Abstract
Molecular epidemiological assessments, drug treatment optimization, and development of immunological interventions all depend on understanding pathogen adaptation and genetic variation, which differ for specific pathogens. Mycobacterium tuberculosis is an exceptionally successful human pathogen, yet beyond knowledge that this bacterium has low overall genomic variation but acquires drug resistance mutations, little is known of the factors that drive its population genomic characteristics. Here, we compared the genetic diversity of the bacteria that established infection to the bacterial populations obtained from infected tissues during murine M. tuberculosis pulmonary infection and human disseminated M. bovis BCG infection. We found that new mutations accumulate during in vitro culture, but that in vivo, purifying selection against new mutations dominates, indicating that M. tuberculosis follows a dominant lineage model of evolution. Comparing bacterial populations passaged in T cell-deficient and immunocompetent mice, we found that the presence of T cells is associated with an increase in the diversity of the M. tuberculosis genome. Together, our findings put M. tuberculosis genetic evolution in a new perspective and clarify the impact of T cells on sequence diversity of M. tuberculosis.
Author Summary
Mycobacterium tuberculosis is amongst the most successful and enigmatic pathogens that has burdened humanity for thousands of years. The success of this pathogen depends on unique strategies employed to adapt during infection. Understanding these strategies is key to decipher the complexity of M. tuberculosis and crucial for epidemiological predictions and drug treatments. However, little is known on the impact of transmission population bottlenecks and host immune pressures on sequence diversity of mycobacteria. By combining deep sequencing and parallel evolution, we characterized the evolution of M. tuberculosis genetic diversity during the course of pulmonary infection in mice, and in disseminated M. bovis BCG infection in humans. We found that under in vitro rich culture conditions, multiple adaptive mutations arise, but none of these generate lasting allele diversity in vivo. We found that this phenomenon was not uniquely dictated by transmission bottleneck and that within host evolution contribute to select for a dominant genotype during M. tuberculosis infection. Finally, we used a immuno-deficient mouse model to show that T cells are contributor rather than limitator of genetic diversity during infection. Together, our results emphasize the complexity and uniqueness of M. tuberculosis and contribute understanding M. tuberculosis genetic adaptation.
Introduction
Microbial pathogens adapt to host environments to establish replicative niches and counter immune responses. Adaption, which commonly relies on the generation and transmission of genetic variants with increased fitness, is especially critical for obligate pathogens that must infect, replicate, and be transmitted to new hosts to survive and propagate. While mutations emerge randomly in the genome, the accumulation and loss of variants is the result of both genetic drift and natural selection, and is influenced by transmission population bottlenecks within and between hosts. The large reductions in population size during bottlenecks can have strong evolutionary effects, leading to erosion of genetic diversity and reduction in evolutionary potential and individual fitness through random fixation of slightly deleterious alleles[1, 2]. The severe nature of bottlenecks implies that the success of infection depends on 1) the genetic diversity of the pathogen population in the donor host and, 2) the selection of adapted genotypes during transmission. For HIV and other RNA viruses, founder particles are biased to favor the transmission of variants associated with increased fitness[3, 4]. The corresponding increase in fitness results from the selection and contribution of low frequency variants, and not to the effect of a single, dominant genome. The same principle applies also to other pathogens, including certain bacteria and parasites[5] [6].
Understanding within-host pathogen population diversity has important implications for drug treatment and resistance[7],[8], and for inferring transmission networks[9],[10],[11] and evolutionary processes[12]. Two opposing models of within-host microbial evolution have been described[6]: a ‘dominant-lineage’ model, in which beneficial mutations lead to unique genotypes to establish and maintain infection, and a ‘diverse-community’ model, in which minor variants rise to intermediate frequencies and coexist with major variants in the microbial population. Here, we distinguish between these models and determine the roles of drift, bottlenecks, and selection in genetic variation of Mycobacterium tuberculosis during infection.
Members of the M. tuberculosis complex (MTBC) cause tuberculosis, a chronic infection transmitted by aerosol that remains a deadly disease, despite the availability of drug treatment[13]. M. tuberculosis is an obligate pathogen, and has no natural ecological niche other than its human hosts, with which it has coevolved for thousands of years[14, 15]. Immunological control of M. tuberculosis depends on T lymphocytes, which recognize peptide fragments of bacterial proteins bound to polymorphic MHC (HLA in humans) molecules[16]. The MTBC is characterized by a largely clonal population structure classified into 7 main human-adapted phylogenetic lineages[15],[17]. Despite thousands of years after divergence from a common ancestor, all MTBC lineages share identical 16S rRNA sequences and 99.9% nucleotide identity at the whole genome level[18]. However, M. tuberculosis can generate diversity over short and long time courses. For example, most drug resistance determinants in the MTBC represent chromosomal mutations selected by drug exposure[18]. Moreover, population genetic analyses have highlighted that nonsynonymous single nucleotide polymorphisms (nSNPs) tend to accumulate in the M. tuberculosis genome at a higher rate than in related organisms[19],[17]. Because nSNPs are often deleterious, it was suggested that M. tuberculosis undergoes random genetic drift associated with serial population bottlenecks[17]. Yet, M. tuberculosis does not show the typical signs associated with increased genetic drift[18]. Indeed, its genome has moderate numbers of insertion sequences, few pseudogenes, and no obvious other signals of extensive genome degradation.
Several findings support the notion that within-host selection plays a critical role in shaping the genome of the MTBC. Comparison of a globally representative sample of M. tuberculosis isolates yielded evidence of strong purifying selection, with different patterns of selection related to gene function[20]. We also found that antigen conservation dominates in this pathogen, and that the vast majority of the currently known T cell epitopes are more conserved than any other part of the genome, indicating that these sequences are under strong selective pressure[21]. Although the factors driving epitope conservation in M. tuberculosis are still undetermined, these observations led to the hypothesis that human T cell recognition could play an important role by limiting genetic diversity[21]. In the context of drug exposure, although within-host selection is a source of heterogeneity, the frequency of mutations is low and their locations in the genome highly specific[22].
To better understand conservation and diversity of M. tuberculosis during infection, and to delineate the contribution of T cell selection, we combined high-density whole-genome sequencing (WGS) and mathematical modeling to assess the evolution of genetic diversity of M. tuberculosis in murine pulmonary infection, and in human disseminated M. bovis BCG infection.
Results
Genetic diversity in a H37Rv stock targets key metabolic proteins
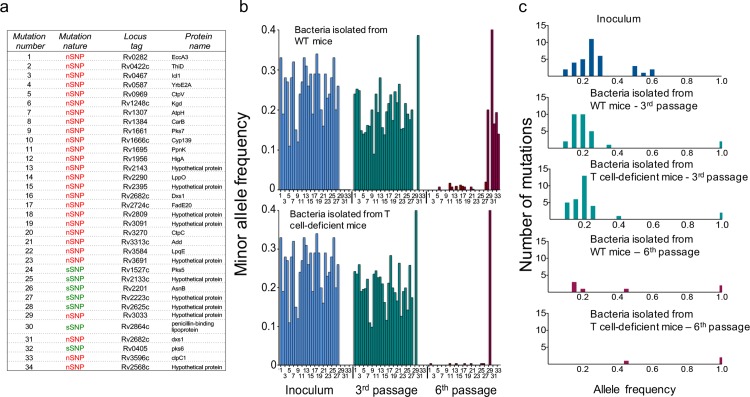
In the widely-studied virulent strain of M. tuberculosis, H37Rv, genomic differences among stocks from individual laboratories have been reported, indicating that in vitro culture generates M. tuberculosis diversity[23]. To further understand this observation, we sequenced our laboratory stock and compared it to that of the H37Rv reference (NC_000962)[24]. Deep sequencing revealed that the stock population was heterogeneous and contained 34 polymorphisms present at various frequencies (S1A Table). Among those mutations, 25 were nonsynonymous single nucleotide changes affecting 25 proteins (Table 1); nonsynonymous mutations were overrepresented in genes whose products are involved in metabolic pathways with potentially important in vivo functions (observed = 12, expected = 7; χ2, p<0.05; Table 1). Notably, we observed that 28% of the bacterial population had a nSNP in the gene encoding Isocitrate lyase 1 (Icl1), an enzyme essential for allowing net carbon gain by diverting acetyl-CoA from β-oxidation of fatty acids into the glyoxylate shunt pathway[25],[26]. Disruption of icl1 attenuates M. tuberculosis persistence and virulence in mice without affecting bacterial growth during the acute phase of infection. An additional metabolic gene, kgd, contained an nSNP in 28% of our stock and encodes an α-ketoglutarate decarboxylase involved in an alternative pathway that generates succinate for the tricarboxylic acid cycle, which may help the pathogen cope with hypoxia[27]. Since Icl1 and Kgd are employed in oxygen- or glucose-deprived environments, we hypothesized that they do not play a key role during growth in rich culture media. This hypothesis was supported by the predicted impact of the identified nSNPs on protein function and/or stability: amino acid substitutions in 17 (68%) of the 25 mutated proteins, including Icl1 and Kgd, were predicted to be deleterious (Table 1). Taken together, these data indicate that during in vitro growth, MTBC accumulates mutations in genes which are not relevant for in vitro growth but might be key for in vivo growth and/or transmission.
Table 1. Characteristics of nSNP-containing genes identified in initial M. tuberculosis inoculum population.
| Mutation nature | Locus tag | Protein name | Amino acid substitution | Putative function | Functional Category | Amino acid change tolerance |
|---|---|---|---|---|---|---|
| nSNP | Rv0282 | EccA3 | V482G | ESX-3 secretion system protein | Transport | Deleterious |
| nSNP | Rv0422c | ThiD | Q21K | Thiamine metabolism | Metabolism | Deleterious |
| nSNP | Rv0467 | Icl1 | A148G | Glyoxylate - dicarboxylate metabolism | Metabolism | Deleterious |
| nSNP | Rv0587 | YrbE2A | V146G | ABC - phospholipid transporters | Transport | Deleterious |
| nSNP | Rv0969 | CtpV | A490P | Cation-transporting ATPase | Transport | Deleterious |
| nSNP | Rv1248c | Kgd | T675P | Alternate tricarboxylic acid cycle | Metabolism | Deleterious |
| nSNP | Rv1307 | AtpH | V241G | Oxidative phosphorylation | Metabolism | Deleterious |
| nSNP | Rv1384 | CarB | A860G | Pyrimidine metabolism - alanine, aspartate - glutamate metabolism | Metabolism | Deleterious |
| nSNP | Rv1661 | Pks7 | R808P | Lipid biosynthesis proteins - polyketide synthase | Metabolism | Deleterious |
| nSNP | Rv1666c | Cyp139 | V1G | Bisphenol, aminobenzoate, limonene and pinene degradation | Metabolism | Neutral |
| nSNP | Rv1695 | PpnK | V294G | Nicotinate - nicotinamide metabolism | Metabolism | Deleterious |
| nSNP | Rv1956 | HigA | V96G | Antitoxin | Detoxification | Neutral |
| nSNP | Rv2143 | Hypothetical protein | H106P | Putative phosphoribosyl transferase | Metabolism | Neutral |
| nSNP | Rv2290 | LppO | Y125S | Lipoprotein | Cell wall | Deleterious |
| nSNP | Rv2395 | Hypothetical protein | S625P | Unknown | Transport | Deleterious |
| nSNP | Rv2682c | Dxs1 | A424P | Thiamine metabolism - terpenoid biosynthesis | Metabolism | Neutral |
| nSNP | Rv2724c | FadE20 | N245T | Oxidoreductases - geraniol degradation | Metabolism | Deleterious |
| nSNP | Rv2809 | Hypothetical protein | N57T | Unknown | Unknown | Deleterious |
| nSNP | Rv3091 | Hypothetical protein | H217P | Unknown | Unknown | Neutral |
| nSNP | Rv3270 | CtpC | A166E | Manganese/zinc-exporting P-type ATPase | Transport | Deleterious |
| nSNP | Rv3313c | Add | S325G | Purine metabolism | Metabolism | Neutral |
| nSNP | Rv3584 | LpqE | V95G | Lipoprotein | Cell wall | Deleterious |
| nSNP | Rv3691 | Hypothetical protein | A62G | Unknown | Unknown | Neutral |
| nSNP | Rv2157c* | murF | L399V | Lysine biosynthesis - peptidoglycan biosynthesis - vancomycin resistance | Cell wall | Deleterious |
| nSNP | Rv0272c* | Hypothetical protein | H91R | Putative serine aminopeptidase | Unknown | Neutral |
Genomic selection in M. tuberculosis during serial passage in immunocompetent or T cell deficient mice
Based on our observation that T cell epitopes are the most conserved regions of the MTBC genome, we tested whether T cell immunity constrains genetic diversity during infection. We used WGS to compare the frequency of variants in M. tuberculosis after six in vivo passages of the H37Rv stock containing sequence variants in immunocompetent wild-type (WT) or in T cell-deficient (TCR β/δ-/-) mice (Fig 1). Following the third and sixth passages, a pool of M. tuberculosis bacteria from the lungs of each mouse was sequenced and compared to the sequence of the initial aerosol inoculum.
Fig 1. Mouse passage protocol.
5 Wild type (C57BL/6; purple) or 5 T cell-deficient (TCR β/δ-/-; orange) mice were each infected with 100 CFU of M. tuberculosis H37Rv stock. After 6 weeks of infection, the lungs of each mouse were homogenized to prepare an inoculum used to infect the next set of mice of the same phenotype (see Methods). Genomic DNA samples of the initial inoculum and of the bacterial populations after the 3rd and 6th mouse passages were examined by WGS.
In vivo selection in mice promotes reversion towards consensus sequence of M. tuberculosis
In vivo passaging allowed 14 and 18 doublings of the bacteria in lungs of WT and T cell deficient mice, respectively, followed by a ~10,000-fold bottleneck during infection of the next group of mice, so we were able to characterize the impact of transmission bottlenecks and in vivo selection on the genetic diversity within the inoculum population. We monitored the frequencies of the 34 pre-existing polymorphisms in the M. tuberculosis population after the third and sixth passages in the two groups of mice. Following three passages, 4 variants were purified out, and 2 achieved fixation in the isolates from both immunocompetent and T cell-deficient mice. These 6 variants were present at the highest frequency (≥50%) in the initial inoculum population (S1A Table) and the majority (4/6) were sSNPs or intergenic mutations. In contrast, 82% (23/28) of the minor variants present in the initial inoculum and that were maintained after three passages were nSNPs (Fig 2A). The frequency of these 28 variants was decreased compared to that in the inoculum population (average total frequencies: 25% in the inoculum,19% in the WT mouse isolates and 20% in the T cell deficient mouse isolates; multiple t test of coverage, p<0.01; Fig 2). These observations indicated that mixed populations of M. tuberculosis continued to exist in both groups of mice after three passages. However, after six passages, all of the minor variants (28/28) were lost (polymorphic reads <5%) in both mouse groups and each mouse was infected with a clonal M. tuberculosis population closer to the H37Rv reference sequence (Fig 2). To infer whether some variants identified in the initial inoculum might be linked, we analyzed the changes in frequencies of each SNP after the three first passages in WT and T cell-deficient mice. We found 5 sets of variants containing 2 to 3 SNPs each that evolved in parallel (multiple t test of coverage, p<0.05) during in vivo passage in both mouse groups, suggesting that these mutations were linked (S2 Table).
Fig 2. Sequence variants present in the initial inoculum are purified out during mouse passages.
(a) List of SNPs present in the initial inoculum population and the mouse isolates at a frequency <50%. (b) Evolution of SNPs found in the initial inoculum (blue), following the third (green) and sixth (red) mouse passages in WT and T cell deficient mice. Mutation numbers (from the list in panel a) are indicated on the x axis. (c) Number of variant alleles with high, intermediate, and low frequencies identified in the initial M. tuberculosis inoculum (blue) and in the M. tuberculosis populations isolated after the third (green) and sixth (red) mouse passages.
Mathematical modeling shows that in vivo selection of M. tuberculosis follows a dominant lineage model of evolution independent of T cells
The data are not consistent with a simple dilutional bottleneck, since minor sequence variants were not progressively lost from the population. Instead, the frequency of the minor variants was unchanged or only slightly reduced after the third passage, but were absent after the sixth passage (S1 Table). In contrast, the sequence variants that were present as a higher fraction of the initial inoculum were purified within the first 3 passages, resulting in fixation or loss (S1 Table). This pattern was independent of the presence of T cells. To explain the observed purification dynamics, we developed a mathematical model using a system of ordinary differential equations, analyzed by dynamical system theory[28]. The model parameters were estimated from the data by the generalized profiling method[29]. The model results strongly supported that in vivo selection of M. tuberculosis populations is driven by a deterministic process for which the kinetics depend on the frequencies of variants in the initial inoculum population (Fig 3). When the variant allele frequency in the inoculum population was less than 50%, the purifying process consisted of two steps: the first step was characterized by the competitive co-existence of minor genotypes. This co-existence continued for a minimum of three passages. The second step was a dynamic process, occurring during or after passage 4, resulting in fixation of polymorphisms and purification of a single M. tuberculosis population. In contrast, when the percentage of variants in the inoculum was equal to or greater than 50%, the bacterial population underwent a rapid purification process that occurred within the first 3 passages, leading to either elimination of these mutants or their fixation as the dominant genotype. These results show evidence that 1) M. tuberculosis evolution is deterministic and follows a “dominant lineage” model in vivo and 2) is a dynamic process largely independent of the presence of T cells that cannot be explained by passive transmission bottlenecks.
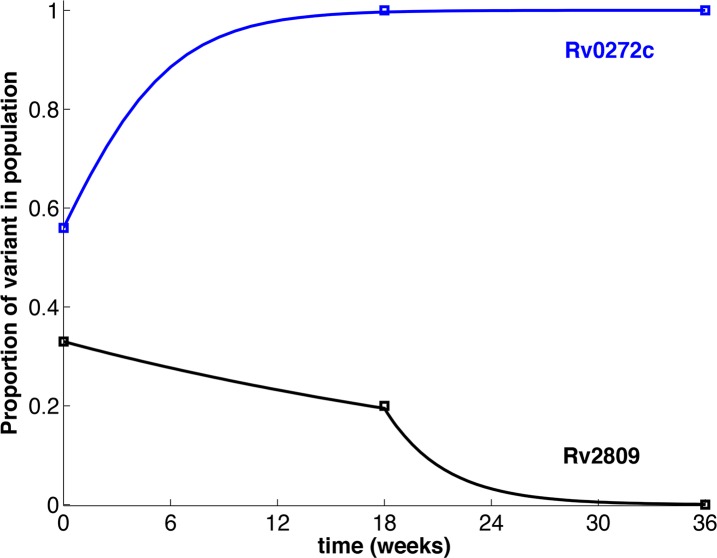
Fig 3. Fixation of variant alleles in the bacterial population progresses by a deterministic process.
The squares indicate the proportion of variants in the inoculum and at specific recovery timepoints following serial infection of mice. The curves represent the modelled progression as influenced by a decision marker. Rv0272c (blue line) represents a variant that undergoes rapid purification leading to fixation after 3 passages; Rv2809 (black line) represents two-stage purification leading to fixation after 6 passages. Rapid purification resulting in either full or zero fixation within 3 passages occurred for all variant alleles present at ≥ 50% in the original inoculum; two-stage fixation occurred for all variant alleles present at <50% in the inoculum (S1 Table).
T cells increase the diversity of M. tuberculosis during prolonged infection
To determine whether within-host selection at specific loci can also generate variation, and to examine the contribution of T cells in generating this diversity, we characterized the polymorphisms that appeared de novo in the M. tuberculosis population after infection of wild-type and T cell-deficient mice. Genome sequencing of bacterial populations following the third mouse passages revealed 2 minor variants that were undetectable in the initial inoculum and present in the WT (1 mutation) or T cell deficient mouse isolates (1 mutation) (Table 2 and S1B Table). While no additional polymorphisms were detected in the T cell deficient isolates after the 6th passage, 4 mutations appeared in the genomes of the WT mouse isolates. Among the polymorphisms identified in WT mouse isolates, 4 were nSNPs (Table 2). By contrast, the unique mutation identified in the bacterial populations from T cell deficient mice was a synonymous SNP (Table 2). Together, these results indicate that T cell-dependent immunity can contribute to sequence diversity in M. tuberculosis.
Table 2. Characteristics of mutated genes identified in the M. tuberculosis mouse lung populations.
| Mouse group | Mutation nature | Locus tag | Protein name | Amino acid substitution | Putative function | Functional category | Amino acid change tolerance |
|---|---|---|---|---|---|---|---|
| WT | nSNP | Rv2682c | Dxs1 | R163L | Thiamine metabolism & Terpenoid biosynthesis | Metabolism | Deleterious |
| WT | nSNP | Rv3596c | ClpC1 | K186E | Regulatory ATPase | Metabolism | Deleterious |
| WT | nSNP | Rv3033 | Hypothetical protein | V42A | Unknown | Unknown | Neutral |
| WT | nSNP | Rv2568c | Hypothetical protein | A313S | Unknown | Unknown | Neutral |
| WT | sSNP | Rv0405 | Pks6 | - | Membrane bound polyketide synthase | Metabolism | - |
| T cell deficient | sSNP | Rv2864c | PBL | - | Penicillin-binding lipoprotein | Cell wall and cell processes | - |
To further evaluate the impact of T cells on the genome diversity of M. tuberculosis during infection, we determined the average mutation rates of M. tuberculosis in both mouse groups. Considering a similar bacterial generation time of 20 hours for both groups, we estimated that the average mutation rate of M. tuberculosis was highly reduced during prolonged infection in T cell deficient mice (3.8 x 10-9 in WT mice versus 7.7 x 10-10 in T cell deficient mice) and this, despite a higher bacterial burden in T cell deficient mice (2 logarithms). Thus, the reduction in the mutation rate calculated for the T cell deficient mouse isolates strongly supports a role for T cells in generating diversity in M. tuberculosis during prolonged infection.
Genetic diversity of M. bovis BCG in disseminated human infection support a dominant lineage model of evolution
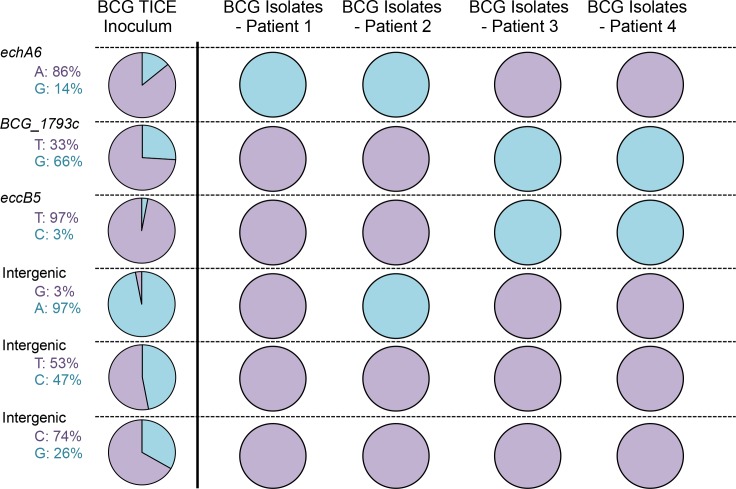
To determine whether the results obtained in mice are also relevant in human infections, we examined the extent and nature of within-host selection of slow-growing mycobacteria isolated from human patients. Since it is not possible to determine the sequence of the inhaled inoculum that establishes human tuberculosis, we took an alternative approach. Invasive carcinoma of the urinary bladder in immunocompetent patients is commonly treated by instillation of a standardized preparation of mycobacteria (M. bovis BCG). In rare cases, disseminated BCG infection develops, and involves tissues beyond the bladder[30]. Since the BCG inoculum is defined and prepared according to pharmaceutical standards, we could be confident in the identity of the bacteria to which a patient was exposed. We first determined the genome sequence of the inoculum used to treat bladder cancer in the United States, M. bovis BCG Tice. Similarly to what we observed for our H37Rv inoculum, the BCG Tice inoculum population was heterogeneous and contained 6 variants present at frequencies from 3% to 66% (S3 Table and Fig 4). Two of these variants were nSNPs present in echA6 or eccB5. The gene echA6 encodes a putative enoyl-CoA hydratase capable of supplying energy and carbon from fatty acid β-oxidation during starvation[31]. EccB5 is a membrane protein of the ESX-5 type VII secretion system involved in secretion of proteins and uptake of nutrients[32]. Both proteins are thus implicated in metabolic pathways used in potentially hostile environments.
Fig 4. Frequencies of SNPs identified in M. bovis BCG inoculum and in the disseminated M. bovis BCG isolated from patients treated for bladder cancer.
Frequencies of variants are indicated and represented in the pies in light blue. Sequences corresponding to the published sequence of M. bovis BCG Tice are indicated and represented in purple.
To characterize the evolution of these minor variants during dissemination in humans, we sequenced M. bovis BCG isolates from 4 patients. We found that the heterogeneity of the inoculum population was eliminated in vivo, as the four variants were either purified out or achieved fixation (Fig 4). Together, these results reinforce the conclusion that M. tuberculosis populations follow a dominant lineage model of evolution during infection.
Discussion
The combination of genomics and mathematical modeling to determine the impact of in vivo evolution on sequence diversity of M. tuberculosis yielded evidence that conservation of the M. tuberculosis genome is driven by a deterministic process leading to the selection of a dominant genotype in vivo. By examining evolution of a heterogeneous population of M. tuberculosis from in vitro passaged bacteria during prolonged infection in mice, we found that conservation of M. tuberculosis in vivo is driven by selection forces limiting genome diversity acquired in vitro. Even though the variants in the initial inoculum population could replicate in vivo, within-host selection limited their ability to compete with the rest of the population, and a single dominant genome eventually emerged. These results indicate that specific mutants are purged in vivo and that a dominant lineage establishes and maintains long–term infection. This has important implications for understanding the mechanisms of evolution of M. tuberculosis and is in contrast to what is observed during infection with the opportunistic pathogen Burkholderia dolosa6. After entering the airways of people with cystic fibrosis, B. dolosa establishes long-term colonization during which emergence of mutations in the population leads to diversification rather than genetic fixation, with potential for cooperative action among subclones. Because M. tuberculosis is a human obligate pathogen that has co-evolved for thousands of years with the same unique host species, the need for variation to establish successful infections in the lung may be reduced and could even be deleterious for the pathogen population [33].
Our results help explain the global phylogenetic structure of the MTBC. Although the MTBC is considered clonal, each lineage is distinct and associated with a specific human population[15], leading to the hypothesis that the genetic conservation of each lineage is the result of an enduring sympatric relationship with their host (same geographical origin). Several observational studies provide evidence supporting this model. Notably, a Swiss cohort study found that MTBC lineages tend to transmit preferentially among sympatric host populations[34]. Two other studies in Ghana offered a potential explanation for the geographical restriction of lineage 5 in West Africa, by showing association between these lineages and specific human ethnicities[35, 36]. A dominant lineage model of evolution can also help explain the various clinical outcomes resulting from infections with strains from distinct lineages of the MTBC. This would explain the finding that individuals from the Gambia that were infected with 'modern' lineages 2 and 4 were more likely to progress to active disease than individuals infected with 'ancient' lineage 6 which is endemic and likely evolved in that region[37].
The finding that within-host selection contributes to shape the M. tuberculosis genome is also supported by the increased diversity observed during M. tuberculosis growth in nutrient-rich culture media. For decades, laboratories have maintained M. tuberculosis H37Rv. Originally derived from a clinical strain (H37), it was recovered in 1905 from a patient with pulmonary tuberculosis. Early records showed the capacity of H37 to adapt to different in vitro conditions leading to phenotypic dissociation between virulent and avirulent derivatives[38]. Although the mechanisms by which this dissociation occurred were unknown, it highlighted the plasticity of M. tuberculosis metabolism and its central role for virulence. We found that in vitro culture conditions represent permissive environments for genetic drift and that these changes targeted enzymes involved in metabolic pathways dispensable during in vitro culture, but essential for optimal growth under nutrient-limiting conditions in vivo. These results demonstrate the capacity of M. tuberculosis to generate variation under permissive conditions.
A significant finding of this study is the impact of T cells in generating diversity in M. tuberculosis populations in vivo. By comparing genome sequences of M. tuberculosis populations isolated from WT and T cell deficient mice, we found that the presence of T cells was associated with the appearance of unique variants of the M. tuberculosis genome during prolonged in vivo infection. The findings reported here are in contrast with the hypothesis that T cells are strictly driving M. tuberculosis epitope conservation. Although the present results could either be due to direct recognition of specific peptide sequences by clonotypic T cells or to the indirect consequence of T cell activation and effector mechanisms, the possibility that direct recognition by T cells can contribute to sequence diversity is consistent with our recent finding that naturally occurring sequence variation in specific M. tuberculosis epitopes affects human T cell recognition[21]. In addition, the findings emphasize the point that the impact of T cell recognition, whether to promote conservation or diversity, is a function of the specific antigen/epitope and genetic locus, and that T cell recognition of distinct antigens can have different outcomes that may favor the pathogen or the host.
Overall, the results reported here reveal that purifying selection and increased genomic diversity are not two mutually exclusive processes during M. tuberculosis infection. While the impact of purifying selection was apparent shortly after the initial infection, increased genomic diversity occurred progressively and was observed after serial infection-transmission cycles over the course of months. Similar results were recently published that reveal evidence of both purifying selection and genome diversification in M. tuberculosis isolates obtained from distinct lesions and organs of HIV-coinfected humans that succumbed to infection[39]. Thus, in another context, prolonged infection leads to increased M. tuberculosis genetic diversity in humans despite overall purifying selection pressure. Together, the findings in immunocompetent humans, HIV-infected humans, and mice, all indicate that although selection favors overall sequence conservation in M. tuberculosis, there are also long term forces that favor diversifying selection, most likely to adapt to a new environment or to counter new stresses. Our findings will guide deeper investigation of the mechanisms used by M. tuberculosis to adapt and to continue to be a globally successful pathogen.
In conclusion, our findings indicate that M. tuberculosis genetic selection is driven by a deterministic process imposed by both genetic drift and within-host selection, leading to a dominant lineage mode of evolution. Although M. tuberculosis is not rapidly mutating, our results indicate that this pathogen is capable of genetic plasticity dictated by environmental changes. The necessity to adapt leads to selection and the contribution of dominant genotypes determined by the host. Our results also demonstrate for the first time the impact of T cells on sequence diversity of M. tuberculosis and indicates that T cell responses are a force that can promote diversity at specific sites rather than to only maintain conservation during infection.
Materials and Methods
Ethics statement
All animal experiments were done in accordance with procedures approved by the NYU School of Medicine Institutional Animal Care and Use Committee (IACUC - Laboratory Animal Care Protocol #160426–01). These IACUC regulations conformed to the national guidelines provided by the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Mouse infection and sample collections
In an ABSL3 facility, M. tuberculosis H37Rv was cultured and used to infect 5 WT (C57BL/6) or 5 T cell-deficient (TCR β/δ-/-) mice by the aerosol route with ~100 CFU/ mouse. After 42 days of infection, mice were euthanized and their lungs homogenized in 5 ml of 7H9 culture medium and the bacterial population was allowed to expand during a minimal period (~1 week) in vitro. The culture from one mouse was then diluted to prepare an aerosol inoculum of 100 CFU, which was used to infect a new set of mice of the same background. At each passage, CFU were calculated and frozen stocks were made. Following the third (21 weeks) and sixth (42 weeks) mouse passages, a pool of M. tuberculosis from each mouse was used for DNA extraction and sequencing.
M. bovis BCG isolates from 4 patients were provided from the collection of the New York State Mycobacteriology Laboratory in the form of solid agar cultures. The samples were de-identified, but were known to have been obtained from 4 different adult patients treated for bladder cancer. For each culture, frozen stocks were made before entire plates were swept for gDNA preparation. The pharmaceutical grade M. bovis BCG Tice strain was obtained from Theracys.
Illumina sequencing
Genomic DNA was extracted using a standard kit (Qiagen), and sequenced by GATC-Biotech. Illumina single read sequencing was performed with single-read of 51 bases and a target coverage of at least 3 million high-quality bases. On average, 9.5 million reads were obtained per isolate. We used Burrows-Wheeler Aligner (BWA)[40] to map the reads from the genome sequences against the H37Rv NC_000962 reference sequence. BWA outputs were analyzed and annotated using SAMtools[41], and ANNOVAR [42]. SNPs in genes annotated as PE/PPE genes, integrases, transposases, resolvases, maturases, or phages were removed from the analysis.
Mutation identification, deep population sequencing
Bacteria from entire plates were pooled from each lung sample and sequenced with deep coverage. The reads were aligned to H37Rv NC_000962 reference sequence and we identified fixed mutations, appearing in all reads, and polymorphisms, appearing in only a fraction of the reads. For study of disseminated M. bovis BCG isolates, bacteria on entire plates were pooled and the sequence reads were aligned to the M. bovis BCG Tice reference sequence (SAMN03023974). To remove false positive polymorphic sites caused by systematic sequencing or alignment errors[43], we developed a set of thresholds and statistical tests that rejected polymorphic sites where the mutated and ancestral reads had significantly different properties. The population sequencing approach reliably detected polymorphisms where the minor allele frequency was greater than 10%, while decreasing the cost and labor required per sample. We considered a position to be polymorphic if it met the following quality thresholds in the given sample: minor allele frequency: more than 10% of reads supported a particular minor allele; minor allele coverage: at least 50 reads aligned in both the forward and reverse direction, and the total number of reads aligning is below the 99th percentile of covered positions in that sample; base quality: average base quality (provided by sequencer) was greater than 20 for both the major and minor allele calls on both the forward and reverse strand; mapping quality: average mapping quality (provided by aligner) was greater than 19 for reads supporting both the major and minor alleles on both the forward and reverse strand; indels: no reads aligning to that position support an indel at any position along that read; isogenic control: More than 98.0% of reads aligning to this genomic position in the isogenic control support a major allele; strand bias: A p-value < 0.01 supporting a null hypothesis that the minor allele frequency for the SNPs identified in the mouse isolates is the same for reads aligning to both the forward and reverse strand (Fisher’s exact test)[6]. For some mutations identified in the inoculum population, the strand bias criterion was not taken into account if the polymorphism was confirmed after the 3rd mouse passage and purified out after the 6th mouse passage.
Mutation confirmation
Minor variants identified at high frequency (>30%) in the bacterial populations were confirmed by Sanger sequencing of PCR amplification products using primers anchoring unique regions flanking the mutated genes. PCR was performed using the FastStart High fidelity PCR system (Roche). The purified product was diluted and submitted with the forward and reverse primers to Genewiz for dideoxy chain termination sequencing. BLAST was used to align the resulting sequences against the corresponding genes to confirm the presence of multiple peaks at the polymorphic positions.
Characterization of protein functions and amino acid tolerance prediction
To characterize the families and functions of nSNP-encoding proteins, we used the following databases: Tuberculist (http://tuberculist.epfl.ch/), KEGG (http://www.genome.jp/kegg/), UniProt (http://www.uniprot.org/) and ModBase (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi). To predict the impact of amino acid changes on protein function, we used 3 algorithms in parallel: Sift (http://sift.jcvi.org/), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) and Provean (http://provean.jcvi.org/index.php). A mutations was considered deleterious if the outputs obtained from the 3 algorithms converged toward the same conclusion.
Model description
According to the experimental results, there are two fixation states of polymorphisms for the frequency of mutated reads: one with zero representation (denoted as F0) and the other with full representation (denoted as F1). We introduced a variable y into the system to characterize the level of a decision marker that depicts the competitions between two fixation states. We assumed that y satisfies y' =a (1-y)-b y, where a represents the transition rate from F0 to F1, and b represents the transition rate from F1 to F0. Write y* = a/(a+b) and this quantity is indeed the equilibrium solution of the equation above. Furthermore, we assumed that the closer the value of y to y*, the more it favors F1. Meanwhile, the initial condition of y was set as follows: y(0) = 0 when the percentage of variant alleles in the inoculum population is below 50%, whereas y(0) = y* when the percentage of variant alleles in the inoculum population is above or equal to 50%. The percentage of M. tuberculosis variant alleles was defined as x. We assumed that x obeys the following dynamics: x' = r(t)x(1-x)-d(t) x, where the growth of x is assumed to be logistic, and r(t) and d(t) represent the time dependent per capita birth and death rates, respectively. Let er and ed (or Δr and Δd) denote the baseline (or elevation of) birth and death rates of x, respectively. Let Td be the decision time. Then r(t) and d(t) can be written as: (1) r (t) = er+Δr, if |y*-y(Td)|< c and t> = Td, and r(t)= er otherwise; (2) d(t) = ed+Δd, if |y*-y(Td)|> = c and t> = Td, and d(t) = ed otherwise. Here c defines a prescribed decision threshold. Particularly, the fixation (achieved at the end) will be in favor of F1 if the distance between the value of y and y* at the decision time is close enough and is less than c, and F0 otherwise.
Model fitting procedure
The proposed system of ordinary differential equations is analytically solvable. However, only the part of the system (i.e. x) is observable. Thus, the method of nonlinear least squares is not applicable to fit the model to the data. So, we estimated the model parameters from data by employing the general profiling procedure proposed by Ramsay et al.[29]. These parameter estimates along with estimated initial values of x component allowed us to solve the ordinary differential equations. We assumed a Gaussian error distribution with constant variance for all data points. This assumption was reasonable because the errors introduced during measurement of the data were not estimated explicitly and were likely to be the same for all data points. The general profiling procedure was implemented as two nested levels of optimization. In the first level, the solution (x(t),y(t)) of our model was approximated with smooth curves by penalized smoothing with ordinary differential equations defined penalty, conditional on the model parameter vector. In the second level, the model parameter vector is estimated by minimizing the weighted sum of square errors.
Estimation of in vivo mutation rate from WGS data
Average mutation rate was estimated as in Ford, et al. 2011. In short, mutation rate (μ) was estimated using the equation[44]:
where the number of SNPs (m) was divide by the genome size (N = 4.4Mb) times the number of generations (t/g). We opted for a generation time of 20h for both mouse groups. M is defined by the number of SNPs observed, N is determined based on 93% coverage of a 4.4Mb genome (N = 4×106), t is the total duration of each infection in hours, and g is the generation time in hours (20h).
Supporting Information
(XLSX)
(XLSX)
(XLSX)
Data Availability
All genomic data are are available at BioProject NCBI, accession PRJNA312221. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
Supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases: R01 AI090928 and U19 AI111211 (JDE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4(6):457–69. 10.1038/nrg1088 [DOI] [PubMed] [Google Scholar]
- 2.Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet. 2013;14(12):827–39. 10.1038/nrg3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031 10.1126/science.1254031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borderia AV, Isakov O, Moratorio G, Henningsson R, Aguera-Gonzalez S, Organtini L, et al. Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype. PLoS pathogens. 2015;11(5):e1004838 10.1371/journal.ppat.1004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberle M, Balmer O, Brun R, Roditi I. Bottlenecks and the maintenance of minor genotypes during the life cycle of Trypanosoma brucei. PLoS pathogens. 2010;6(7):e1001023 10.1371/journal.ppat.1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46(1):82–7. 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun G, Luo T, Yang C, Dong X, Li J, Zhu Y, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206(11):1724–33. 10.1093/infdis/jis601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44(1):106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr., et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–80. 10.1038/ng.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seed KD, Bodi KL, Kropinski AM, Ackermann HW, Calderwood SB, Qadri F, et al. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. MBio. 2011;2(1):e00334–10. 10.1128/mBio.00334-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13(2):137–46. 10.1016/S1473-3099(12)70277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alizon S, Luciani F, Regoes RR. Epidemiological and clinical consequences of within-host evolution. Trends Microbiol. 2011;19(1):24–32. 10.1016/j.tim.2010.09.005 [DOI] [PubMed] [Google Scholar]
- 13.WHO. Global tuberculosis report Geneva: World Health Organization, 2014. [Google Scholar]
- 14.Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514(7523):494–7. 10.1038/nature13591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45(10):1176–82. 10.1038/ng.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philips JA, Ernst JD. Tuberculosis pathogenesis and immunity. Annual review of pathology. 2012;7:353–84. 10.1146/annurev-pathol-011811-132458 [DOI] [PubMed] [Google Scholar]
- 17.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6(12):e311 10.1371/journal.pbio.0060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagneux S. Genetic diversity in Mycobacterium tuberculosis. Curr Top Microbiol Immunol. 2013;374:1–25. 10.1007/82_2013_329 [DOI] [PubMed] [Google Scholar]
- 19.Coscolla M, Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol. 2014;26(6):431–44. 10.1016/j.smim.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepperell CS, Casto AM, Kitchen A, Granka JM, Cornejo OE, Holmes EC, et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS pathogens. 2013;9(8):e1003543 10.1371/journal.ppat.1003543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coscolla M, Copin R, Sutherland J, Gehre F, de Jong B, Owolabi O, et al. M. tuberculosis T Cell Epitope Analysis Reveals Paucity of Antigenic Variation and Identifies Rare Variable TB Antigens. Cell Host Microbe. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders NJ, Trivedi UH, Thomson ML, Doig C, Laurenson IF, Blaxter ML. Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background. J Infect. 2011;62(3):212–7. 10.1016/j.jinf.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 23.Ioerger TR, Feng Y, Ganesula K, Chen X, Dobos KM, Fortune S, et al. Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol. 2010;192(14):3645–53. 10.1128/JB.00166-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 25.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406(6797):735–8. 10.1038/35021074 [DOI] [PubMed] [Google Scholar]
- 26.Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr., et al. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000;7(8):663–8. 10.1038/77964 [DOI] [PubMed] [Google Scholar]
- 27.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci U S A. 2005;102(30):10670–5. 10.1073/pnas.0501605102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guckenheimer J, Holmes P. Nonlinear Oscillations, Dynamical Systems, and Bifurcations of Vector Fields. Edition 6 ed. New York: Springer-Verlag; 2002. [Google Scholar]
- 29.Ramsay JO, Hooker G, Campbell D, Cao J. Parameter estimation for differential equations: a generalized smoothing approach. J R Statist Soc. 2007;69:741–96. [Google Scholar]
- 30.Gan C, Mostafid H, Khan MS, Lewis DJ. BCG immunotherapy for bladder cancer--the effects of substrain differences. Nat Rev Urol. 2013;10(10):580–8. 10.1038/nrurol.2013.194 [DOI] [PubMed] [Google Scholar]
- 31.Tsuge T, Sato S, Hiroe A, Ishizuka K, Kanazawa H, Shiro Y, et al. Contribution of the distal pocket residue to the acyl-chain-length substrate specificity of (R)-specific enoyl-CoA hydratases from Pseudomonas: site-directed mutagenesis, chimeragenesis, X-ray crystallographic analysis, and homology modeling. Appl Environ Microbiol. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ates LS, Ummels R, Commandeur S, van de Weerd R, Sparrius M, Weerdenburg E, et al. Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria. PLoS Genet. 2015;11(5):e1005190 10.1371/journal.pgen.1005190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brites D, Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev. 2015;264(1):6–24. 10.1111/imr.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenner L, Egger M, Bodmer T, Furrer H, Ballif M, Battegay M, et al. HIV infection disrupts the sympatric host-pathogen relationship in human tuberculosis. PLoS Genet. 2013;9(3):e1003318 10.1371/journal.pgen.1003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asante-Poku A, Yeboah-Manu D, Otchere ID, Aboagye SY, Stucki D, Hattendorf J, et al. Mycobacterium africanum is associated with patient ethnicity in Ghana. PLoS Negl Trop Dis. 2015;9(1):e3370 10.1371/journal.pntd.0003370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asante-Poku A, Otchere ID, Osei-Wusu S, Sarpong E, Baddoo A, Forson A, et al. Molecular epidemiology of Mycobacterium africanum in Ghana. BMC Infect Dis. 2016;16:385 10.1186/s12879-016-1725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jong BC, Adetifa I, Walther B, Hill PC, Antonio M, Ota M, et al. Differences between tuberculosis cases infected with Mycobacterium africanum, West African type 2, relative to Euro-American Mycobacterium tuberculosis: an update. FEMS Immunol Med Microbiol. 2010;58(1):102–5. 10.1111/j.1574-695X.2009.00628.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steenken W, Gardner L. History of H37 strain of tubercle bacillus. American Review of Tuberculosis. 1946;54:62–6. [DOI] [PubMed] [Google Scholar]
- 39.Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat Med. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39(13):e90 10.1093/nar/gkr344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43(5):482–6. 10.1038/ng.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All genomic data are are available at BioProject NCBI, accession PRJNA312221. All other relevant data are within the paper and its Supporting Information files.