G protein to the lipid bilayer. G has neither sequence nor structural homology with the attachment protein of viruses in the Paramyxoviridae family [11].
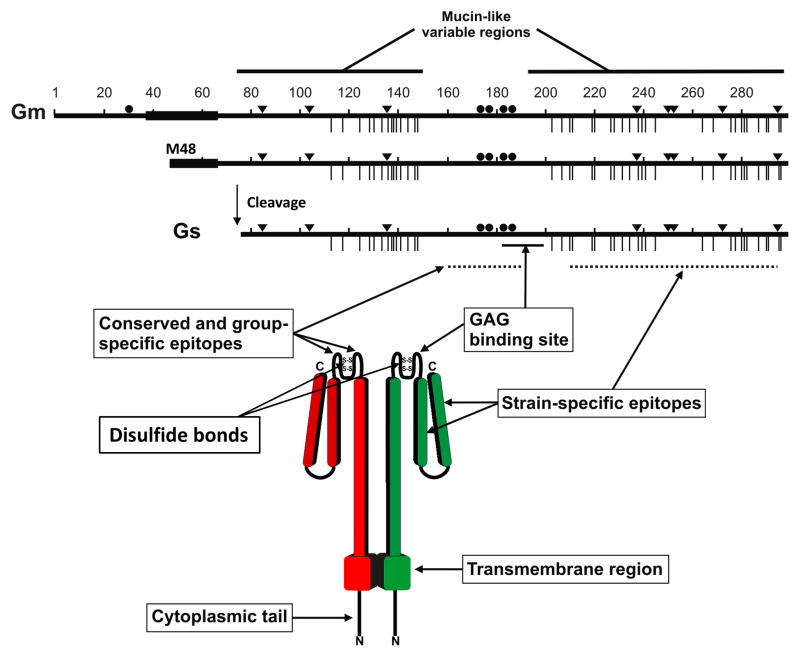
Figure 1. Human respiratory syncytial virus G glycoprotein.
The full length, 298 amino acid membrane-anchored G protein (Gm) and the 233 amino acid soluble G protein (Gs) are shown (Long strain). Hydrophobic regions are denoted by thick lines. Gs is formed by alternative translation initiation at M48, followed by cleavage after residue 65. Inverted triangles represent N-linked glycosylation sites and vertical lines indicate O-linked glycosylation sites. Cysteine residues overlapping the central conserved domain are represented by solid circles. The lower part of the figure depicts a model of the 3-dimensional structure of Gm. While Gm is probably tetrameric [38], a dimer is shown for simplicity. The mucin-like regions are depicted as extended rod-like structures due to the presence of multiple O-linked sugars that have a tendency to stretch the polypeptide backbone [93] The second hypervariable region is externally located in the model to denote that it harbors multiple epitopes and it is shown as two halves joined by a protease susceptible site [94]. Antibody epitopes and the glycosaminoglycan (GAG) binding site are indicated by arrows. Figure provided by Alfonsina Trento.
The G polypeptide precursor is extensively modified by the addition of both N- and O-linked oligosaccharides and is also palmitoylated at a single cysteine residue in its N-terminal cytoplasmic tail [12]. High-mannose N-linked glycans are co-translationally added to the G protein precursor, followed by the conversion of these sugars to the complex type and addition of O-linked glycans in the Golgi compartment. These modifications convert the 32 kDa precursor into a mature protein of 80–90 kDa (estimated by SDS-PAGE) in most immortalized cell lines [13], whereas a 180 kDa form has been described in human airway epithelial (HAE) cultures [14] that is postulated to represent either a dimer of the 90 kDa G protein or the 90 kDa form with additional or more extensive O-linked carbohydrate chains.
The G protein ectodomain consists of two large heavily glycosylated “mucin-like” domains, rich in serine, threonine and proline residues (characteristic of mucins), connected by a short central region devoid of carbohydrates (Fig. 1) [15]. The sequence of the two mucin-like domains is extremely variable among viral strains [16] but they all have several potential sites for N-glycosylation and multiple serines and threonines that are predicted to be O-glycosylated by the NetOGlyc software [17]. This sequence variability has been used in numerous studies of molecular epidemiology and evolution of hRSV [18]. Thus, hRSV strains have been classified into two genetic groups, A and B, that correlate with the antigenic groups, initially identified by reactivity with certain monoclonal antibodies (mAbs) [19;20]. Within each group numerous clades or genotypes have been identified. Viruses of different genotypes and even different antigenic groups frequently co-circulate in each yearly outbreak. The dominance of these genotypes changes in successive epidemics and replacement of certain genotypes by others has been noticed at the global level [21].
The central conserved region of hRSV G (aa 163–189) has four cysteines (residues 173, 176, 182 and 186) that are conserved in all viral strains. Within this region there is a stretch of 13 amino acids (164–176) that is strictly maintained in all strains while the remaining sequence of the central region is somewhat group-specific. Disulfide bridges are formed between Cys173 and Cys186, and between Cys176 and Cys182, resulting in a cystine noose motif which resembles the structure found in the 55 kDa tumour necrosis factor receptor [22;23]. The Cys-rich motif is missing in the highly related G protein of human metapneumovirus (hMPV), which otherwise shares the overall amino acid composition and sequence variability of hRSV G [24;25]. The ectodomains of both hRSV and hMPV G are predicted to be disordered (except for the hRSV cystine noose), consistent with the high content of serine, threonine, and proline residues and extensive O-glycosylation [26].
The central region of hRSV G contains the CX3C motif (aa 182–186) that can bind to CX3CR1—the specific receptor of the fractalkine chemokine—and hence induce leukocyte chemotaxis [27]. Several authors have reported hRSV binding to differentiated HAE cells by the interaction of the G protein with CX3CR1 in the apical surface of ciliated cells [28–30]. Inhibition of CX3CR1 binding reduces but does not entirely suppress infection of HAE cultures, indicating that CX3CR1is an important but not the only hRSV receptor in these cells. It has been reported that hRSV uses cell surface proteoglycans for attachment to established cell lines [31–33] mainly by interactions of the G protein with glycosaminoglycans (GAGs) [34;35]. Whether proteoglycans may also act as an hRSV G receptor in HAE cells is still controversial [28;30].
In addition to the membrane bound form of hRSV G, infected cells also produce a soluble form of G (sG) [36] by internal initiation of translation at a second AUG codon (Met48) located in the middle of the transmembrane region and subsequent cleavage after residue Asn66 (Fig. 1) [37]. While sG is monomeric, membrane bound G is oligomeric (probably a tetramer) emphasizing the relevance of the transmembrane domain for oligomerization [38]. The actual role of sG in hRSV biology is not known, although it has been postulated to help evade the antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc-receptor-bearing leukocytes [39].
Antigenicity and immunogenicity of the hRSV G glycoprotein
Three types of epitopes have been identified in the G protein by murine mAbs: i) conserved epitopes which are present in all viral strains and that map within the conserved 13 amino acid stretch of the unglycosylated central region, ii) group-specific epitopes that partially overlap with the conserved epitopes but are shared only by strains of the same antigenic group and iii) strain-specific epitopes that are present only in certain strains of the same antigenic group and have been mapped in the C-terminal hypervariable region of the G protein ectodomain (Fig. 1) [40]. These variable epitopes are influenced by cell-type-specific glycosylation [13].
The majority of murine mAbs specific for the G glycoprotein have minimal effects on virus infectivity in classical complement-independent neutralization assays performed with immortalized cell lines [41;42]. However, pools of antibodies binding to different epitopes of G showed a synergistic effect on this type of neutralization [43], suggestive of hRSV inhibition by steric hindrance.
Recent studies have demonstrated that 131-2G [41], a murine mAb which binds to an epitope located in the central region of hRSV G and that is conserved in all viral strains tested so far, reduces hRSV binding to CX3CR1 in HAE cell cultures [28;29]. Antibody 131-2G reduces several disease manifestations in hRSV challenged mice, including pulmonary inflammation [44] and mucus production [45]. Mice inoculated with G protein polypeptides or peptides spanning the central conserved region of G elicited antibodies that blocked the interaction of the G protein with CX3CR1 and had reduced pathogenesis mediated by hRSV infection [46]. Likewise, mice vaccinated with recombinant influenza virus carrying a chimeric HA protein containing the conserved domain of hRSV G [47] or nanoparticles carrying the CX3C motif of hRSV G [48] had reduced virus titers and pathology in the lungs after a hRSV challenge. These results extend those previously obtained with a BBG2Na vaccine that comprised residues 130 to 230 of hRSV G fused to the albumin-binding region of the streptococcal protein B [49]. This vaccine was tested in humans but these trials were halted after two individuals in a phase II trial developed type III hypersensitivity, likely attributable to the bacterial component. Nevertheless, the results cited in this paragraph unlock new possibilities for hRSV vaccine development based on the G glycoprotein.
Structure and function of the hRSV F glycoprotein
The F protein is a type I glycoprotein which shares structural motifs with the F proteins of other Pneumoviridae (e.g., hMPV) and Paramyxoviridae (e.g., parainfluenza virus type 5, PIV5) viruses, despite limited sequence identity, suggesting that they all function through similar mechanisms. The F glycoprotein is synthesized as an inactive precursor (F0) of 574 amino acids that has three hydrophobic peptides (Fig. 2): i) the N-terminal signal peptide (aa 1–21), which directs translocation of the nascent polypeptide to the lumen of the endoplasmic reticulum and is not present in the mature molecule, ii) the transmembrane region (aa 525–550) near the C-terminus, which anchors F to the cell and viral membranes, and iii) the so-called fusion peptide (aa 137–155), which inserts into the target cell membrane during the fusion process. F0 is post-translationally cleaved after two polybasic furin sites at residues 109 (cleavage site I) and 136 (cleavage site II), separated by 27 amino acids (pep27), to become fusion competent [50]. The double proteolytic cleavage is shared with the homologous F protein of bovine RSV but it is a unique feature among the Pneumoviridae and Paramyxoviridae F proteins, which are cleaved only once. Once cleavage of hRSV F is completed, the intervening pep27 is released from the mature protein [51] and two chains are generated (F2 N-terminal to F1) which remain covalently linked by two disulfide bridges (Cys70–Cys212 and Cys37–Cys439). The newly created N-terminus of the F1 chain contains the fusion peptide. There are two N-linked glycosylation sites in F2 and one in F1, and these are conserved in all hRSV strains. The F1 chain has a central cysteine-rich region flanked by two heptad repeats: HRA is located C-terminal to the fusion peptide and HRB precedes the transmembrane region. The mature hRSV F glycoprotein is a homotrimer of F1+F2 subunits.
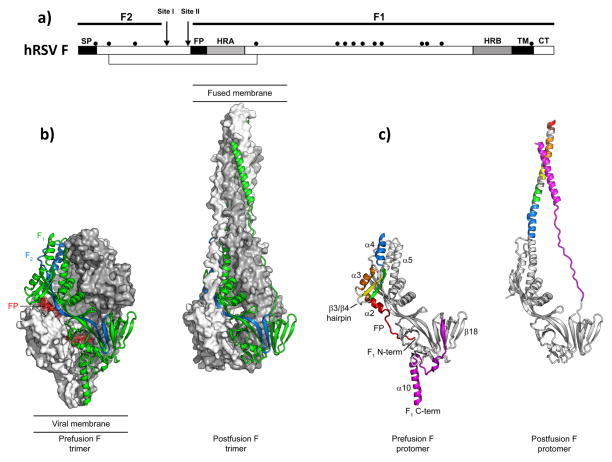
Figure 2. Human respiratory syncytial virus F glycoprotein.
(a) Diagram of the F protein precursor denoting the signal peptide (SP), the fusion peptide (FP) and the transmembrane region (TM), as well as cleavage sites I and II and the cysteine residues (black dots). (b) Structure of the F protein trimer folded in the prefusion (left) and postfusion (right) conformation. One protomer is shown as ribbons and colored blue (F2 chain), green (F1 chain) and red (fusion peptide). Molecular surfaces are shown for the other two F protomers, colored grey and white. (c) A single hRSV F protomer is displayed as ribbon, folded in the prefusion (left) and postfusion (right) conformation. The same colors are used for the secondary structure elements (indicated in the prefusion protomer) in the two conformations. Note that the structure colored grey is essentially unchanged in the prefusion and postfusion conformation.
The main function of hRSV F is to promote fusion of the viral and cell membranes; however, there have been reports of spontaneous deletion mutants [52] and recombinant viruses [53] in which F is the only viral glycoprotein. These mutants replicate in established cell lines but not in HAE cultures [14] and are attenuated in animal models [54]. Therefore, at least in ΔG viruses, F has to assume the virus binding function of the G protein, in addition to its membrane fusion activity. Indeed, F has been found to bind proteoglycans [55] and other cell-surface molecules, such as nucleolin [56], compatible with its role as a substitute attachment protein. However, whether wild-type virus requires F binding to cells for infectivity is still not entirely clear.
The relevance of the hRSV F double cleavage for membrane fusion is also uncertain. Grafting of the double cleavage site of hRSV F in Sendai virus F resulted in a dramatic increase of cell-cell fusion mediated by the chimeric protein in transfected cells, as well as a decrease in dependence of hemagglutinin-neuraminidase (HN) co-expression for cell-cell fusion [57]. Furthermore, replacement of Sendai virus F by the chimeric protein reduced virus thermostability and decreased dependence on HN binding to sialic acid for infection, mimicking the unique ability of hRSV to fuse and infect cells in the absence of a separate attachment protein [58]. Therefore, the presence of two cleavage sites in hRSV F seems to modulate its membrane fusion activity by still ill-defined mechanisms.
The F trimer is assembled in the virus particle in a metastable conformation, called pre-fusion. During membrane fusion, F experiences a series of conformational changes that result in a highly stable structure, called post-fusion (see later). Important knowledge about these conformational changes has been recently gained by solving the atomic structures of soluble forms of hRSV F folded in either the prefusion or postfusion conformation [59–62], as shown in Fig. 2.
One of the main hurdles in these studies was the stabilization of a soluble form of hRSV F in its prefusion conformation. Initially, it was found that the expression of the F protein ectodomain led to formation of soluble trimers (sF) that retained epitopes recognized by certain neutralizing mAbs [63]. Partial deletion of the fusion peptide prevented aggregation of sF after cleavage [64] and allowed its crystallization in the absence of detergents [59;60]. The X-ray structures determined from these crystals demonstrated that sF was folded in the postfusion conformation, indicating that the F ectodomain assembles spontaneously into the highly stable postfusion form when expressed without the transmembrane region. Of note, the full-length F also refolds into the postfusion form if extracted with detergents from the cell or viral membranes. Therefore, a central challenge was to obtain a soluble hRSV F ectodomain stabilized in the prefusion form, amenable to crystallization. This was initially achieved by co-expression of the hRSV F ectodomain in complex with the Fab fragment of a neutralizing mAb (D25) which did not bind to postfusion F and hence was presumably specific for the prefusion conformation [61]. Indeed, the structure of F in that complex differed substantially from the previously described postfusion hRSV F and resembled the structure of the paramyxovirus PIV5 prefusion F, reported by Yin et al., [65]. Based on that structure, several mutants of the hRSV F ectodomain were made to stabilize it in the prefusion conformation in the absence of antibodies [62]. One of the most stable mutants (DS-Cav1) had two serines (155 and 290) substituted by cysteines to create an intrasubunit disulfide bond, two cavity filling mutations (S190F and V207L) in the F1 chain to help stabilization, and a foldon trimerization domain at the C-terminus [62]. DS-Cav1 has been extensively used in several studies but other mutants, stabilized in the prefusion conformation by alternative strategies (SC-TM), have been obtained with enhanced stability properties [66].
Fig. 2 shows a comparison of the hRSV F prefusion and postfusion structures. Most of the secondary and tertiary structure is preserved in the pre- and postfusion forms. In contrast, the N- and C-termini of the F1 chain undergo substantial conformational changes. During the fusion process, the fusion peptide and the first five secondary-structure elements at the N-terminus of F1 rearrange and fuse with the α5 helix to form an extended helix of >100 Å in length. Near the C-terminus of F1, parallel strand β22 dissociates as the C-terminal helix rearranges to form the outer helix of the postfusion six-helix bundle (6HB). Similar rearrangements had been inferred by comparison of the related prefusion PIV5 and postfusion PIV3 F structures [67,68], suggesting that all these viruses share a similar membrane fusion mechanism.
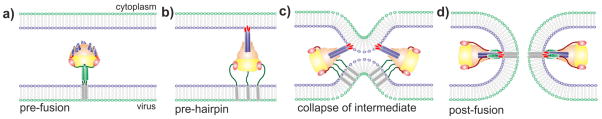
Fig. 3 illustrates the model of membrane fusion mediated by hRSV F, taken from the initial model proposed for PIV5 [65]. After binding of the incoming virus to the cell surface, prefusion F is activated by an ill-defined process which apparently does not require the attachment G glycoprotein, a major difference with the F protein of the Paramyxoviridae. After activation, F initiates a series of conformational changes which probably involve separation of the HRB helices and formation of the long HRA α-helix, as mentioned above. These changes relocate the fusion peptide—which is buried deep inside the prefusion globular head—towards the end of the newly formed helix, which probably assembles into a coiled-coil trimer of HRA sequences. Since the fusion peptide is hydrophobic, it avoids the hydrophilic environment by inserting into the outer layer of the cell membrane, leading to formation of the so-called pre-hairpin intermediate. At this stage, the cell and viral membranes are connected by sequences of the F1 subunit which lie between the fusion peptide and the transmembrane region, respectively. This unstable pre-hairpin intermediate refolds by zipping the HRB helix towards the HRA coiled-coil, bringing the two membranes into proximity. Finally, the fusion peptide and the transmembrane domain of F end up in the same membrane after formation of the 6HB, in which an internal core of three HRA α-helices is surrounded by three antiparallel HRB helices. Completion of the 6HB assembly leads to exchange of lipids between the two membranes, formation of the initial fusion and expansion of this pore to complete membrane fusion.
Figure 3. Model of membrane fusion mediated by the hRSV F glycoprotein.
(a) A single prefusion F protein trimer is depicted inserted into the viral membrane through a HRB stalk (green). (b) Upon activation, the short α-helices of HRA (blue) refold into a long trimeric coiled-coil (blue) and the fusion peptide of each subunit (red) is inserted into the target membrane, forming the so-called pre-hairpin intermediate. (c) Collapse of this unstable intermediate approaches the two membranes. (d) Assembly of the six-helix-bundle (6-HB), formed by a core of three HRA α-helices surrounded by three antiparallel HRB α-helices, results in formation of the fusion pore.
Antigenicity and immunogenicity of the hRSV F glycoprotein
The first panels of mAbs raised against hRSV F were obtained from immunized mice using the hybridoma technology [41;69;70]. These antibodies identified several epitopes that were eventually mapped in the F protein primary structure by isolation and characterization of escape mutants [69;71;72] or by reactivity of antibodies with peptides or F protein fragments [73;74]. Binding of at least some of these antibodies to F could be competed with human sera, indicating that the matching epitopes were relevant in a natural infection. However, the F proteins used in these studies were likely folded in the postfusion conformation since, as noted before, F folds spontaneously into this structure when it is either expressed as a soluble ectodomain or detergent-extracted from cell membranes. It was thus relatively unsurprising that as reported [75;76] most of the neutralizing activity present in human immunoglobulin preparations could not be depleted by adsorption to immobilized preparations of postfusion F. It was therefore concluded that most natural human neutralizing antibodies recognize epitopes preserved only in the prefusion conformation of hRSV F. This was further corroborated by the isolation of mAbs from immortalized human lymphocytes [77] that were specific for prefusion F. These mAbs had higher neutralizing potency [61] than those originally described that reacted with both prefusion and postfusion F [76].
Other human neutralizing antibodies have been recently reported that either recognize neutralizing epitopes exclusive to prefusion F, such as AM14 [78], or show preferential binding to prefusion over postfusion F, such as MPE8 [79]. Interestingly, MPE8 cross-neutralizes not only hRSV but additionally three other Pneumoviridae: bovine RSV, hMPV and pneumonia virus of mice (PVM). Another mAb, 54G10, raised against hMPV F has also shown cross-neutralization with hRSV [80].
Fig. 4 shows the location of antibody epitopes and antigenic sites identified so far in the prefusion and postfusion conformations of hRSV F. Some of these sites are found only in the prefusion (e.g., site Ø) or postfusion (e.g., site 6HB) conformation, while others are present in both conformations (e.g., site II) since, as mentioned above, an extensive area of the protein surface is shared by the prefusion and postfusion conformations (Fig. 2) [61]. In general, the antibodies that bind preferentially to prefusion F are better neutralizers than those that recognize epitopes shared by prefusion and postfusion F. As expected, antibodies which recognize epitopes specific to postfusion F are non-neutralizing.
Figure 4. Antigenic sites of hRSV F glycoprotein.
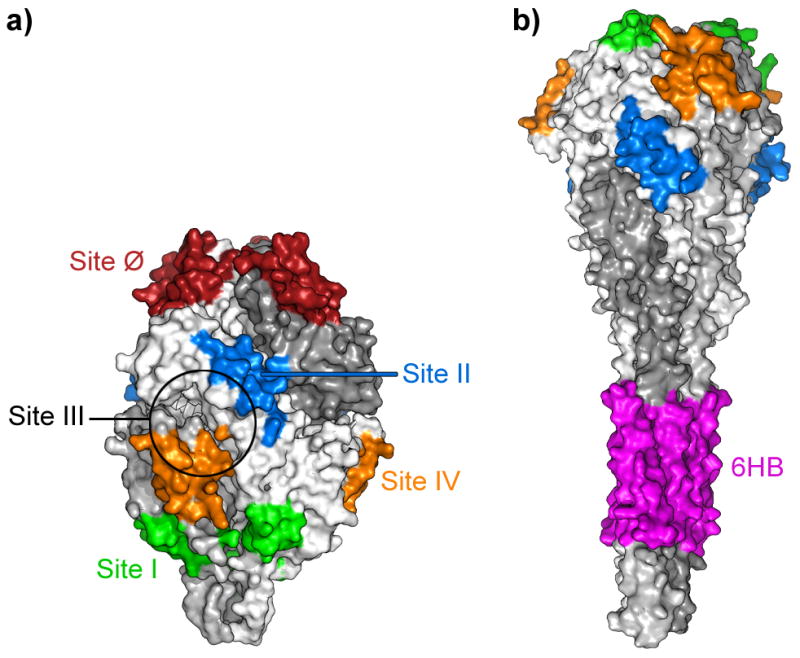
The location of the different antigenic sites is shown in both the prefusion (a) and postfusion (b) conformation of hRSV F. Antigenic site III is delineated by a circle which includes residues identified by mutagenesis to be essential for binding of mAb MPE8 [79], since no other structural information is available yet for this site.
It is assumed but not formally demonstrated that neutralizing antibodies bind to prefusion F and block the initiation of the conformational changes that lead to membrane fusion [81]. Concurring with the neutralizing potency shown by the different mAbs, a detailed analysis of the antibodies present in individual human sera demonstrated that the majority of their neutralizing activity was due to prefusion-specific antibodies directed against antigenic site Ø [82].
Several studies have compared the immunogenic and protective efficacy of purified prefusion and postfusion soluble F in animal models. For instance, the prefusion stabilized DS-Cav1 protein was found to induce ten times higher levels of neutralizing antibodies in mice and rhesus macaques than postfusion F [62]. Similarly, the alternatively stabilized prefusion SC-TM protein also elicited 10–20-fold higher levels of neutralizing antibodies than postfusion F in cotton rats that were additionally shown to be protected against a hRSV challenge [66]. A recent comparison of prefusion, postfusion and a monomeric form of F that shares antigenic properties with prefusion F [83] also demonstrated the superiority of prefusion F in inducing neutralizing antibodies and protection against a hRSV challenge in mice without perceptible pathology [84]. However, protection against a hRSV challenge has also been achieved with postfusion F, likely by induction of neutralizing antibodies that recognize epitopes shared with prefusion F [59;85]. It is worth noting that postfusion F is a highly stable molecule, a valuable characteristic from the point-of-view of vaccine production and distribution.
Stabilized full-length prefusion hRSV F has also been incorporated as an extra gene into PIV3 recombinants [86]. These viruses showed an enhanced neutralizing antibody response against hRSV compared with PIV3 recombinants expressing a soluble postfusion form of hRSV F. Prefusion stabilized hRSV F has also been expressed at the surface of virus-like particles (VLPs) made in cells that expressed the nucleoprotein (NP) and the matrix (M) protein of Newcastle disease virus (NDV). The purified VLPs were inoculated i.m. in either mice [87] or cotton rats [88], and these animals elicited a serum neutralizing antibody response and were protected against an hRSV challenge. Thus, immunization with pre-fusion F, either as a subunit vaccine or incorporated to recombinant viruses or VLPs, seems to be a promising approach for hRSV vaccine development.
Expression of individual F protein epitopes grafted onto different protein scaffolds has also been reported, including the helix-loop-helix motif of antigenic site II [89;90]. Structural analysis of these proteins indicated that they could faithfully reproduce the structural and antigenic features of hRSV F site II. Some of these constructs were able to induce strong neutralizing antibody responses in macaques but only after several immunizations, suggesting that further optimizations of these scaffolds are required before being considered a practical approach to an hRSV vaccine. These scaffolds do, however, represent an interesting alternative for obtaining simplified vaccines with improved production and stability.
In summary, advances in understanding the structure of hRSV glycoproteins, especially the F glycoprotein, have brought new stimulus for development of long-awaited hRSV vaccines which will help to control one of the most important causes of infant hospitalization [91] and one of the leading global causes of infant mortality [92].
Acknowledgments
Work in the Madrid lab is currently funded by grant SAF2015-67033-R from Plan Nacional de I+D+I. J.S.M is supported in part by award P20GM113132 from the National Institute of General Medical Sciences of the National Institutes of Health.
Reference List
- 1.Afonso CL, Amarasinghe GK, Banyai K, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016 Aug;161(8):2351–60. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011 Dec;162(1–2):80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine S, Klaiber-Franco R, Paradiso PR. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987 Sep;68(Pt 9):2521–4. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 4.Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983 Jul;47(1):171–7. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez M, Garcia-Barreno B, Melero JA, Carrasco L, Guinea R. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology. 1997 Sep 1;235(2):342–51. doi: 10.1006/viro.1997.8696. [DOI] [PubMed] [Google Scholar]
- 6.Gan SW, Tan E, Lin X, et al. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem. 2012 Jul 13;287(29):24671–89. doi: 10.1074/jbc.M111.332791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis. 1991 Apr;163(4):693–8. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- 8.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986 Jun;140(6):543–6. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 9.Falsey AR, Walsh EE. Relationship of serum antibody to risk of respiratory syncytial virus infection in elderly adults. J Infect Dis. 1998 Feb;177(2):463–6. doi: 10.1086/517376. [DOI] [PubMed] [Google Scholar]
- 10.Graham BS, Anderson LJ. Challenges and opportunities for respiratory syncytial virus vaccines. Curr Top Microbiol Immunol. 2013;372:391–404. doi: 10.1007/978-3-642-38919-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wertz GW, Collins PL, Huang Y, Gruber C, Levine S, Ball LA. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4075–9. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins PL, Mottet G. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 1992 Apr;73(Pt 4):849–63. doi: 10.1099/0022-1317-73-4-849. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Beato R, Martinez I, Franci C, Real FX, Garcia-Barreno B, Melero JA. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology. 1996 Jul 15;221(2):301–9. doi: 10.1006/viro.1996.0379. [DOI] [PubMed] [Google Scholar]
- 14.Kwilas S, Liesman RM, Zhang L, Walsh E, Pickles RJ, Peeples ME. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J Virol. 2009 Oct;83(20):10710–8. doi: 10.1128/JVI.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson PR, Spriggs MK, Olmsted RA, Collins PL. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5625–9. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melero JA, Moore ML. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity. Curr Top Microbiol Immunol. 2013;372:59–82. doi: 10.1007/978-3-642-38919-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998 Feb;15(2):115–30. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 18.Cane PA. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol. 2001 Mar;11(2):103–16. doi: 10.1002/rmv.305. [DOI] [PubMed] [Google Scholar]
- 19.Anderson LJ, Hierholzer JC, Tsou C, et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985 Apr;151(4):623–33. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 20.Mufson MA, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985 Oct;66(Pt 10):2111–24. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 21.Houspie L, Lemey P, Keyaerts E, et al. Circulation of HRSV in Belgium: from multiple genotype circulation to prolonged circulation of predominant genotypes. PLoS One. 2013;8(4):e60416. doi: 10.1371/journal.pone.0060416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langedijk JP, Schaaper WM, Meloen RH, van Oirschot JT. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J Gen Virol. 1996 Jun;77(Pt 6):1249–57. doi: 10.1099/0022-1317-77-6-1249. [DOI] [PubMed] [Google Scholar]
- 23.Langedijk JP, de Groot BL, Berendsen HJ, van Oirschot JT. Structural homology of the central conserved region of the attachment protein G of respiratory syncytial virus with the fourth subdomain of 55-kDa tumor necrosis factor receptor. Virology. 1998 Apr 10;243(2):293–302. doi: 10.1006/viro.1998.9066. [DOI] [PubMed] [Google Scholar]
- 24.van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002 Mar 30;295(1):119–32. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- 25.van den Hoogen BG, Herfst S, Sprong L, et al. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004 Apr;10(4):658–66. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leyrat C, Paesen GC, Charleston J, Renner M, Grimes JM. Structural insights into the human metapneumovirus Glycoprotein ectodomain. J Virol. 2014 Jul 16; doi: 10.1128/JVI.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM, Anderson LJ. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat Immunol. 2001 Aug;2(8):732–8. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SM, McNally BA, Ioannidis I, et al. Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS Pathog. 2015 Dec;11(12):e1005318. doi: 10.1371/journal.ppat.1005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong KI, Piepenhagen PA, Kishko M, et al. CX3CR1 Is Expressed in Differentiated Human Ciliated Airway Cells and Co-Localizes with Respiratory Syncytial Virus on Cilia in a G Protein-Dependent Manner. PLoS One. 2015;10(6):e0130517. doi: 10.1371/journal.pone.0130517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chirkova T, Lin S, Oomens AG, et al. CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. J Gen Virol. 2015 Sep;96(9):2543–56. doi: 10.1099/vir.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallak LK, Spillmann D, Collins PL, Peeples ME. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol. 2000 Nov;74(22):10508–13. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallak LK, Collins PL, Knudson W, Peeples ME. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000 Jun 5;271(2):264–75. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- 33.Martinez I, Melero JA. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J Gen Virol. 2000 Nov;81(Pt 11):2715–22. doi: 10.1099/0022-1317-81-11-2715. [DOI] [PubMed] [Google Scholar]
- 34.Techaarpornkul S, Collins PL, Peeples ME. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002 Mar 15;294(2):296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 35.Feldman SA, Hendry RM, Beeler JA. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999 Aug;73(8):6610–7. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendricks DA, McIntosh K, Patterson JL. Further characterization of the soluble form of the G glycoprotein of respiratory syncytial virus. J Virol. 1988 Jul;62(7):2228–33. doi: 10.1128/jvi.62.7.2228-2233.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts SR, Lichtenstein D, Ball LA, Wertz GW. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J Virol. 1994 Jul;68(7):4538–46. doi: 10.1128/jvi.68.7.4538-4546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escribano-Romero E, Rawling J, Garcia-Barreno B, Melero JA. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J Virol. 2004 Apr;78(7):3524–32. doi: 10.1128/JVI.78.7.3524-3532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bukreyev A, Yang L, Fricke J, et al. The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J Virol. 2008 Dec;82(24):12191–204. doi: 10.1128/JVI.01604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez I, Dopazo J, Melero JA. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J Gen Virol. 1997 Oct;78(Pt 10):2419–29. doi: 10.1099/0022-1317-78-10-2419. [DOI] [PubMed] [Google Scholar]
- 41.Anderson LJ, Bingham P, Hierholzer JC. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J Virol. 1988 Nov;62(11):4232–8. doi: 10.1128/jvi.62.11.4232-4238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh EE, Hall CB, Schlesinger JJ, Brandriss MW, Hildreth S, Paradiso P. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J Gen Virol. 1989 Nov;70(Pt 11):2953–61. doi: 10.1099/0022-1317-70-11-2953. [DOI] [PubMed] [Google Scholar]
- 43.Martinez I, Melero JA. Enhanced neutralization of human respiratory syncytial virus by mixtures of monoclonal antibodies to the attachment (G) glycoprotein. J Gen Virol. 1998 Sep;79(Pt 9):2215–20. doi: 10.1099/0022-1317-79-9-2215. [DOI] [PubMed] [Google Scholar]
- 44.Radu GU, Caidi H, Miao C, Tripp RA, Anderson LJ, Haynes LM. Prophylactic treatment with a G glycoprotein monoclonal antibody reduces pulmonary inflammation in respiratory syncytial virus (RSV)-challenged naive and formalin-inactivated RSV-immunized BALB/c mice. J Virol. 2010 Sep;84(18):9632–6. doi: 10.1128/JVI.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyoglu-Barnum S, Gaston KA, Todd SO, et al. A respiratory syncytial virus (RSV) anti-G protein F(ab′)2 monoclonal antibody suppresses mucous production and breathing effort in RSV rA2-line19F-infected BALB/c mice. J Virol. 2013 Oct;87(20):10955–67. doi: 10.1128/JVI.01164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Choi Y, Haynes LM, et al. Vaccination to induce antibodies blocking the CX3C-CX3CR1 interaction of respiratory syncytial virus G protein reduces pulmonary inflammation and virus replication in mice. J Virol. 2010 Jan;84(2):1148–57. doi: 10.1128/JVI.01755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee YN, Hwang HS, Kim MC, et al. Recombinant influenza virus carrying the conserved domain of respiratory syncytial virus (RSV) G protein confers protection against RSV without inflammatory disease. Virology. 2015 Feb;476:217–25. doi: 10.1016/j.virol.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorquera PA, Choi Y, Oakley KE, et al. Nanoparticle vaccines encompassing the respiratory syncytial virus (RSV) G protein CX3C chemokine motif induce robust immunity protecting from challenge and disease. PLoS One. 2013;8(9):e74905. doi: 10.1371/journal.pone.0074905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Power UF, Plotnicky-Gilquin H, Huss T, et al. Induction of protective immunity in rodents by vaccination with a prokaryotically expressed recombinant fusion protein containing a respiratory syncytial virus G protein fragment. Virology. 1997 Apr 14;230(2):155–66. doi: 10.1006/viro.1997.8465. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9859–64. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Begona Ruiz-Arguello M, Gonzalez-Reyes L, Calder LJ, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002 Jul 5;298(2):317–26. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 52.Karron RA, Buonagurio DA, Georgiu AF, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997 Dec 9;94(25):13961–6. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001 Aug;75(15):6825–34. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy BR, Collins PL. Live-attenuated virus vaccines for respiratory syncytial and parainfluenza viruses: applications of reverse genetics. J Clin Invest. 2002 Jul;110(1):21–7. doi: 10.1172/JCI16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crim RL, Audet SA, Feldman SA, Mostowski HS, Beeler JA. Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J Virol. 2007 Jan;81(1):261–71. doi: 10.1128/JVI.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011 Sep;17(9):1132–5. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 57.Rawling J, Garcia-Barreno B, Melero JA. Insertion of the two cleavage sites of the respiratory syncytial virus fusion protein in Sendai virus fusion protein leads to enhanced cell-cell fusion and a decreased dependency on the HN attachment protein for activity. J Virol. 2008 Jun;82(12):5986–98. doi: 10.1128/JVI.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rawling J, Cano O, Garcin D, Kolakofsky D, Melero JA. Recombinant sendai viruses expressing fusion proteins with two furin cleavage sites mimic the syncytial and receptor-independent infection properties of respiratory syncytial virus. J Virol. 2011 Mar;85(6):2771–80. doi: 10.1128/JVI.02065-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swanson KA, Settembre EC, Shaw CA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9619–24. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of the Respiratory Syncytial Virus Fusion Glycoprotein in the Post-fusion Conformation Reveals Preservation of Neutralizing Epitopes. J Virol. 2011 May 25;85(15):7788–96. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLellan JS, Chen M, Leung S, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013 May 31;340(6136):1113–7. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLellan JS, Chen M, Joyce MG, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013 Nov 1;342(6158):592–8. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, et al. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000 May 25;271(1):122–31. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 64.Martin D, Calder LJ, Garcia-Barreno B, Skehel JJ, Melero JA. Sequence elements of the fusion peptide of human respiratory syncytial virus fusion protein required for activity. J Gen Virol. 2006 Jun;87(Pt 6):1649–58. doi: 10.1099/vir.0.81715-0. [DOI] [PubMed] [Google Scholar]
- 65.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006 Jan 5;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krarup A, Truan D, Furmanova-Hollenstein P, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005 Jun 28;102(26):9288–93. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16672–7. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beeler JA, Van Wyke CK. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989 Jul;63(7):2941–50. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Barreno B, Palomo C, Penas C, Delgado T, Perez-Brena P, Melero JA. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J Virol. 1989 Feb;63(2):925–32. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arbiza J, Taylor G, Lopez JA, et al. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1992 Sep;73(Pt 9):2225–34. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 72.Lopez JA, Bustos R, Orvell C, et al. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J Virol. 1998 Aug;72(8):6922–8. doi: 10.1128/jvi.72.8.6922-6928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lounsbach GR, Bourgeois C, West WH, Robinson JW, Carter MJ, Toms GL. Binding of neutralizing monoclonal antibodies to regions of the fusion protein of respiratory syncytial virus expressed in Escherichia coli. J Gen Virol. 1993 Dec;74(Pt 12):2559–65. doi: 10.1099/0022-1317-74-12-2559. [DOI] [PubMed] [Google Scholar]
- 74.Langedijk JP, Meloen RH, van Oirschot JT. Identification of a conserved neutralization site in the first heptad repeat of the fusion protein of respiratory syncytial virus. Arch Virol. 1998;143(2):313–20. doi: 10.1007/s007050050288. [DOI] [PubMed] [Google Scholar]
- 75.Sastre P, Melero JA, Garcia-Barreno B, Palomo C. Comparison of affinity chromatography and adsorption to vaccinia virus recombinant infected cells for depletion of antibodies directed against respiratory syncytial virus glycoproteins present in a human immunoglobulin preparation. J Med Virol. 2005 Jun;76(2):248–55. doi: 10.1002/jmv.20349. [DOI] [PubMed] [Google Scholar]
- 76.Magro M, Mas V, Chappell K, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):3089–94. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwakkenbos MJ, Diehl SA, Yasuda E, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010 Jan;16(1):123–8. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gilman MS, Moin SM, Mas V, et al. Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein. PLoS Pathog. 2015 Jul;11(7):e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corti D, Bianchi S, Vanzetta F, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013 Sep 19;501(7467):439–43. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 80.Schuster JE, Cox RG, Hastings AK, et al. A Broadly Neutralizing Human Monoclonal Antibody Exhibits In Vivo Efficacy Against Both Human Metapneumovirus and Respiratory Syncytial Virus. J Infect Dis. 2014 May 26; doi: 10.1093/infdis/jiu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Magro M, Andreu D, Gomez-Puertas P, Melero JA, Palomo C. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J Virol. 2010 Aug;84(16):7970–82. doi: 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ngwuta JO, Chen M, Modjarrad K, et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med. 2015 Oct 14;7(309):309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swanson KA, Balabanis K, Xie Y, et al. A monomeric uncleaved respiratory syncytial virus F antigen retains prefusion-specific neutralizing epitopes. J Virol. 2014 Oct;88(20):11802–10. doi: 10.1128/JVI.01225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palomo C, Mas V, Thom M, et al. Influence of Respiratory Syncytial Virus F Glycoprotein Conformation on Induction of Protective Immune Responses. J Virol. 2016 Jun 1;90(11):5485–98. doi: 10.1128/JVI.00338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith G, Raghunandan R, Wu Y, et al. Respiratory syncytial virus fusion glycoprotein expressed in insect cells form protein nanoparticles that induce protective immunity in cotton rats. PLoS One. 2012;7(11):e50852. doi: 10.1371/journal.pone.0050852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liang B, Surman S, Amaro-Carambot E, et al. Enhanced Neutralizing Antibody Response Induced by Respiratory Syncytial Virus Prefusion F Protein Expressed by a Vaccine Candidate. J Virol. 2015 Sep;89(18):9499–510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGinnes CL, Schmidt MR, Kenward SA, Woodland RT, Morrison TG. Murine immune responses to virus-like particle-associated pre- and postfusion forms of the respiratory syncytial virus F protein. J Virol. 2015 Jul;89(13):6835–47. doi: 10.1128/JVI.00384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cullen LM, Blanco JC, Morrison TG. Cotton rat immune responses to virus-like particles containing the pre-fusion form of respiratory syncytial virus fusion protein. J Transl Med. 2015;13:350. doi: 10.1186/s12967-015-0705-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McLellan JS, Correia BE, Chen M, et al. Design and Characterization of Epitope-Scaffold Immunogens That Present the Motavizumab Epitope from Respiratory Syncytial Virus. J Mol Biol. 2011 Apr 27; doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Correia BE, Bates JT, Loomis RJ, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014 Mar 13;507(7491):201–6. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nair H, Nokes DJ, Gessner BD, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010 May 1;375(9725):1545–55. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012 Dec 15;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990 Aug;15(8):291–4. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 94.Garcia-Beato R, Melero JA. The C-terminal third of human respiratory syncytial virus attachment (G) protein is partially resistant to protease digestion and is glycosylated in a cell-type-specific manner. J Gen Virol. 2000 Apr;81(Pt 4):919–27. doi: 10.1099/0022-1317-81-4-919. [DOI] [PubMed] [Google Scholar]