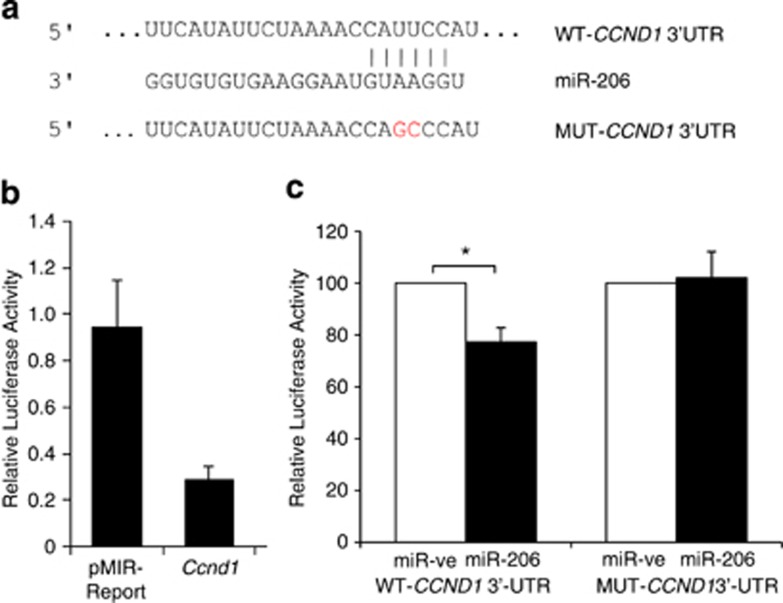
Figure 1.
miR-206 directly targets CCND1 at a specific 3′-UTR site. (a) Schematic of putative miR-206 target site within the human CCND1 3′-UTR as predicted by TargetScan and PicTar algorithms. Mutations introduced into the full-length human CCND1 3′-UTR are highlighted in red. (b) Relative luciferase activity was measured in 3T3-L1 cells co-transfected with a luciferase reporter construct containing a fragment of the mouse Ccnd1 3′-UTR encompassing the miR-206 binding site or empty pMIR-Report vector, along with miR-ve or miR-206. Data represent relative Firefly values as determined by the ratio of Firefly to Renilla luciferase activity with control miR-ve set as 1. Values are the mean±s.e.m. of three separate experiments. (c) Relative luciferase activity was measured in H1299 cells co-transfected with reporter constructs containing human full-length wild-type (WT)-CCND1 3′-UTR or mutant (MUT)-CCND1 3′-UTR along with miR-ve or miR-206. Firefly luciferase units were normalized against Renilla luciferase units to control for transfection efficiency. Percentage Firefly luciferase values were determined with control miR-ve set at 100%. Data represent the mean+s.e.m. of three independent experiments. A paired t-test was used to determine significance between miR-ve and miR-206 transfected cells (* indicates P<0.05).