Abstract
The Fabaceae, the third largest family of plants and the source of many crops, has been the target of many genomic studies. Currently, only the grasses surpass the legumes for the number of publicly available expressed sequence tags (ESTs). The quantity of sequences from diverse plants enables the use of computational approaches to identify novel genes in specific taxa. We used BLAST algorithms to compare unigene sets from Medicago truncatula, Lotus japonicus, and soybean (Glycine max and Glycine soja) to nonlegume unigene sets, to GenBank's nonredundant and EST databases, and to the genomic sequences of rice (Oryza sativa) and Arabidopsis. As a working definition, putatively legume-specific genes had no sequence homology, below a specified threshold, to publicly available sequences of nonlegumes. Using this approach, 2,525 legume-specific EST contigs were identified, of which less than three percent had clear homology to previously characterized legume genes. As a first step toward predicting function, related sequences were clustered to build motifs that could be searched against protein databases. Three families of interest were more deeply characterized: F-box related proteins, Pro-rich proteins, and Cys cluster proteins (CCPs). Of particular interest were the >300 CCPs, primarily from nodules or seeds, with predicted similarity to defensins. Motif searching also identified several previously unknown CCP-like open reading frames in Arabidopsis. Evolutionary analyses of the genomic sequences of several CCPs in M. truncatula suggest that this family has evolved by local duplications and divergent selection.
Legumes constitute a large plant family that presents humans with a treasure trove of resources for a variety of uses. Throughout the world, legumes provide important sources of protein, oil, mineral nutrients, and nutritionally important natural products (Graham and Vance, 2003). Grain legume species, including pea (Pisum sativum), common bean (Phaseolus vulgaris), and lentil (Lens culinaris), account for over 33% of human dietary protein. Other legumes, including clovers (Trifolium spp.) and medics (Medicago spp.), are widely used as animal fodder. Refined oils, such as soybean (Glycine max) oil, have industrial applications in paint, diesel fuel, electrical insulation, and solvents. Legumes also accumulate phytochemicals, including isoflavonoids, which impact human health through pharmaceutical use and as dietary supplements (Dixon and Sumner, 2003).
An important feature of legumes is their ability to obtain nutrients via symbioses with soil microbes. The formation of nitrogen-fixing nodules via interaction with bacteria collectively know as rhizobia is virtually unique to legumes, although some species in eight families of the eurosid I clade of dicots can form nodules in association with nitrogen-fixing actinomycetes (Soltis et al., 1995; Doyle and Luckow, 2003). An exchange of specific signal molecules between host and microbe triggers many developmental events in the host, including extensive modulation of gene expression (for review, see Downie and Walker, 1999; Gage and Margolin, 2000; Cullimore et al., 2001). Although there has been recent rapid progress in identification of genes regulating early stages of nodule formation (Parniske and Downie, 2003; Szczyglowski and Amyot, 2003), much is still to be learned about nodule organogenesis. Legumes can also form symbiotic associations with arbuscular mycorrhizal fungi, which aid the acquisition of minerals from the soil. Unlike the formation of nitrogen-fixing nodules, symbioses with mycorrhizal fungi are found in the majority of higher plants. However, some of the same plant genes are required for both symbioses, so that legumes have emerged as major study systems for mycorrhizal interactions (Albrecht et al., 1999; Liu et al., 2003). While these symbioses are beneficial to the plant, many fungal and bacterial pathogens have a large negative impact on crop yields of legumes.
The many uses of legumes and the variety of symbiotic and pathogenic interactions found provide numerous targets for functional genomics research. Currently, there are nearly 700,000 nucleotide sequences representing the Fabaceae available from the National Center for Biotechnology Information (NCBI taxonomy browser, http://www.ncbi.nlm.nih.gov/Taxonomy, November, 2003). In particular, significant strides have been made in the functional genomics of the model legumes M. truncatula and Lotus japonicus and the crop legume soybean (G. max and Glycine soja; VandenBosch and Stacey, 2003). There are over 185,000, 36,000, and 340,000 Expressed Sequence Tags (ESTs) available for these species, respectively (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). Key outputs from EST projects are public databases of sequences that allow widely distributed investigators to access information on gene structure, expression, and putative function (Asamizu et al., 2000; Endo et al., 2000; Bell et al., 2001; Journet et al., 2002; Poulsen and Podenphant, 2002; Shoemaker et al., 2002; Lamblin et al., 2003). The Institute for Genomic Research (TIGR) has analyzed ESTs from a variety of plant species, including the three legume species, clustering the ESTs into tentative consensus sequences (TCs) that represent the minimally redundant set of a species' expressed genes, also called unigenes (Quackenbush et al., 2001; http://www.tigr.org/tdb/tgi/plant.shtml).
The wide availability of ESTs and other gene-containing sequences from plants, including the near complete rice (Oryza sativa) and Arabidopsis genomes, provides an opportunity for comparative sequence analysis. For example, the well-characterized Arabidopsis genome is an asset to identify orthologous genes in other, less well-characterized species. However, comparative approaches may also be used to identify genes that are taxon specific.
In the case of legumes, it is clear that many of the genes involved in the hallmark legume functions of nodulation or isoflavonoid biosynthesis likely evolved from pathways shared among many plant species. For example, several classes of broadly conserved receptor kinases have been found to be required for nodule formation or regulation of nodule numbers (Parniske and Downie, 2003; Szczyglowski and Amyot, 2003), including a homolog of CLAVATA1, which controls shoot meristem size in Arabidopsis (Clark et al., 1997). Using Boolean searches of TIGR M. truncatula Gene Index 4, Federova et al. (2002) identified 340 TCs with nodule-specific expression patterns. Many of the genes identified shared sequence homology to sequences from nonlegumes. In addition to the recruitment of broadly conserved genes for novel legume functions, we hypothesize that legumes may have novel genes involved in legume-specific functions. These genes may be truly unique to legumes, or may have diverged so much from their progenitors that they appear to be unique. If so, a comparative analysis of expressed sequences would bring to light novel genes whose function would be difficult to study in other taxa.
The goal of this project was to use available sequence databases to quickly and efficiently identify and characterize sequences from M. truncatula, soybean, and L. japonicus that lack close homologs in other nonlegume taxa. As an operational definition, legume-specific genes have no BLAST (Altschul et al., 1997) sequence homology, below a specified threshold, to any publicly available nonlegume sequences. Note that homologs of genes identified as legume specific may later be found in nonlegume taxa. Following our definition, we identified a pool of novel sequences representing about 6% of the legume TCs. Because most of these sequences lacked significant homology to genes of known function, computational means were used to group homologous sequences and predict conserved motifs among them. The results of motif analysis could have been used to further eliminate sequences that were not legume specific. However, this would have removed those sequences that have diverged significantly from nonlegume ancestral genes. Further, it would have provided few biological insights into the function of the remaining genes. By using motif analysis, we have identified novel gene families including Pro-rich proteins, Cys cluster proteins, and F-box related proteins. The results of this bioinformatic study will enable laboratory assessment of gene function and analysis of evolutionary patterns of these and other unique gene families.
RESULTS
Identification of Legume-Specific Genes
A series of BLAST analyses was used to identify and remove broadly conserved sequences from the set of expressed sequences from legumes. A summary of the approach and the results are presented in Table I. Initially, the TIGR M. truncatula, L. japonicus, and soybean (G. max and G. soja) TCs were compared to the TIGR maize (Zea mays, ZmGI), tomato (Lycopersicon esculentum, LeGI), rice (OsGI), and Arabidopsis (AtGI) gene indices (Quackenbush et al., 2001; http://www.tigr.org/tdb/tgi) using BLASTN and TBLASTX. While the gene indices included both TC and singleton EST sequences, only legume TCs were analyzed because they were more likely to represent full-length transcripts. By using a limited dataset of four nonlegume species, many legume TCs with sequence homology to nonlegume sequences could be identified with minimal computer processing time. Perl scripts were used to parse the BLASTN and TBLASTX reports and to identify legume TCs with homology to nonlegume sequences. If a legume TC had BLAST homology to any nonlegume sequence with an E-value more significant than 10−4, it was not considered legume specific. Only TCs without significant hits or with hits only to other legumes species were considered as putatively legume specific. Roughly 90% of legume TCs were eliminated using this approach.
Table I.
Computational identification of legume-specific genes
| BLAST Algorithm, E-Value Cutoff | Data Set | Total Sequences in Data Set | Legume-Specific G. max/soja TCs Retained | Legume-Specific M. truncatula TCs Retained | Legume-Specific L. japonicus TCs Retained |
|---|---|---|---|---|---|
| Legume TCsa | 45,783 | 24,750 | 17,243 | 3,790 | |
| BLASTN, 10−4 | TIGR AtGI, LeGI, OsGI, ZmGIb | 142,492 | 7,938 | 5,309 | 886 |
| TBLASTX, 10−4 | TIGR AtGI, LeGI, OsGI, ZmGI | 142,492 | 2,412 | 1,417 | 144 |
| BLASTX, 10−4 | GenBank nonredundant database | 1,335,905 | 2,230 | 1,267 | 136 |
| TBLASTX, 10−4 | Remaining TIGR Plant GIsc | 334,347 | 2,101 | 1,141 | 110 |
| TBLASTX, 10−4 | Arabidopsis and rice genomesd | – | 2,081 | 1,128 | 106 |
| Tera-BLASTN, 10−4 | EST_others | 6,985,891 | 2,020 | 1,046 | 103 |
| Tera-TBLASTX, 10−4 | EST_others | 6,985,891 | 1,997 | 1,028 | 101 |
| TBLASTX, 10−20 | Eliminated Legume TCse | 42,657 | 1,572 | 861 | 92 |
Legume TCs from the G. max/soja, M. truncatula, and L. japonicus gene indices were used as query sequences for sequential BLAST analyses against various sequence data sets from nonlegumes. The TCs retained as legume specific from each analysis served as the query for the subsequent BLAST analysis.
The initial dataset contains all TCs from the TIGR soybean (GmGI, v9), M. truncatula (MtGI, v6), and L. japonicus (LjGI, v1) gene indices.
The data set includes TCs and singletons from TIGR's Arabidopsis (AtGI, v9), tomato (LeGI, v8), rice (OsGI, v11), and maize (ZmGI, v11) gene indices.
Cotton (CGI, v4), C. reinhardtii (ChrGI, v2), sunflower (HaGI, v1), barley (HvGI, v5), lettuce (LsGI, v1), ice plant (McGI, v3), Pinus spp. (PGI,v1), rye (RyeGI, v2), sorghum (SbGI, v4), potato (StGI, v6), wheat (TaGI, v6), and grape (VvGI, v1).
TIGR Arabidopsis genome (4.0) and TIGR rice genome (version from February 18, 2003).
Legume TCs eliminated in the previous BLAST analyses were blasted against the remaining legume-specific TCs.
With the remaining subset of putatively legume-specific TCs, increasingly stringent and computationally intensive searches were used to identify homologous sequences from nonlegume species. Stringency levels were raised by increasing the number of sequences present in the nonlegume data set and/or by changing the BLAST analysis from BLASTN to BLASTX or TBLASTX. As before, legume TCs with homology to nonlegume sequences with an E-value more significant than 10−4 were eliminated.
BLASTX was used to compare the remaining 3,973 legume-specific TCs to the GenBank protein database (NR). Three hundred and forty additional legume TCs with homology to nonlegume sequences were identified and removed. TBLASTX was then used to compare the remaining 3,633 legume-specific TCs to the remaining nonlegume plant gene indices available at TIGR: barley (Hordeum vulgare), Chlamydomonas reinhardtii, cotton (Gossypium spp.), grape (Vitis vinifera), ice plant (Mesembryanthemum crystallinum), lettuce (Lactuca sativa), Pinus spp., potato (Solanum tuberosum), rye (Secale cereale), sorghum (Sorghum bicolor), sunflower (Helianthus annuus), and wheat (Triticum aestivum). An additional 283 legume TCs with homology to nonlegume sequences were eliminated.
Until this point, all of the described BLAST searches involved nonlegume sequences that most likely represented real genes, including those that have been identified experimentally and others that have been predicted from genomic sequences. Since gene prediction programs are fallible (Zhang, 2002), additional genes or portions of genes surely reside within the genomic data. Therefore, TBLASTX was used to compare the remaining 3,352 legume-specific genes against the available genomic sequences of Arabidopsis (The Arabidopsis Genome Initiative, 2000; Bevan et al., 2001) and rice (Goff et al., 2002; Yuan et al., 2003). Thirty-seven TCs were identified with sequence homology more significant than 10−4 to the rice or Arabidopsis genomes. A final TBLASTX search against NCBI's EST_others database narrowed the list of putative legume-specific genes to 1,028; 1,997; and 101 from M. truncatula, soybean, and L. japonicus, respectively.
One area of concern was that some of the putative legume-specific TCs might be too short to yield informative BLAST hits with nonlegume sequences. One example would be if the TC corresponded to the untranslated portions of a transcript. Since untranslated regions do not code for protein, they are less likely to be conserved among species. In order to identify sequences falling into this category, we used TBLASTX to compare the remaining legume-specific genes to the original set of legume TCs. If a legume-specific TC showed homology to a lengthier legume TC that was not legume specific, it was unlikely to be legume specific. However, if a legume-specific TC had homology only to itself or other remaining legume-specific TCs, it was retained in the legume-specific category. After this analysis, 2,525 total legume-specific TCs remained from all three species (Table I and Supplemental Table I, which can be viewed at www.plantphysiol.org).
To confirm that the identified M. truncatula legume-specific TCs were of plant origin and not that of the nitrogen-fixing symbiont, the M. truncatula legume-specific TCs were compared to the complete genome of Sinorhizobium meliloti (Galibert et al., 2001) and a portion of the M. truncatula genome. No sequences matches were found to the S. meliloti genome. However, 63 M. truncatula TCs had exact matches to the available M. truncatula genome (Supplemental Table I).
Characterization of Legume-Specific TCs
Following the analyses above, 92; 861; and 1,572 legume-specific TCs remained in L. japonicus, M. truncatula, and G. max/soja, respectively. A comprehensive list of all TCs identified is presented in Supplemental Table I, while a summary of TC length and the number of ESTs per TC in the legume-specific TCs can be found in Table II. BLASTX analysis to the GenBank NR database revealed that less than three percent of the identified legume-specific TCs had homology to other legume sequences with an E-value less than 10−4 (Supplemental Table II). Of the five most highly expressed TCs in M. truncatula, two (MtTC59272 and MtTC60015) had sequence homology (E-value ≤ 10−15) to seed albumins (pea, GenBank accession no. AAC61879; G. max, GenBank accession no. Q9ZQX0), while the other three (MtTC68206, MtTC59435, and MtTC60038) had sequence homology (E-value ≤ 10−4) to a hypothetical protein from Galega orientalis (GenBank accession no. CAB51773). In soybean, GmTC120439, the most heavily expressed TC (76 ESTs), lacked sequence homology to any available sequence in the GenBank NR database. GmTC120738, which was made up of 53 ESTs, showed greatest homology to the phloem-specific Vein1 protein from Vicia faba (GenBank accession no. S66340). Twelve percent of legume-specific TCs had sequence homology to sequences present in dbEST from other legume species. Previous data suggests the lack of homology among legume species may be due to tissue bias in sequencing projects rather than true species differences (Graham et al., 2004). A complete description and analysis of all the legume-specific TCs is provided in Supplemental Table I.
Table II.
Summary statistics of legume-specific TCs
| G. max/soja | M. truncatula | L. japonicus | |
|---|---|---|---|
| Number of legume-specific TCs | 1,572 | 861 | 92 |
| Size range of TCs (bp) | 103–2,962 | 104–1,706 | 156–854 |
| Average size of all TCs (bp) | 476 | 580 | 418 |
| Average size of all TCs ≥ 300 bp | 527 | 610 | 460 |
| Maximum number of ESTs per TC | 76 | 67 | 8 |
| Average number of ESTs per TC | 2.4 | 3.3 | 2.8 |
Single Linkage Clustering and Motif Analysis
One approach for hypothesizing function for legume-specific TCs lacking significant BLAST hits was to scan the TCs for conserved motifs identified from other proteins. InterProScan (European Bioinformatics Institute, Hinxton, UK; Mulder et al., 2003) identified 46 TCs containing 55 conserved motifs. Of these motifs, 41 were rich in specific amino acids. The remaining 14 included hits to a cyclin-like F-box (IPR001810), pectinesterase inhibitors (IPR006501 and TIGR01614), a zinc finger (IPR001878 and PS50158), and a nodulin (IPR003387 and PF02451). These are listed in Supplemental Table I.
A second approach was to mine groups of related legume-specific genes for common, uncharacterized motifs. Motif searching can be more sensitive than BLAST analysis for several reasons. First, highly conserved residues carry more weight in motif analysis. Second, a minimum exact word match is not required. Finally, the information from many homologs can be combined in a single motif. Once a novel motif description is generated, it can be used to scan the public protein databases for matches. Common motifs among target proteins suggest possible functions.
In order to identify families of legume-specific TCs for motif analysis, single linkage clustering analysis of legume-specific TCs was performed. Including as many diverse sequences as possible within a cluster increases the likelihood of identifying conserved motifs. Therefore, the 2,525 legume-specific genes were combined with 50, 672, and 688 homologous singletons from L. japonicus, G. soja/max, and M. truncatula, respectively. Clustering identified 665 groups that corresponded to potential gene families or cross-species homologs. The groups are identified in Supplemental Table I, with their size distribution shown in Supplemental Figure 1. The majority of TCs did not cluster with other sequences and are denoted as Group 0 in Supplemental Table I. Sixty-seven groups were chosen for motif analysis based on their size and/or tissue-specific expression patterns. For 17 of the groups, no obvious open reading frame with shared motifs could be found, suggesting that the sequences in these groups were not full length, represented unusually long untranslated regions, or that the gene products themselves were RNAs. Nine additional groups were almost certainly not legume specific. In these cases, TCs clustered with singletons that had strong hits to nonlegumes. Motif analyses in many of the remaining groups were quite fruitful. Thirteen groups had Motif Alignment Search Tool (MAST) or hidden Markov model (HMM) hits (E-value < 10−4) to nonlegume Swiss-Prot/TrEMBL sequences or to the Arabidopsis genome. The member sequences, motifs, and sequence alignments for each group are provided in additional supplemental data available at www.medicago.org/documents/Publications/Graham04_supplement. Three classes of families are particularly noteworthy and are described in detail below: (1) novel F-box proteins, (2) Pro-rich cell wall, and (3) small Cys-rich proteins. Tables III and IV and Figures 1 and 2 demonstrate the general features of groups in these categories.
Table III.
Characteristics of predicted F-box, Pro-rich, and Cys-rich proteins
| Protein Descriptiona | Groupb | Lengths of Predicted Proteins (aa)c | Species Distribution of Sequencesd | Predominant Expression Patterne | Total Number of ESTs in Group |
|---|---|---|---|---|---|
| F-box associated | 640 | 150–400 | Mt(5) | No specificity | 6 |
| F-box | 630 | 400 | Mt(4) | No specificity | 17 |
| Pro-rich | 5 | 200–500 | Mt(57) Gm(7) | No specificity | 65 |
| Pro-rich | 485 | 95 | Gm(2) Mt(1) | Above-ground | 22 |
| Pro-rich | 669 | 100 | Mt(3) | No specificity | 5 |
| Cys-rich | 31 | 60 | Mt(333) | Nodules | 899 |
| Cys-rich | 645 | 70 | Mt(12) | Seeds | 41 |
| Cys-rich | 38 | 110 | Gm(11) | Seeds | 36 |
| Cys-rich | 36 | 85 | Gm(3) Mt(1) | Seeds | 19 |
| Cys-rich | 40 | 70 | Gm(3) | Seeds | 8 |
| Cys-rich | 41 | 80 | Gm(3) | Seeds | 13 |
| Cys-rich | 655 | 85 | Mt(3) | Seeds flowers | 8 |
| Cys-rich | 666 | 130 | Mt(6) Gm(4) | No specificity | 109 |
Further descriptions of predicted proteins are provided in the text
Group numbers are arbitrary identifiers assigned during single-linkage clustering analysis of homologous sequences.
Amino acid (aa) lengths of predicted proteins were determined using ClustalW alignments with gaps removed.
Mt refers to sequences from M. truncatula, Gm refers to sequences from G. max/soja, and Lj refers to sequences from L. japonicus. The number of sequences from each species is shown in parentheses.
Tissue specificity was determined by analyzing the cDNA libraries of origin.
Table IV.
Motif analysis of predicted F-box, Pro-rich, and Cys-rich proteins
| Protein Descriptiona | Groupb | Representative Motifsc | Species Hits from Swiss-Prot/TrEMBLd | Hits to the Arabidopsis Genomee | Hits to the M. truncatula |
|---|---|---|---|---|---|
| F-box associated | 640 | FXIX{2}LGEXGX{2}ESWXKXF | – | 2 | 22 |
| F-box | 630 | [KR]XV[CS]KXW | – | 3 | 1 |
| M. truncatula (3) | |||||
| G. max (3) | |||||
| M. sativa (1) | |||||
| Pro-rich | 5 | SP… (PPV[VYE]K){3,} | T. repens (1) | 0 | 0 |
| Pea (1) | |||||
| C. arietinum (1) | |||||
| Bean (1) | |||||
| Carrot (1) | |||||
| Pro-rich | 485 | SP…PX{5}PX{4}PPXPPX{4}PX{4}P…PX{5}P…PP…P | – | 0 | 0 |
| Pro-rich | 669 | SP…PK…GXIXYQPPT…PXTPX{3}PX{3}PX…PXRPP | – | 0 | 0 |
| SP…CX{5}C…CX{4}C SP…CX{5}C…C…CX{4}CXC | G. orientalis (1) | ||||
| V. faba (5) | |||||
| Cys-rich | 31 | Pea (8) | 8 | 37 | |
| T. repens (1) | |||||
| Scorpion (5) | |||||
| Cys-rich | 645 | SP…C…CX{5}CX{3}C…GXC…CCC | – | 8 | 1 |
| Cys-rich | 38 | SP…CX{3}CX{3}C…CX{3}CX{3}C…CX{3}C | – | 2 | 1 |
| Cys-rich | 36 | SP…CX{5}C…C…CXC…CX{4}CXC | – | 0 | 0 |
| Cys-rich | 40 | SP…C…C…CX{3}C…GXC…CCC | – | 0 | 0 |
| Cys-rich | 41 | SP…C…CX{6}CX{3}C…CX{6}CXCX{2}C | – | 0 | 0 |
| Cys-rich | 655 | SP…CX{6}C…CX{6}C | – | 0 | 1 |
| G. max (1) | |||||
| G. soja (1) | |||||
| M. truncatula (2) | |||||
| Cys-rich | 666 | SP…C…C…C…CXC…C…CX{5}CX{8}CX{12}C | Pea (5) | 6 | 0 |
| L. angustifolius (1) | |||||
| P. vulagaris (1) |
Further descriptions of predicted proteins are provided in the text.
Group numbers are arbitrary identifiers assigned during single-linkage clustering analysis of homologous sequences.
In the representative motifs, single-letter amino acid abbreviations are used. The letters SP in bold designate the signal peptide. X denotes any amino acid. Numbers in curly brackets designate the number of repeats of the previous element. Letters in brackets denote a choice for a single amino acid residue. Three periods designate a gap introduced to optimize alignments. Due to its sequence diversity, two different motifs were used to model members of group 31.
For MAST and HMM analysis against the Swiss-Prot/TrEMBL database an E-value cutoff of 10−4 was used. Regular expression patterns were used to find hits to the Swiss-Prot/TrEMBL database for group 5. The following new abbreviations were used: Trifolium repens (T. repens) and Lupinus angustifolius (L. angustifolius).
Hits to the Arabidopsis and Medicago genomes were identified using either HMM or MAST motifs with an E-value cutoff of 10−4.
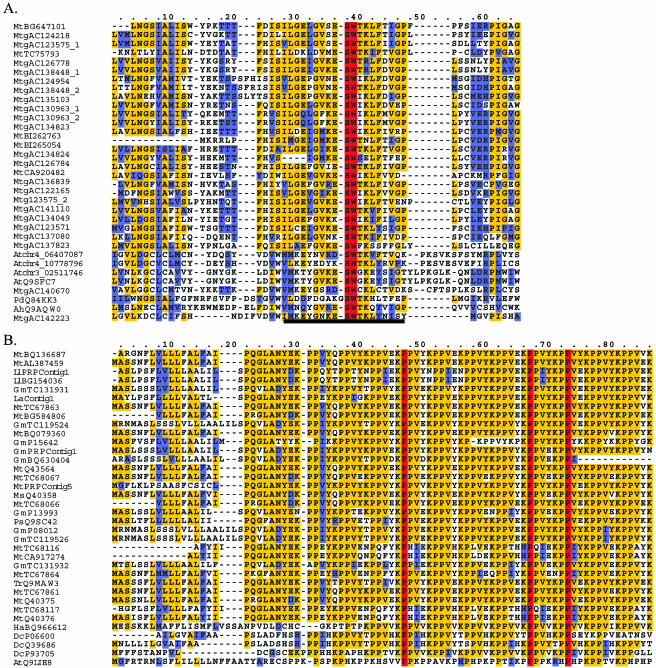
Figure 1.
Motif analysis of clustered sequences reveals similarity to F-box-associated domains and Pro-rich proteins. Residues that are identical throughout the proteins are shown in red. Residues conserved in more than 50% of the proteins are shown in yellow. Similar amino acid residues are shown in blue. Gaps (-) were introduced to optimize the alignments. Given the large number of sequences in the alignments, not all sequences are shown. All sequence names are preceded by a two-letter abbreviation representing the species name: At (Arabidopsis), Pd (Prunus dulcis), Ah (Antirrhinum hispanicum), Ll (L. luteus), La (L. albus), Ms (M. sativa), Ps (pea), Ha (sunflower), and Dc (carrot). Mt (GenBank accession) and Gm (GenBank accession) refer to EST singletons from M. truncatula and G. max/soja, respectively. MtTC and GmTC refer to TCs from M. truncatula and G. max/soja, respectively. Mtg (GenBank accession) refers to M. truncatula genomic sequence. Atchr (number) refers to unannotated Arabidopsis chromosomal sequence. The approximate position of the predicted start site follows the underscore. This number is based on the analysis of the Arabidopsis genome sequence (TIGR 3.0). A, Motif analysis of group 640 revealed similarity to a core 20 amino acid region within the larger F-box-associated domain. This region has been underlined. The alignment demonstrates the variability found outside this core domain. Sequences PdQ84KK3, AtQ9SFC7, and AhQ9AQW0 were identified from Swiss-Prot/TrEMBL. B, Regular expression pattern analysis of group 5 revealed similarities to Pro-rich cell wall proteins. LlPRPContig1, LaPRPContig1, GmPRPContig1, and MtPRPContig5 represent contigs assembled using the Sequencher software. GmP15642, GmP13993, GmP08012, MtQ43564, MtQ40375, MtQ40376, MsQ40358, PsQ9SC42, DcP06600, DcQ39686, DcP93705, and AtQ9LIE8 were identified from Swiss-Prot/TrEMBL.
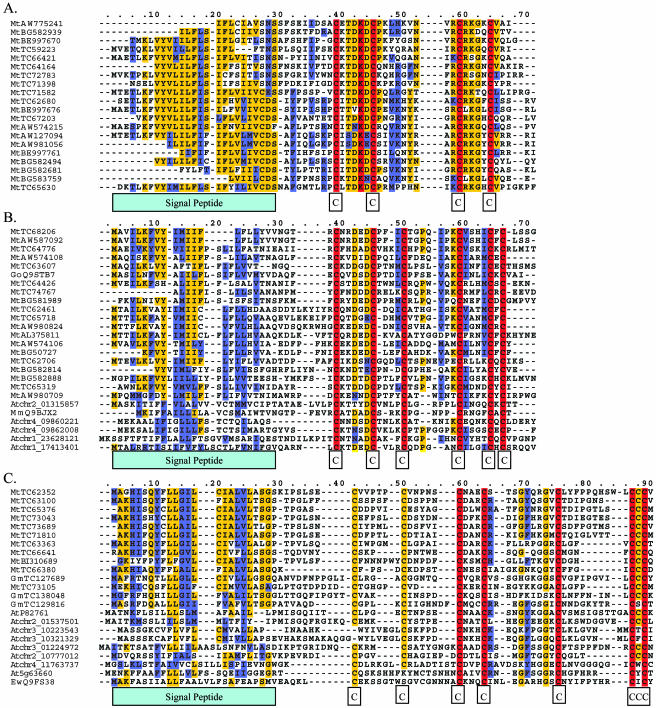
Figure 2.
Motif analysis of different groups of Cys cluster proteins. Small figures below each of the alignments depict the pattern of conserved residues. Residue coloring and sequence nomenclature are the same as used in Figure 1. Gaps (-) were introduced to optimize the alignments. Given the large number of sequences in the alignments, not all sequences are shown. A, Motif analysis of nodule-specific CCP group 31.01 identified no homologous sequences from other species. B, Motif analysis of nodule-specific CCP group 31.02 identified sequences from unannotated regions of the Arabidopsis genome and from Swiss-Prot/TrEMBL. MmQ9BJX2 is a neurotoxin protein identified from scorpion; GoQ9STB7 is a hypothetical protein from G. orientalis. C, Prior to motif analysis, seed-specific CCP groups 645 and 40 were merged into a single alignment. The resulting motif identified sequences from the Arabidopsis genome and from Swiss-Prot/TrEMBL. Hits to Arabidopsis came from both annotated (At5g63660) and unannotated regions of the genome. Hits from Swiss-Prot/TrEMBL included a putative gamma-thionin from Eutrema wasabi (EwQ9FS38) and putative self-incompatibility factor LCR46 from Arabidopsis (AtP82761).
F-Box Proteins
Two groups (640 and 630) of sequences without significant BLAST similarity with one another clearly shared motifs with discrete families of F-box proteins. Group 640 had five sequences from M. truncatula, from a variety of root and shoot libraries (Table III). Multiple EM for Motif Elicitation (MEME; Bailey and Elkan, 1994) identified two ordered motifs (widths 35 and 21 amino acids) shared by all five sequences. MAST (Bailey and Gribskov, 1998) was unable to find any significant hits (E-value < 10−4) to other proteins using these motifs. However, an HMM trained on the trimmed alignments containing these motifs found two hits to the Arabidopsis genome (At4g12560 and At4g22390) and 22 hits to the partial M. truncatula genome (Table IV). When the 22 genomic hits from M. truncatula were added to the original five sequences in the group, the resulting MEME motifs and HMMs identified about 90 proteins from a variety of legumes and nonlegumes in Swiss-Prot/TrEMBL. Many of the top-scoring hits were annotated as S locus F-box proteins, primarily from Prunus spp. and Arabidopsis. A portion of the amino acid alignment is shown in Figure 1A. Interestingly, the primary conserved motifs did not include the F-box domain itself as determined by the InterProScan (Zdobnov and Apweiler, 2001). Rather, homology was confined to a core 20 amino acid sequence within the F-box-associated domain whose function is presently unknown (Fig. 1A; Winston et al., 1999).
One area of concern was that members of group 640 were not full-length genes. The missing portions of these genes could encode F-box domains. Therefore, the 22 hits from the M. truncatula genome, which were identified by a domain outside of the F-box, were scanned for F-box domains using InterProScan (Zdobnov and Apweiler, 2001). Of the 22 hits from the M. truncatula genome, four had predicted F-box domains (IPR0018 and/or PF00646) with E-values between 10−6 and 10−11. Twelve additional M. truncatula genomic sequences also had weak (E-value > 10−3) hits. These results suggest that members of group 640 would likely encode weak homologs of F-box proteins, if they had been full length.
A second group (group 630) of F-box-containing proteins from M. truncatula was also found among the legume-specific groups. While no BLAST hit was found for this group, InterProScan identified a cyclin-like F-box (IPR001810) for one of the members of the group, but not the others, which may not have been full length. Using HMMs created for this group, we found several hits to the Arabidopsis genome (At1g20790, At5g18160, and At1g15680; Table IV). All of the significant hits were annotated as hypothetical F-box-containing proteins at the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org; Huala et al., 2001). Indeed, in this case, the common element between the group and the hits in Arabidopsis appears to be the F-box domain itself.
Pro-Rich Proteins
Group 5 included 57 sequences from M. truncatula (one TC and 56 singletons) and seven singletons from soybean (Table III). Sequences within this group were composed of pentameric and hexameric repeats and had homology to Pro-rich cell wall proteins from a variety of legumes. BLAST analyses with sequences from this family were problematic. Using a low-complexity filter during BLAST effectively screened out the mature protein sequences because they were repetitive. However, removing the filter resulted in hits to numerous Pro-rich proteins in which the order and arrangement of Pro residues was not conserved. Similar problems were encountered using MEME. Therefore, perl regular expression patterns were used, as described in the methods section. The first pattern identified proteins containing at least three subunits of PPVEK, PPVYK, or PPVVK, in any combination. This pattern identified 11 sequences in Swiss-Prot/TrEMBL from a diverse set of legumes, plus a single sequence from carrot (Daucus carota; Table IV).
As a more exhaustive test, we scanned dbEST for the nucleic acid equivalent of our motif. This search identified 1,824 ESTs from a variety of legumes and sunflower. Many of these corresponded to TCs or singletons in our original list, but the frequency of misassembly for this repetitive Pro-rich family at TIGR was high, based on visual inspection of contigs. Therefore, all the identified ESTs were assembled into contigs. In the end, 59 unique consensus sequences were identified: 27 from M. truncatula, 15 from G. max/soja, 1 from L. japonicus, 3 from Lupinus luteus, 3 from Lupinus albus, 1 from Phaseolous coccineus, and 9 from sunflower. The carrot and sunflower sequences were significantly different from their legume counterparts, having exclusively valines and/or histidines in the fourth position of the repeat (Fig. 1B). Among the legumes, valines were found in this position infrequently, and histidines were rare. Noting the variations observed in the carrot and sunflower sequences, we generalized the motif to (PP[ILMV][EYVHNA][KT]){3}. This search identified two additional sequences from carrot and one from Arabidopsis. The Arabidopsis sequence is significantly different from all other sequences identified; it is several times longer, contains hexameric in addition to the usual pentameric repeats, and it frequently has Thr rather than Lys in the last position (Fig. 1B).
Two other groups of Pro-rich proteins were also identified (group 485 and group 699). Group 485 was made up of two soybean sequences and one M. truncatula sequence. Group 669 was made up of three M. truncatula sequences. Neither group had any apparent tissue specificity (Table III). Like group 5, members of groups 485 and 669 also encoded a signal peptide and Pro-rich mature peptide (Table IV). However, groups 485 and 699 lacked the repetitive Pro-rich pentamers and encoded much smaller peptides. Motif analysis using these small groups was unable to identify similar sequences in the M. truncatula or Arabidopsis genomes.
Cys-Rich Proteins
A large fraction of the legume-specific genes encoded Cys-cluster proteins (CCPs). These share several common features: (1) an N-terminal signal sequence, (2) a small, highly charged or polar-mature protein sequence, (3) a characteristic arrangement of 4, 6, 8, or 10 Cys residues likely involved in disulfide bridges, (4) an apparent tissue-specific expression profile, and (5) low similarity to other expressed CCPs (Table III). The largest group of CCPs, group 31, is made up almost entirely of M. truncatula sequences. The group contains 197 M. truncatula singleton sequences and 136 TCs, all of which are expressed almost exclusively in nodules. A small fraction (less than 1%) of EST sequences from this group were also found in other root tissues. While the majority of sequences in this group had no BLAST homology to sequences within NR, approximately 20% had low homology (approximately equal to 10−4) hits to the same group of sequences: a hypothetical protein from G. orientalis (CAB51773; Kaijalainen et al., 2002), late nodulins from V. faba (CAB96471, CAB96474, CAB96476; Frühling et al., 2000), and a nodule-specific protein and ENOD3 (Scheres et al., 1990) from pea (BAB40944 and AAB23537, respectively).
The MEME program, when applied to the nodule-specific CCPs (group 31), generated three motifs of 21, 15, and 11 amino acids, respectively. The first motif encoded the signal peptide, and the remaining two contained pairs or triplets of Cys residues and their surrounding residues (Table IV). Scans of Swiss-Prot/TrEMBL using MAST recovered all of the GenBank BLAST hits of individual TCs mentioned above, plus several new hits with significance better than 10−4 (Table IV). The new hits included several more genes from legumes annotated as early and late nodulins, plus a set of five potassium channel-blocking neurotoxins from Manchurian scorpion (Mesobuthus martensii.) Further, the dozen hits with E-value between 0.1 and 10−4 were almost all K+ channel-blocking neurotoxins from a variety of scorpion species. All scorpion toxins hit only motifs 2 and 3, having their own distinctive signal peptide.
The extensive divergence among the nodule-specific CCPs made the creation of an accurate multiple-sequence alignment impossible for the whole group. However, an accurate alignment is a prerequisite for building an effective HMM. Hence, 261 sequences from group 31 were distributed among 11 distinct subgroups. A final set of HMMs was created to describe each of the 11 subgroups of nodule-specific CCPs. Alignments of the largest two subgroups, 31.01 and 31.02, are displayed in Figure 2A. For HMM generation, the subgroups were deliberately modeled without the signal peptide, since only the mature peptide itself proved to be similar to the scorpion toxins in the MEME/MAST analysis. Since HMMs were specifically designed for each subgroup, none of the 11 HMMs picked up as many significant hits as had the MEME motifs when scanning Swiss-Prot/TrEMBL. However, they proved to be useful in subdividing the large diverse family. For example, only subfamily two picked up any hits to scorpion toxins.
The 11 HMMs were then used to scan the Arabidopsis genome, which was translated into all six reading frames. These searches yielded eight hits with E-values more significant than 10−4, but seven of these hits were accounted for by subgroups two (subgroup 31.02; Fig. 2B) and nine (subgroup 31.09). Seven of the eight hits to the Arabidopsis genome lie in regions with no predicted genes on chromosomes one, two, and four of TIGR Arabidopsis sequence 4.0 (TAIR). The remaining hit, At1g43720, was a predicted hypothetical gene. In contrast to Arabidopsis, the rice genome had no significant hits to any of the 11 subgroups.
In addition to the group of nodule-specific CCPs, we also identified several groups of predominantly seed-specific CCPs (Table III). Group 645 was composed of 10 M. truncatula TCs and two singletons corresponding to the pods with seeds and immature seed libraries. Groups 38 (5 TCs, 6 singletons), 40 (2 TCs and a singleton), and 41 (2 TCs and a singleton) were soybean specific and were composed of ESTs from immature seed coats, seed coats, and very young seeds. Group 36 contains one M. truncatula singleton, one soybean singleton, and two soybean TCs composed of ESTs from mature and immature seed coats, mature pods, and immature cotyledons. Group 655 (2 TCs and a singleton) corresponded to developing flowers and pods with seeds. Unlike the nodule-specific CCPs, none of these groups had significant BLAST hits in the NR database. However, if a lower E-value cutoff of 10−4 were used, many would cluster with the nodule-specific CCPs.
The first round of MEME motif building did not yield any hits more significant than 10−4 for any of the seed-specific Cys-rich protein groups. However, the first round analysis for group 645 had a weak (E-value > 10−4) hit to a protease inhibitor from pear (Pyrus communis) that conserved the exact Cys positions (Table IV). Adding this single sequence to the second round of MEME analysis resulted in a set of motifs that had significant hits to nearly 100 proteins in Swiss-Prot/TrEMBL. Collectively, these included sequences annotated as gamma thionins, protease inhibitors, insect and plant defensins, and sodium channel-blocking scorpion toxins. A few examples of these can be seen in Figure 2C.
A single HMM was created from an alignment of all 12 original members from group 645, the largest seed-specific CCP group. Like the first iteration of the MEME motif, the HMM also did not have any significant hits to Swiss-Prot/TrEMBL. However, it had eight hits to the Arabidopsis genome and one hit to the existing M. truncatula genomic bacterial artificial chromosome (BAC) sequence (E-value < 10−4). An alignment of these sequences is shown in Figure 2C. At5g63660 was predicted to be a plant defensin by TAIR. P82761 (Swiss-Prot/TrEMBL) is the putative self-incompatibility protein LCR46. The remaining six hits lie in regions not predicted to encode genes on chromosomes two, three, and four (Fig. 2C; TAIR).
Group 666 also appeared to be Cys-rich and had strong sequence homology to the soybean albumin 1 precursor (GenBank accession BAA04219). This group was composed of one TC and three singletons from soybean and three TCs and singletons from M. truncatula. Two of the TCs from M. truncatula had high levels of expression. MtTC59272 and MtTC60015 were composed of 61 and 36 ESTs, respectively, mainly from roots and mycorrhizal roots. Unlike the CCPs in group 31, members of group 666 are longer, contain more conserved Cys residues, and show diminished tissue specificity.
Organization and Evolution of CCPs
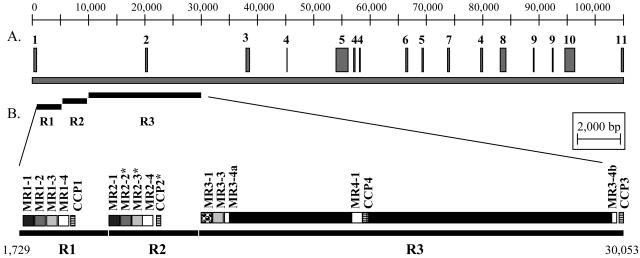
In order to examine the genomic organization of all the CCPs, BLASTN was used to search the available M. truncatula genome sequences. Thirteen BACs were identified that had exact matches to expressed members of the nodule-specific CCP family. Seven of these BACs contained at least two predicted CCPs. Only BAC clone Mth2-34P9 (GenBank accession AC121238) was completely assembled. This BAC contained four predicted CCPs, only one of which had corresponding ESTs. CCP2 corresponded to TC78513, which contained 10 ESTs from four different nodule libraries spanning four to 60 d postinoculation with S. meliloti. Comparison of the EST sequences of CCP2 with the genomic sequence revealed it did not contain an intron. Given the structural similarity to CCP1 and CCP3, it is unlikely that these have introns. However, CCP4 appears to contain a 314-bp insertion or intron relative to the other CCPs. Without EST confirmation, it is impossible to tell if CCP4 actually contains an intron or is a pseudogene.
Dot plot analysis was used to examine the organization of CCPs within BAC Mth2-34P9 (Fig. 3). Initially three tandem repeats were identified, each of which corresponded to one CCP. Repeat R1 was 4,184 bp in length and contained CCP1. Repeat R2 was 4,192 bp in length and corresponded to CCP2. Repeat R3 was 19,948 bases in length and corresponded to CCP3 and CCP4. The large size of R3 and the extra CCP were due to a large insertion of 17,916 bases. Repeats R1 and R2 shared more than 97% nucleotide identity with each other and 92% nucleotide identity with R3.
Figure 3.
Genomic organization of CCPs on BAC Mth2-34P9. A, Regions of BAC Mth2-34P9 with BLASTX homology less than 10−10 to GenBank sequences are as shown: 1, Ta11 non-Long Terminal Repeat retroelement (Arabidopsis, gi|15226160); 2, Putative retrotransposon gag protein (pea, gi|31126675); 3, POLX_ TOBAC retrovirus-related Pol polyprotein (Nicotiana tabacum, gi|130582); 4, Vesicle-associated membrane protein (Arabidopsis, gi|15225415); 5, Putative polyprotein (N. tabacum, gi|20161451); 6, Albumin 1 (M. truncatula, gi|3238736); 7, Protein T31J12.4 (Arabidopsis, gi|25402534); 8, Mariner transposase (G. max, gi|7488706); 9, Expressed protein (Arabidopsis, gi|18410357); 10, Scarecrow-like transcription factor (Arabidopsis, gi|15236725); and 11, Transposable element Tnp2 (Antirrhinum majus, gi|1345502. B, Organization of CCPs, repeats (R), and MRs. Each thick black line represents a repeat and is named accordingly. MRs and CCPs are shown as boxes above each repeat. MRs are shaded similarly to show that MRs in conserved positions between repeats are more similar to each other than are MRs within a repeat. The only exception is MR3-1, which is shaded differently. Expressed sequences can be identified by the asterisk following the name.
Closer inspection of the large repeat units revealed a number of tandemly duplicated mini repeats (MRs). Repeats R1 and R2 each contained four tandemly duplicated MRs (MR1-1, MR1-2, MR1-3, MR1-4, and MR2-1, MR2-2, MR2-3, MR2-4, in order, respectively) that ranged in size from 502 to 523 bases. MR1-4 ended 25 bases before the predicted translation start site of CCP1, while 54 bp separated MR2-4 from CCP2. Repeat R3 contained three tandemly duplicated MRs (MR3-1, MR3-3, and MR3-4) ranging in size from 513 bases to 524 bases. MR3-4 was split by the 17,916-bp insertion. The insertion itself also contained a 617-bp MR (MR4-1). MR3-4 and MR4-1 each ended 54 bp upstream of the predicted translation start site of CCP3 and CCP4.
Interestingly, we were able to identify a TC identical to portions of MR2-2 and MR2-3. TC82353 corresponded to bases 76 through 522 of MR2-2 and bases 1 through 376 of MR2-3. The TC was composed of two ESTs from senescing nodules. Further experimental analysis will be performed to determine if the MRs are being expressed as a single transcript with CCP2.
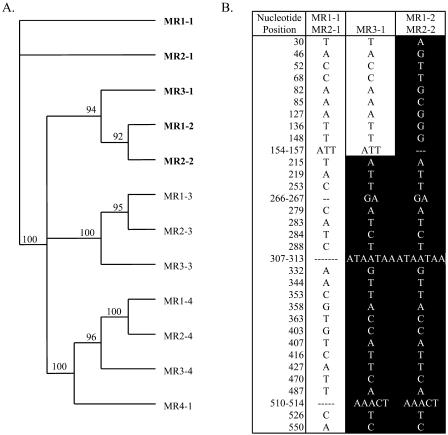
Phylogenetic analyses of repeats and MRs were performed to determine how this complex arrangement of repeats might have developed (Fig. 4A). Sequence identity was highest between MRs located at the same relative position within the different larger repeats. For example, MR1-2 and MR2-2 (from the larger repeat regions R1 and R2, respectively) showed greater similarity to each other than to other MRs from their own larger repeats. This confirmed that the three tandem repeats all originated from a single ancestral sequence that likely contained four MRs and a CCP coding sequence.
Figure 4.
Analysis of MRs from BAC Mth2-34P9. A, Phylogenetic analysis was performed on the aligned nucleotide sequences of MRs. Bootstrap scores are provided at each branch of the tree. In general, MRs in conserved positions across the larger repeats share greater similarity to each other than to other MRs in the same repeat. The only exception is MR3-1, which appeared to have combined features of MR1-1, MR2-1, MR1-2, and MR2-2 (shown in bold). B, The sequences of MR1-1 and MR2-1 were compared to the sequences of MR1-2 and MR2-2 to identify conserved polymorphic sites. The position and sequence of the polymorphism in each group of sequences is indicated. Dashes indicate gaps in the alignment. The sequence of MR3-1 is shown at each of these positions. From polymorphic positions 30 through 157, MR3-1 matches the sequences of MR1-1 and MR2-1. However, from polymorphic positions 215 through 550, MR3-1 matches the sequences of MR1-2 and MR2-2. This indicates that an unequal recombination event somewhere between nucleotides 158 and 214 fused two progenitors MRs together to form MR3-1.
The large number of repeats within this BAC and their high degree of sequence similarity suggested this region might be prone to unequal recombination. The organization of MRs in repeats R1 and R2 suggested that region R3 was missing an MR. Phylogenetic analysis revealed clustering between MR1-3, MR2-3, and MR3-3 and between MR1-4, MR2-4, and MR3-4 (Fig. 4A). The MR3-1 sequence did not clearly cluster with MR1-1 and MR2-1, or with MR1-2 and MR2-2. However, if MR3-1 was divided in half, the first half clustered with MR1-1 and MR2-1, while the other half clustered with MR2-2 and MR3-2. This suggested that unequal recombination between repeats might have deleted portions of two ancestral MRs to form a new MR (MR3-1), which was a mixture of both. To determine if this had occurred, polymorphic sites were identified by comparing conserved nucleotide positions shared between MR1-1 and MR2-1 to conserved nucleotide positions shared by MR1-2 and MR2-2 (Fig. 4B). Polymorphic sites included base changes and insertions/deletions. Once these polymorphic sites were identified, they were compared to the sequence of MR3-1. At the 10 polymorphic sites identified from consensus position 30 through 157, MR3-1 matched the sequences of MR1-1 and MR2-1. However, at the 23 polymorphic sites identified from consensus positions 215 to 550, MR3-1 matched the sequences of MR1-2 and MR2-2. Therefore, a recombination event, somewhere between consensus positions 158 and 215, combined two ancestral MRs to form MR3-1. The 17,916-bp insertion in MR3-4 is also likely due to unequal recombination.
As a final step in the analysis of CCPs in this region, the ratio (Ka/Ks) of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitutions was determined. If a gene is under purifying selection, the Ka/Ks ratio should be less than one (Hughes, 1999). A Ka/Ks value greater than one suggests the gene is under diversifying selection. All six possible pairwise comparisons were made between the CCPs on BAC Mth2-34P9. The predicted signal peptide, which would not be present in the mature protein, was removed prior to analysis as was the insertion/intron in CCP4. CCP4 was not discounted as a pseudogene, because the position and size of the insertion was consistent with introns observed in other CCPs from other BACs. Ka/Ks ratios ranged from 4.215 to 7.35, suggesting divergent selection. The Ka/Ks ratio was not determined for the comparison of CCP1 and CCP2, since no synonymous substitutions were identified. Two-by-two contingency tables were used to identify gene comparisons in which the Ka/Ks ratio was significantly different from neutral selection (Ka/Ks = 1). All five comparisons were significantly different (P < .005) from a model of neutral selection.
To determine if MRs were associated with CCPs elsewhere in the genome, the different MRs were used as BLAST queries against MtGI (version 7) and the available M. truncatula genome sequence. Two TCs and a singleton that did not match the sequence of BAC Mth2-34P9 were identified. TC85293 showed homology to all the MRs and encoded a CCP. It was composed of 10 ESTs from mature nodules. TC79662 also had homology to a small portion of an MR and contained a CCP. It was composed of four ESTs from three different nodule libraries. A singleton (BE998091) from the senescing nodule library was most similar to bases 1 through 183 of MR1-2.
To determine if the pattern of duplications observed was unique to BAC Mth2-34P9, another BAC with multiple CCPs was examined. BAC Mth2-10A20 (GenBank accession AC138527) contains five CCPs, four of which contain ESTs exclusively from mature and senescing nodules (TC64692, TC75960, TC64231, and TC73971). Comparisons of the ESTs with the BAC sequence revealed that the four expressed CCPs and likely the fifth CCP, whose expression has not been detected, contain introns. The introns range in length from 107 to 108 bp and are located 70 to 73 bp downstream of the translation start site. A complete genomic analysis of the CCPs on BAC Mth2-10A20 was impossible, since the remaining six sequence contigs have not been assembled. However, three CCPs are present in a single contig. This contig contains a tandem duplication, 4,629 bp in length. A second duplication, at least 1,570 bp in length, is also present. However, the extent of this duplication cannot be determined without further sequence assembly.
DISCUSSION
Using increasingly stringent BLAST searches, we have identified over 2,500 legume-specific genes from M. truncatula, L. japonicus, and G. max/soja. The analyses included comparisons to the GenBank NR and EST_others databases, as well as comparisons to the rice and Arabidopsis genomes. By the very nature of the analysis, only a small subset of the legume-specific genes identified have homology to previously characterized genes or gene products. The observed results are consistent with the representation of legume sequences in the GenBank NR database relative to those of better characterized groups. For example, within NR, there are over 117,000 protein entries for Arabidopsis alone (NCBI taxonomy browser, http://www.ncbi.nih.nlm.gov/Taxonomy, November, 2003). In contrast, for all of the legume species within the Fabaceae, there are only 11,300 protein sequences. While Arabidopsis has proven to be a useful model for many aspects of plant biology, it is not a good model for studying nodule development. Legume-specific TCs are especially enriched in transcripts from nodules; almost 56% of the ESTs corresponding to legume-specific genes in M. truncatula come from nodules. In contrast, only 22% of the ESTs corresponding to all Medicago TCs come from nodules. Future analysis of legume-specific genes expressed in nodules may therefore provide insights into novel symbiotic functions.
Given the low representation of legume sequences in the GenBank databases, several approaches were taken in order to assign putative functions to the legume-specific genes. All of the legume-specific genes and homologous singletons were grouped together into families of related sequences. The sequences within a group were then mined for conserved motifs that could be used to scan the protein databases at Swiss-Prot/TrEMBL. Proteins of known function that shared these motifs could provide a hint of function. Using this technique, we identified several groups of interest. However, the most interesting were families of F-box related proteins, Pro-rich proteins, and Cys-rich proteins.
A Diverse Family of F-Box-Related Genes From Legumes
F-box-related proteins have been identified with a wide array of cellular functions. F-box proteins are involved in transcriptional regulation and signal transduction through Skp1, Cdc53/Cullin1, F-box protein ubiquitin-ligase complexes, transcript elongation, cell cycle transition, and self-incompatibility in plants (Kipreos and Pagano, 2000; Ushijima et al., 2003). The range in F-box protein functions is due in part to the diversity of secondary motifs. Comprehensive analyses of the Arabidopsis genome identified between 560 and 690 predicted F-box proteins (Gagne et al., 2002; Kuroda et al., 2002). Leu-rich and kelch repeats were the most abundant secondary motifs identified. Other motifs such as WD-40, Armadillo, and tetratricopeptide repeats were also found (Gagne et al., 2002). A large number of F-box proteins had associated regions with no similarity to previously characterized motifs.
In our analysis, we identified two groups with distant homology to F-box proteins and related domains. In Arabidopsis, roughly 35% of genes encoding F-box-related proteins exist in clusters of two to seven F-box genes (Gagne et al., 2002). This arrangement would allow shuffling of domains among F-box genes. The presence of F-box proteins with similar N-terminal and dissimilar C-terminal domains support the idea that motif shuffling has lead to further F-box protein diversification (Gagne et al., 2002). Of the 22 M. truncatula genome hits from group 640, 10 came from BACs containing two or more F-box proteins (data not shown). Motif shuffling between clusters of F-box-related proteins would allow sequence diversification to occur, possibly leading to the evolution of novel functions in legumes.
A Family of Pentameric Pro-Rich Repeats Overabundant in Legumes
The plant cell wall has several important functions, including structural support, defense against pathogens, and signaling between the plant cell and the outside world. The Hyp-rich glycoprotein superfamily includes extensins, repetitive Pro-rich proteins (RPRPs), and arabinogalactan proteins that are proposed to represent a phylogenetic continuum (Kieliszewski and Lamport, 1994). According to these authors, the presence of a cross-linking motif, Val Tyr Lys (VYK), separates RPRPs and extensins from the rest of the family. These two groups differ due to the presence of a repeated SerHyp4 motif in extensins, which is lacking from RPRPs, and by the extensive glycosylation of Ser and Hyp residues in extensins, in comparison to the relatively rare glycosylation of Hyp residues in RPRPs.
We have identified a group (group 5) of sequences encoding proteins composed of pentameric repeats with homology to RPRP wall proteins from a variety of legumes (Fig. 4B), including the well-characterized PRP1 and PRP2 from G. max (Hong et al., 1987, 1990; Datta et al., 1989). The Pro-rich proteins in this group are constructed in large part from PPVYK, PPVVK, and PPVEK subunits and, while individual proteins may show striking patterns of tissue- and organ-specific accumulation, the group as a whole lacks tissue-specific expression. Using motif analysis, we were able to identify similar RPRP sequences from carrot, sunflower, and Arabidopsis. Unlike the legume sequences however, the repeat units in nonlegume RPRPs were composed of different amino acids. These results suggest that legume Pro-rich proteins favor particular subunits that are not well represented in other taxa sequenced to date.
If one considers the pentameric Pro-rich repeat to be a functional unit within RPRPs, it is probable that legume RPRPs may have novel functions related to their novel pentameric motifs. Most notably, the PPVYK motif appears to be rare in RPRPs known to date from plants outside the legume family, and is therefore clearly not diagnostic of the RPRPs as a group. This motif functions in peroxide-mediated cross linking involving Tyr residues in both extensins and RPRPs, which renders the proteins insoluble and confers rigidity to the wall (Bradley et al., 1992; Brisson et al., 1994; Frueauf et al., 2000). The restricted distribution of the PPVYK motif indicates that covalent cross linking of RPRPs may be the exception rather than the rule. Though the functional significance of other pentamers remains to be elucidated, the RPRPs present an excellent problem for phylogenetic consideration of structure/function relationships of cell wall proteins.
A Diverse Family of Cys Cluster Proteins with Similarity to Defensins
The first nodule-specific CCP homolog, ENOD3, was identified by Scheres et al. (1990) in pea. Homologs were later identified in V. faba (Frühling et al., 2000) and G. orientalis (Kaijalainen et al., 2002). Györgyey et al. (2000) identified nineteen members of the family from a small collection of M. truncatula ESTs. Originally, this group of genes was thought to be a small gene family. The extent of the family was simultaneously recognized by ourselves (Graham et al., 2004), by Fedorova et al. (2002) who identified the family by its nodule-specific expression pattern, and by Mergaert et al. (2003), who used BLAST to query the M. truncatula ESTs for sequences with homology to Cys cluster proteins identified by Györgyey et al. (2000; Fig. 2, A and B). Since nodule-specific CCPs were detected only in certain legumes, Mergaert et al. (2003) hypothesized the family was specific to legumes in the galegoid group and had a role in indeterminate nodule formation. We have identified seed-specific CCPs from both M. truncatula and soybean, which suggests this is not the case (Fig. 2C). Hybridization of M. truncatula nodule-specific CCPs to soybean BACs suggests homologous sequences are present in the soybean genome (data not shown). Failure to identify nodule-specific CCPs from soybean or L. japonicus, both of which form determinate nodules, could be due to differences in the timing of tissue harvest relative to the stage of nodule development or in the limited representation of ESTs from nodules in these two species.
Motif analysis of the CCPs has revealed similarity to plant defensins, whose conserved Cys residues are important in the formation of the knottin fold (Thomma et al., 2002). Like defensins, the CCPs can be characterized by an N-terminal signal sequence, a small highly charged or polar mature protein sequence, and a tissue-specific expression pattern (Thomma et al., 2002). In addition, the amino acids sequences are highly divergent with the exception of the conserved Cys residues (Fig. 2; Thomma et al., 2002; Ganz, 2003). In Arabidopsis, the intron sizes of the putative defensins identified by Thomma et al. (2002) range from 74 to 377 bases (data from TAIR: http://www.arabidopsis.org). The start position of the intron ranges from 56 to 139 bases after the translation start site. Similar intron positions and sizes were observed in the genomic sequence corresponding to the M. truncatula CCPs.
The similarity of CCPs to defensins is intriguing because of their potential utility in crop improvement. Defensin expression in plants can typically be found in leaves (Terras et al., 1995), seeds (Almeida et al., 2000; Chen et al., 2002), flowers (Park et al., 2002; Lay et al., 2003) and can be induced by stress (Koike et al., 2002) or pathogen inoculation (Manners et al., 1998; Berrocal-Lobo et al., 2002). Plant defensins have been identified that confer resistance to fungal (Gao et al., 2000; Park et al., 2002; Cabral et al., 2003), bacterial (Koike et al., 2002), or insect pathogens (Chen et al., 2002; Lay et al., 2003). Overexpression of defensins has been used in a few crop species to develop lines resistant to particular pathogens. In rice, overexpression of the wasabi defensin gene conferred resistance to rice blast (Magnaporthe grisea; Kanzaki et al., 2002). A defensin isolated from Medicago sativa was used to confer resistance to Verticillium dahliae in potato (Gao et al., 2000). Unlike typical R-gene-mediated defense responses, a single defensin can provide resistance to multiple pathogens (Lay et al., 2003) or races of pathogens (Kanzaki et al., 2002). Identification and utilization of defensins could have broad impacts on current agricultural practices.
Defensins have been identified throughout the animal and plant kingdoms and are thought to be members of small (15–40 members) gene families (Boman, 2003). In the human, mouse, and Arabidopsis genomes, defensins are present in clusters (Linzmeier et al., 1999; Thomma et al., 2002; Maxwell et al., 2003). The number of defensins identified in other species is far lower than the large number of CCPs identified in M. truncatula. Analysis of EST data revealed at least 300 different CCPs are expressed (Graham et al., 2004; Mergaert et al., 2003). Analysis of available genomic data revealed additional CCP homologs that are not found in current EST libraries.
Why are there so many more CCPs in M. truncatula? It's possible there aren't, but rather additional undiscovered defensins do exist in other species. The sequence diversity of defensins has made them difficult to identify experimentally and computationally. For example, using CCP motifs developed from M. truncatula, we were able to identify nine putative defensins from Arabidopsis. Only one of these had been previously identified, while the other eight were identified in regions that were not predicted to contain genes (TAIR). Advances in computational biology will likely lead to the discovery of additional defensins (Schutte et al., 2002). At the same time, the fact that most Medicago CCPs are expressed in the nodule suggests that at least this particular class of defensins has evolved only recently, with the evolution of nodules.
How have so many diverse CCPs originated? Analysis of the available genomic data suggests an evolutionary model similar to that of nucleotide binding site (NBS), Leu-rich repeat (LRR) family of disease resistance genes. Like the CCPs and defensins, NBS/LRR genes have been found both as single genes and clustered throughout plant genomes (for review, see Hulbert et al., 2001). Completion of the Arabidopsis genome has facilitated analysis of NBS/LRR evolution at a genome level. In Arabidopsis, there are approximately 150 NBS/LRR sequences in the genome (Baumgarten et al., 2003; Meyers et al., 2003). Individual clusters of NBS/LRR genes are thought to have arisen through duplication and unequal recombination. Intergenic recombination between closely related paralogs within a cluster, interallelic recombination, and diversifying selection have driven the evolution of genes within a cluster. Clusters of genes have spread throughout the genome through microscale chromosomal duplications that have been translocated to distant positions (Meyers et al., 2003) and/or through segmental duplication and rearrangement of chromosomal regions (Baumgarten et al., 2003).
Until further sequencing of the M. truncatula genome is complete, it will be difficult to determine all the mechanisms governing the evolution of the CCPs. Our analysis of BACs Mth2-34P9 and Mth2-10A20 clearly reveals evidence of multiple duplication events. Analysis of the repeat regions on BAC Mth2-34P9 has shown that unequal recombination has occurred between repeat units. If the repeats themselves can undergo recombination with paralogous repeats, it is likely their close proximity to the CCPs would allow unequal recombination to occur within the CCPs. The significant levels of nonsynonymous amino acid substitutions could be the result of recombination between paralogous CCPs within a cluster and/or the accumulation of point mutations. The identification of other TCs with MRs and CCPs that do not match BAC Mth2-34P9 suggest that these events occur elsewhere in the genome. Similar phenomena have been seen in mammalian defensin gene clusters. Semple et al. (2003) concluded that the human β-defensin cluster on chromosome 8p23 had evolved by successive rounds of duplication followed by sequence diversification due to positive selection. Positive selection in the β-defensins favors amino acids in the second exon, which encodes the mature protein, and often resulted in a change of charge. Similar results were obtained by Maxwell et al. (2003), who examined a cluster of murine β-defensins syntenic to the cluster on human chromosome 8p23.
Why then are so many CCPs expressed specifically in nodules? Like seeds, nodules are one of the largest sink tissues in plants. Fifteen to 30 percent of the net photosynthate is transported to the nodule and its surrounding root system (Schubert, 1986). Photosynthate is used in the development and maintenance of the nodule and rhizobia, in nitrogen-fixation reactions, and in the transport of nitrogen-containing compounds from the nodule. In exchange for fixed nitrogen, the plant provides the rhizobia with different carbon compounds. During the early steps of nodule formation, signaling between plant and rhizobia is critical in preventing a defense response in the plant (Mithöfer, 2002; Mitra and Long, 2004). Typical defense responses such as the production of phytoalexins and pathogenesis-related proteins are not induced. If typical defense responses are shut down, how does the plant protect itself and the carbon-rich nodule from a wide array of pathogenic organisms? One gram of soil can contain between 6,800 and 34,000 different taxa of bacteria (Curtis et al., 2002). Also present in the soil are fungal and insect pathogens. We believe that CCPs are induced as a secondary defense to protect the nodule from pathogenic organisms, while allowing the symbiosis to continue. Given the broad spectrum of pathogens present in the soil, the secondary defense response must be able to target specific pathogens without harming beneficial rhizobia. This requires an arsenal of different defense compounds, like the CCPs. As pathogen populations evolve, so must plant defense responses.
CONCLUSION
One of our hypotheses was that some of the identified legume-specific genes were derived from nonlegume origins, but have diverged so much they appear unique to legumes. Using single-linkage clustering and motif analysis, we were able to identify gene families with conserved motifs. In some cases, such as the defensin-like CCPs and F-box-related proteins, the motifs identified were clearly represented across diverse taxa. Thus, as hypothesized, these genes may be examples of fast-evolving genes that are so divergent that similarity to their progenitors is not readily detectable by BLAST algorithms. Sequences that are truly novel in legumes may be present among the families that were too small for motif analysis, families where motifs could not be detected, or families whose motifs failed to detect similarity to any known proteins. Experimental analyses and sequence information from a wider diversity of organisms will aid in determining if these genes are indeed novel.
While the function of many legume-specific genes could not be predicted by computational approaches, their expression patterns suggest they are worth investigating experimentally in the future. All of the legume-specific genes we have identified have been made publicly available in the supplemental data, representing a rich resource for legume biologists (see supplemental data for this article and at www.medicago.org/documents/Publications/Graham04_supplement). Among these legume-specific genes, we identified many gene families with nonspecific expression patterns. Additionally, we have identified 10 gene families specifically expressed in roots and nodules, eight in seeds, four expressed only in leaves and flowers, and seven from stressed or pathogen-inoculated tissues. The tissue specificity of these genes suggests they would make excellent candidates for transformation or gene silencing in future analyses of gene function.
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor. Perl scripts will be made available upon request.
BLAST Analyses to Identify Legume-Specific Genes
Unless otherwise stated, all computer analyses were performed on a single Macintosh PowerG4 computer running Mac OS 10.22 with dual 800 MHz processors. Locally installed versions of the NCBI BLASTN, BLASTX, and TBLASTX (Altschul et al., 1997) programs were used to find regions of sequence homology between legume and nonlegume sequences (Table I). Filtering of repetitive sequences was turned off for all BLAST searches to increase the stringency for finding legume-specific genes, unless noted. Perl scripts were used to parse the results of each BLAST analysis described below. Legume TCs with BLAST homology to any nonlegume sequence from a variety of databases with an E-value more significant than 10−4 were not considered legume specific and were removed from the list of putative legume-specific genes. Legume TCs with BLAST homology to nonlegume sequences with an E-value between 10−4 and 10−8 were analyzed by hand, to ensure that homology was not due to the polyA tail or highly repetitive sequences. A summary table of the identified legume-specific TCs is provided in Supplemental Table I.
In the first BLAST iteration, (Table I) the TIGR Medicago truncatula, Lotus japonicus, and soybean (Glycine max and Glycine soja) TCs were compared to the TIGR maize (ZmGI), tomato (LeGI), rice (OsGI), and Arabidopsis (AtGI) gene indices using BLASTN and TBLASTX (Table I; Quackenbush et al., 2001; http://www.tigr.org/tdb/tgi). In the second BLAST iteration, the remaining legume-specific genes were compared to the NR using BLASTX. This was followed by the third BLAST iteration in which TBLASTX comparisons were made to the remaining TIGR plant gene indices from barley (Hordeum vulgare), Chlamydomonas reinhardtii, cotton (Gossypium spp.), grape (Vitis vinifera), ice plant (Mesembryanthemum crystallinum), lettuce (Lactuca Sativa), Pinus spp., potato (Solanum tuberosum), rye (Secale cereale), sorghum (Sorghum bicolor), sunflower (Helianthus annuus), and wheat (Triticum aestivum). BLAST iteration five used TBLASTX to compare legume-specific TCs to the Arabidopsis and rice genomes. Unlike previous searches, the sequences were filtered prior to BLAST to remove low complexity sequences. In the final BLAST iteration, the remaining legume-specific TCs were then compared to EST_others using the DeCypher Bioinformatics Accelerator running the Tera-BLAST hardware accelerated version of BLAST (TimeLogic, Crystal Bay, NV), which was housed in the Center for Computational Genomics and Bioinformatics at the University of Minnesota.
Identification of Contaminating Sequences from Sinorhizobium meliloti
BLASTN analysis against the composite genome of S. meliloti (Galibert et al., 2001) was performed to rule out contaminating sequences. Conversely, BLASTN analysis against available sequences from the M. truncatula genome was performed to confirm sequences were of plant origin (as of May, 2003).
Removal of TCs with Significant Homology to Nonlegume-Specific TCs
TBLASTX analyses were used to compare the remaining legume-specific TCs with the original legume TCs. Putative legume-specific TCs with significant (E-value ≤ 10−20) similarity to TCs not unique to legumes were identified and removed.
Single Linkage Clustering of TCs and Singletons into Homologous Groups
Prior to single linkage clustering, the legume-specific TCs were blasted against the legume singletons using TBLASTX and a 10−6 E-value cutoff. TBLASTX was used to compare the legume-specific TCs and the homologous singletons against themselves using an E-value cutoff of 10−6. Perl scripts were used to identify and cluster TCs or singletons that had overlapping BLAST hits using a significance threshold of E = 10−6. If the BLAST reports of two sequences had at least one TC or singleton in common, the sequences were combined into a group. Additional sequences that had at least one overlapping BLAST hit were then transitively added to the group.
Identification of Conserved Sequence Motifs from InterPro
All of the legume-specific TCs were scanned against a locally installed version of InterProScan (Zdobnov and Apweiler, 2001). InterProScan includes the following motif databases: PROSITE (Falquet et al., 2002), PRINTS (Attwood et al., 2003), Pfam (Bateman et al., 2002), ProDom (Corpet et al., 1999), SMART (Letunic et al., 2002), and TIGRFAMs (Haft et al., 2003).
Construction and Searching of Novel Legume-Specific Motifs
The following procedure was used to identify and refine motifs shared within a group of related legume-specific sequences. Each TC or singleton within a clustered group was first translated in all six frames. The translated sequences were then modeled as a set of ungapped position-specific scoring matrices using the MEME program (Computer Science and Engineering Department, University of California, San Diego; Bailey and Elkan, 1994). An expectation maximum algorithm is used by the program to identify optimal-width motifs conserved among the protein sequences. Only a few reading frames had conserved motifs free of stop codons. All other reading frames were removed from the set, and the MEME motifs were regenerated.
The protein sequences in the proper frame identified above were aligned using ClustalW (European Bioinformatics Institute; Thompson et al., 1994) and manually edited with JALVIEW (European Bioinformatics Institute; Clamp et al., 2004). HMMBUILD and HMMCALIBRATE (HMMER package; School of Medicine, Washington University in St. Louis; Durbin et al., 1998) were used to model the conserved portion of the alignment as an HMM. Note that the HMMs generated by HMMER statistically model the entire alignment including the likelihood of finding insertions and deletions, whereas the profiles generated by MEME model small, ungapped segments within an alignment. Each of these methods has a different level of sensitivity that is dependent on the protein family studied. Hence using both complementary methods increases the likelihood of finding significant hits.
The family models generated by MEME and HMMBUILD were used by the MAST (Computer Science and Engineering Department, University of California, San Diego; Bailey and Gribskov, 1998) and HMMSEARCH (Durbin et al., 1998) programs, respectively, to scan all the sequences in Swiss-Prot/TrEMBL (Boeckmann et al., 2003), a comprehensive nonredundant collection of known protein sequences. The available sequence from the Arabidopsis and M. truncatula genomes (February, 2003) were also scanned in this step. Sequences with significant scores less than 10−4 (unless noted) were added to the original group of legume-specific genes. The motif building and searching procedures were repeated until no additional significant hits were found or until new sequences identified began to dominate the model. In this case, further iterations would be uninformative.
Only groups with 10 or more TCs or singletons were analyzed. Smaller groups were also analyzed if they showed a tissue-specific pattern of expression, based on the library of EST origin. In order to remove redundancy caused by overlapping singletons, the singletons were contigged using Sequencer software (Gene Code, Ann Arbor, MI). Motifs, sequences and sequence alignments for each of the groups analyzed are provided in a downloadable directory structure in the supplemental data (see www.medicago.org/documents/Publications/Graham04_supplement).
One problem encountered during motif analysis was the presence of chimeric clones. A single chimeric clone could bring two unrelated groups of legume-specific genes together into one. Sequence alignments and BLAST analyses were used to identify the chimeric clone. A clone was considered chimeric if: (1) in a multiple sequence alignment, this single sequence was the only link joining two distinct alignments, and (2) alignments of the clone to members of one group do not overlap the sequence boundaries of alignments to members in the other group. Chimerism was confirmed in cases where Medicago genome sequence was available. The chimeric clone was removed from the alignment, and the two groups of sequences were assigned new group numbers.
Motif Analysis of Subgroups of Nodule-Specific CCPs
To create the subgroups, an initial automated alignment of all the CCPs in group 31 was performed using ClustalW. Sequences that did not fit the alignment were removed. Subgroups within the family were identified based on the automated dendrogram from ClustalW and realigned separately. HMMs were then generated for each subgroup and were used to scan MtGI 6.0. Several sequences were identified from MtGI 6.0 that had significant scores within a subgroup, but were not legume specific. In some cases, these sequences had been omitted by our original BLAST filtering procedure because of obvious chimerism with nonlegume-specific sequences. The CCP portion of these TCs was added to the alignment. Sequences within the alignments were shuffled between subgroups until the members in each subgroup scored better in the associated HMM than any sequence outside the group, and the greatest apparent separation among the subgroups was achieved. For CCP-like sequences identified from the Arabidopsis genome, intron sequences were predicted using the NetPlantGene prediction server (http://www.cbs.dtu.dk/services/NetPGene; Hebsgaard et al., 1996).
Pattern Matching Analysis for Pro-Rich Proteins
Several perl regular expression patterns were used to detect Pro-rich protein sequences among the public databases. In the first pattern, (PPV[EYV]K){3}, a triple pentamer repeat was used. In this pattern, square brackets indicate a choice for a single residue and curly braces define the number of times a pattern is repeated. This pattern was used to search Swiss-Prot/TrEMBL (Boeckmann et al., 2003) and identify proteins containing any combination of the following three subunits: PPVEK, PPVYK, or PPVVK. The nucleotide equivalent of this pattern, ((CCXCCXGTX(GTX|GA[AG]|TA[CT])|([GA]TA|[TC]TC|XAC) XACXGGXGG[TC]TT AA[AG])){3}, where X = [ACGTN] was used to search NCBI's dbEST. In this pattern, “|” denotes “or” and square brackets indicate a choice for a single nucleotide. Identified sequences from dbEST were assembled using the Sequencher software to remove redundancy. Noting the pentamer variations found in other nonlegume species, a second pattern was generated. The new pattern, (PP[ILMV][EYVHNA][KT]){3}, was also used to search Swiss-Prot/TrEMBL.
Evolutionary Analysis of CCPs
BLASTN analysis of CCPs against the available M. truncatula genome sequence was used to identify BACs containing CCPs. Identified BACs were then compared to the TIGR MTGI (v7) to get to most recent expression data. In the case of BAC Mth2-34P9, an exact match was found to TC78513, which had homology to the CCPs. The previous version of this TC (MtTC61282), which we used in our successive legume-specific BLAST searches, had been eliminated from our analysis because it did not appear to be legume specific. Comparison of MtTC61282 with BAC Mth2-34P9 revealed the TC was chimeric. The chimera was corrected in the subsequent version of MTGI. TC78513 had no homology to publicly available nonlegume sequences. Therefore, it was considered legume specific.
The program JDotter (Department of Microbiology and Biochemistry, University of Victoria, Canada; Brodie et al., 2004) was used to generate dot plots for comparing genomic sequences and identifying repeats. Evolutionary analyses of BAC sequences were performed using the Wisconsin Package version 10.3 (Accelrys, San Diego). Sequence alignments of CCPs and repeated regions were made using the programs GAP, BESTFIT, and PILEUP. Sequence similarities were determined using OLD_DISTANCES. Phylogenetic trees of the MRs were constructed using the programs PAUPSEARCH and PAUPDISPLAY. PAUPSEARCH was performed with the bootstrap analysis using branch-and-bound search and parsimony options.
Rates of synonymous and nonsynonymous substitutions of CCPs were determined using DIVERGE (Wisconsin Package). Prior to this analysis, the sequences of all four CCPs on BAC Mth2-34P9 were aligned and the insertion/intron sequence was removed from CCP4. In addition, the cleavage site for the CCP signal peptides was predicted using the program SIGNALP (Center for Biological Sequence Analysis, Denmark Technical University, Lyngby, Denmark; Nielsen et al., 1997) and removed. Two-by-two contingency tables were used to determine the statistical significance of nucleotide substitution rates. The nucleotide substitution rates were compared to those expected for neutral selection (Ka/Ks = 1; Hughes, 1999).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AAC61879, Q9ZQX0, CAB51773, S66340, At4g12560, At4g22390, At1g20790, At5g18160, At1g15680, CAB96471, CAB96474, CAB96476, BAB40944, AAB23537, At1g43720, At5g63660, BAA04219, AC121238, and AC138527.
Supplementary Material
Acknowledgments
The authors thank Alan Kilian from TimeLogic and the Center for Computational Genomics and Bioinformatics at the University of Minnesota for allowing access to the DeCypher Bioinformatics Accelerator and for assistance in running analyses. Thanks to Michael Steinbach, Levent Ertoz, and Vipin Kumar from the Department of Computer Science at the University of Minnesota for discussions on sequence clustering.
This work was supported by the National Science Foundation Plant Genome Research Program (award on Medicago truncatula genomics; grant no. 0110206 to D.R.C.) and by the University of Minnesota College of Biological Sciences.
The online version of this article contains Web-only data.
References
- Albrecht C, Geurts R, Bisseling T (1999) Legume nodulation and mycorrhizae formation; two extremes in host specificity meet. EMBO J 18: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MS, Cabral KM, Zingali RB, Kurtenbach E (2000) Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch Biochem Biophys 378: 278–286 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) Generation of 7137 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res 7: 127–130 [DOI] [PubMed] [Google Scholar]
- Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al (2003) PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res 31: 400–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW, et al (2001) The Medicago Genome Initiative: a model legume database. Nucleic Acids Res 29: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Segura A, Moreno M, Lopez G, Garcia-Olmedo F, Molina A (2002) Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol 128: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Mayer K, White O, Eisen JA, Preuss D, Bureau T, Salzberg SL, Mewes HW (2001) Sequence and analysis of the Arabidopsis genome. Curr Opin Plant Biol 4: 105–110 [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman HG (2003) Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254: 197–215 [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie R, Roper RL, Upton C (2004) JDotter: A Java interface to multiple dotplots generated by Dotter. Bioinformatics 20: 279–281 [DOI] [PubMed] [Google Scholar]
- Cabral KM, Almeida MS, Valente AP, Almeida FC, Kurtenbach E (2003) Production of the active antifungal Pisum sativum defensin 1 (Psd1) in Pichia pastoris: overcoming the inefficiency of the STE13 protease. Protein Expr Purif 31: 115–122 [DOI] [PubMed] [Google Scholar]
- Chen KC, Lin CY, Kuan CC, Sung HY, Chen CS (2002) A novel defensin encoded by a mungbean cDNA exhibits insecticidal activity against bruchid. J Agric Food Chem 50: 7258–7263 [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle S, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Corpet F, Gouzy J, Kahn D (1999) Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res 27: 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore JV, Ranjeva R, Bono JJ (2001) Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci 6: 24–30 [DOI] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99: 10494–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Schmidt A, Marcus A (1989) Characterization of two soybean repetitive proline-rich proteins and a cognate cDNA from germinated axes. Plant Cell 1: 945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Sumner LW (2003) Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol 131: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie JA, Walker SA (1999) Plant responses to nodulation factors. Curr Opin Plant Biol 2: 483–489 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg: legume diversity and evolution in a phylogenetic context. Plant Physiol 131: 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R, Eddy SR, Krogh A, Mitchison G (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, UK
- Endo M, Kokubun T, Takahata Y, Higashitani A, Tabata S, Watanabe M (2000) Analysis of expressed sequence tags of flower buds in Lotus japonicus. DNA Res 7: 213–216 [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueauf JB, Dolata M, Leykam JF, Lloyd EA, Gonzales M, VandenBosch K, Kieliszewski MJ (2000) Peptides isolated from cell walls of Medicago truncatula nodules and uninfected root. Phytochemistry 55: 429–438 [DOI] [PubMed] [Google Scholar]
- Frühling M, Albus U, Hohnjec N, Geise G, Pühler A, Perlick AM (2000) A small gene family of broad bean codes for late nodulins containing conserved cysteine clusters. Plant Sci 152: 67–77 [Google Scholar]
- Gage DJ, Margolin W (2000) Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3: 613–617 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293: 668–672 [DOI] [PubMed] [Google Scholar]
- Ganz T (2003) Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 3: 710–720 [DOI] [PubMed] [Google Scholar]
- Gao AG, Hakimi SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CM (2000) Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat Biotechnol 18: 1307–1310 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Graham MA, Silverstein KA, Cannon SB, VandenBosch KA (2004) Computational identification of legume-specific genes. In JP Gustafson, J Snape, RC Shoemaker, eds, Genome Exploitation: Data Mining (Proceedings of the 23rd Stadler Genetics Symposium, March 31–April 2, 2003, University of Missouri, Columbia, MO). Kluwer Academic/Plenum Press, New York (in press)
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györgyey J, Vaubert D, Jimenez-Zurdo JI, Charon C, Troussard L, Kondorosi A, Kondorosi E (2000) Analysis of Medicago truncatula nodule expressed sequence tags. Mol Plant Microbe Interact 13: 62–71 [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O (2003) The TIGRFAMs database of protein families. Nucleic Acids Res 31: 371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24: 3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JC, Nagao RT, Key JL (1987) Characterization and sequence analysis of a developmentally regulated putative cell wall protein gene isolated from soybean. J Biol Chem 262: 8367–8376 [PubMed] [Google Scholar]
- Hong JC, Nagao RT, Key JL (1990) Characterization of a proline-rich cell wall protein gene family of soybean. A comparative analysis. J Biol Chem 265: 2470–2475 [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1999) Adaptive Evolution of Genes and Genomes. Oxford University Press, New York
- Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39: 285–312 [DOI] [PubMed] [Google Scholar]
- Journet E, van Tuinen D, Gouzy J, Crespeau H, Carraeau V, Farmer M, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaijalainen S, Schroda M, Lindström K (2002) Cloning of nodule-specific cDNAs of Galega orientalis. Physiol Plant 114: 588–593 [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Nirasawa S, Saitoh H, Ito M, Nishihara M, Terauchi R, Nakamura I (2002) Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe grisea) in transgenic rice. Theor Appl Genet 105: 809–814 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DT (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M (2000) The F-Box Protein Family. Genome Biol 1: REVIEWS3002. Epub 2000 Nov 10 [DOI] [PMC free article] [PubMed]
- Koike M, Okamoto T, Tsuda S, Imai R (2002) A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem Biophys Res Commun 298: 46–53 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Lamblin AF, Crow JA, Johnson JE, Silverstein KA, Kunau TM, Kilian A, Benz D, Stromvik M, Endre G, VandenBosch KA, et al (2003) MtDB: a database for personalized data mining of the model legume Medicago truncatula transcriptome. Nucleic Acids Res 31: 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay FT, Brugliera F, Anderson MA (2003) Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol 131: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30: 242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzmeier R, Ho CH, Hoang BV, Ganz T (1999) A 450-kb contig of defensin genes on human chromosome 8p23. Gene 233: 205–211 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BP, Broekaert WF (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Maxwell AI, Morrison GM, Dorin JR (2003) Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol Immunol 40: 413–421 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A (2002) Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci 7: 440–444 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Long SR (2004) Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the medicago truncatula/sinorhizobium meliloti symbiosis. Plant Physiol 134: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Park HC, Kang YH, Chun HJ, Koo JC, Cheong YH, Kim CY, Kim MC, Chung WS, Kim JC, Yoo JH, et al (2002) Characterization of a stamen-specific cDNA encoding a novel plant defensin in Chinese cabbage. Plant Mol Biol 50: 59–69 [DOI] [PubMed] [Google Scholar]
- Parniske M, Downie JA (2003) Plant biology: locks, keys and symbioses. Nature 425: 569–570 [DOI] [PubMed] [Google Scholar]
- Poulsen C, Podenphant L (2002) Expressed sequence tags from roots and nodule primordia of Lotus japonicus infected with Mesorhizobium loti. Mol Plant Microbe Interact 15: 376–379 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, van Engelen F, van der Knaap E, van de Wiel C, van Kammen A, Bisseling T (1990) Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell 2: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert KR (1986) Products of biological nitrogen fixation in higher plants: synthesis, transport and metabolism. Annu Rev Plant Physiol 37: 539–574 [Google Scholar]
- Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, Casavant TL, McCray PB Jr (2002) Discovery of five conserved beta -defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA 99: 2129–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple CA, Rolfe M, Dorin JR (2003) Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol 4: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Keim P, Vodkin L, Retzel E, Clifton SW, Waterston R, Smoller D, Coryell V, Khanna A, Erpelding J, et al (2002) A compilation of soybean ESTs: generation and analysis. Genome 45: 329–338 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG (1995) Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA 92: 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Amyot L (2003) Symbiosis, inventiveness by recruitment? Plant Physiol 131: 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J, et al (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7: 573–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Stacey GA (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW (1999) A family of mammalian F-box proteins. Curr Biol 9: 1180–1182 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR (2003) The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31: 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zhang MQ (2002) Computational prediction of eukaryotic protein-coding genes. Nat Rev Genet 3: 698–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.