Abstract
DNA polymorphism is the basis to develop molecular markers that are widely used in genetic mapping today. A genome-wide rice (Oryza sativa) DNA polymorphism database has been constructed in this work using the genomes of Nipponbare, a cultivar of japonica, and 93-11, a cultivar of indica. This database contains 1,703,176 single nucleotide polymorphisms (SNPs) and 479,406 Insertion/Deletions (InDels), approximately one SNP every 268 bp and one InDel every 953 bp in rice genome. Both SNPs and InDels in the database were experimentally validated. Of 109 randomly selected SNPs, 107 SNPs (98.2%) are accurate. PCR analysis indicated that 90% (97 of 108) of InDels in the database could be used as molecular markers, and 68% to 89% of the 97 InDel markers have polymorphisms between other indica cultivars (Guang-lu-ai 4 and Long-te-pu B) and japonica cultivars (Zhong-hua 11 and 9522). This suggests that this database can be used not only for Nipponbare and 93-11, but also for other japonica and indica cultivars. While validating InDel polymorphisms in the database, a set of InDel markers with each chromosome 3 to 5 marker was developed. These markers are inexpensive and easy to use, and can be used for any combination of japonica and indica cultivars used in this work. This rice DNA polymorphism database will be a valuable resource and important tool for map-based cloning of rice gene, as well as in other various research on rice (http://shenghuan.shnu.edu.cn/ricemarker).
Molecular markers are valuable tools in both basic and applied research, such as fingerprinting genotypes, analyzing genetic diversity, determining variety identity, marker-assisted breeding, phylogenetic analysis, and map-based cloning of genes (McCouch et al., 1997; Joshi et al., 2001; Nagaraju et al., 2002; Ni et al., 2002;). In rice (Oryza sativa), more than 10,000 molecular markers have been developed (Saito et al., 1991; Causse et al., 1994; Kurata et al., 1994; Harushima et al., 1998; Wu et al., 2002). Most of them are expressed sequence tags and RFLPs. In addition to expressed sequence tags and RFLPs, simple sequence repeat (SSR) markers and single nucleotide polymorphism (SNP) markers have been developed in recent years. Approximately 2,740 SSR markers have been genetically mapped in rice, about one SSR marker every 157 kb (Chen et al., 1997; Temnykh et al., 2000, 2001; McCouch et al., 2002). The use of SNPs in various rice cultivars is increasing and becoming more significant. A total of 213 SNP markers have been established in rice (Nasu et al., 2002).
The use of rice genetics has become increasingly popular as a model system for the study in monocotyledonous plants. Map-based cloning is one of the most important gene cloning methods used in rice, and it is becoming a common tool in rice molecular genetics. In Arabidopsis, the first flowering plant for which a genome has been completely sequenced, a genome-wide DNA polymorphism database has been constructed (Jander et al., 2002). It allows the development of high-density markers and enables the mapping of a gene to a region as small as 0.16 cM (approximately 40 kb) or less (Lukowitz et al., 2000). Thus, map-based cloning of Arabidopsis genes just requires genetic mapping of a target gene using molecular markers currently available or developed from the genome-wide DNA polymorphism database (Jander et al., 2002) and genetic complementation. Compared with Arabidopsis, the main difficulty in map-based cloning in rice is to narrow a gene in a small region so that a candidate gene can be identified by sequencing and genetic complementation. Relatively low density of molecular markers in rice is available. In addition, some of molecular markers, including RFLP markers in the current rice map, need to be detected by radiolabeling and hybridization, which are expensive and inefficient. Generally, fine-scale mapping in rice narrows down the target interval to about 1 or more cM by using the existing marker map (Yang et al., 2002; Albar et al., 2003; Blair et al., 2003; Wang et al., 2003). Although some rice genes have been cloned using map-based cloning strategy (Ashikari et al., 1999; Yano et al., 2000; Takahashi et al., 2001; Kojima et al., 2002; Yamanouchi et al., 2002; Li et al., 2003), extensive molecular markers, including RFLP and cleaved amplified polymorphic sequence markers, have to be developed to further narrow the target gene in a small genomic region. Thus, a genome-wide map with abundant highly efficient molecular markers will largely facilitate the map-based cloning in rice.
Japonica cv Nipponbare was chosen as a common template for complete genome sequencing by the International Rice Genome Sequencing Project (IRGSP), which used a map-based clone-by-clone shotgun strategy (Sasaki and Burr, 2000; http://rgp.dna.affrc.go.jp/IRGSP/index.html). The genomic sequence was announced to be finished on December 18, 2002, and sequence analysis of chromosomes 1, 4, and 10 was also published (Feng et al., 2002; Sasaki et al., 2002; The Rice Chromosome 10 Sequencing Consortium, 2003). Some indica cultivars were also chosen for genome sequencing. The indica cv 93-11 has been sequenced using the whole-genome shotgun sequencing approach, and 409.76-Mb nonredundant draft sequences have been reported (Yu et al., 2002; http://btn.genomics.org.cn/rice/). As for another typical indica cultivar, Guang-lu-ai 4 (GLA4), about 8.4 Mb of chromosome 4 have been sequenced, and 2.3 Mb of them released (Zhao et al., 2002; http://www.ncgr.ac.cn/english/ep2-i.htm).
The emergence of rice genomic sequences made it possible to construct genome-wide DNA polymorphisms that could be used as genetic markers for high-resolution genetic analysis. Previous studies compared the genome sequences between various rice cultivars. A 493-kb sequence of Nipponbare chromosome 1 and an entire length of chromosome 4 were compared with the indica 93-11 draft sequences. The sequence of 93-11 covered 78% of 493 kb in Nipponbare and 75.42% of chromosome 4, respectively (Feng et al., 2002; Sasaki et al., 2002). In chromosome 10, 96% of a given 1-Mb Nipponbare region was covered by 93-11 sequences (The Rice Chromosome 10 Sequencing Consortium, 2003). Feng et al. (2002) also aligned the sequences of 2.3 Mb of three contiguous segments of indica GLA4 chromosome 4 and 2.4 Mb of its colinear sequences from Nipponbare, and the results showed extensive sequence colinearity. Although segments of genomic sequence between indica and japonica were compared, the genome-wide DNA polymorphisms have not been reported yet.
In this article, we constructed a genome-wide DNA polymorphism database between rice subspecies Nipponbare and 93-11, based on publicly available genomic sequence information, analyzed the database including SNP and Insertion-Deletion (InDel) frequencies and distribution, performed an InDel comparison between Nipponbare/93-11 and Nipponbare/GLA4, and surveyed the association of existing SSRs and InDel polymorphisms in our database. We experimentally validated some InDel polymorphisms, developed a set of InDel markers, and analyzed their applicability for the mapping of other indica/japonica. The DNA polymorphism database and the set of InDel markers developed based on the database will be very useful for rice research community.
RESULTS
Orthologous Region Identification
In order to construct a DNA polymorphism database, genome-wide orthologous regions between rice japonica cv Nipponbare and indica cv 93-11 were identified by the strategy described below. Nipponbare was sequenced by a clone-by-clone shotgun strategy (Sasaki and Burr, 2000) and served as a reference genome. As of September 9, 2003, we downloaded a total of 3,348 bacterial artificial chromosome (BAC)/P1-derived artificial chromosome clones with total size of 457 Mb. Indica cv 93-11 was sequenced using whole-genome shotgun strategy (Yu et al., 2002). A total of 127,551 contigs with a total size of 352 Mb were downloaded. In order to anchor segment of 93-11 contigs to BAC clones of Nipponbare, we used contigs as queries to BLASTN the Nipponbare genomic sequence. In order to correctly anchor contigs of 93-11 onto the Nipponbare genomic sequence, a threshold e-value of 10−100 was applied. Each contig was then anchored to a BAC clone based on the BLAST result. In this case, 37,449 contigs were filtered out by no hits with the BAC clones. The sizes of these contigs are relatively small. Deleted contigs account for 29% of the total number of 93-11 contigs, whereas they only account for about 11.1% of the total size of the genomic sequence of 93-11.
MUMmer3.0 was then used to align the contig sequences of 93-11 with their anchored BAC clones of Nipponbare. The output of the program was further transferred into the database. In some cases, the score values of the BLASTN search do not represent the similarity of an entire contig with corresponding BAC clones. In order to identify the overall similarity of a contig and its corresponding BAC clone, we designed a program to calculate the overall similarity between contigs and their anchored BAC clones and assigned the value to “Identity.” Based on Identity value, we further filtered the contigs from the database with the threshold set as shown in Table I. In this case, the sequence comparison data of 37,183 contigs were removed from the database. Thus, our database contains the sequence comparison between 52,919 contigs of 93-11 with the Nipponbare genomic sequence. These contigs account for 68% of all contig sequences of 93-11 and cover 56% of the entire rice genome.
Table I.
Threshold set to filter out contigs of 93-11 from the DNA polymorphism database
| Contig Length | Threshold of Identity | Number of Contigs Filtered |
|---|---|---|
| kb | % | |
| <2 | 90 | 27,587 |
| 2–5 | 80 | 5,855 |
| 5–10 | 75 | 2,887 |
| >10 | 50 | 854 |
| Total | − | 37,183 |
For different sizes of contigs, thresholds of genomic sequence identity from 50% to 90% were set, and contigs of the identity below the threshold were filtered out.
Analysis of DNA Polymorphism Database
Our database contains four types of polymorphism (PLM_Type) between Nipponbare and 93-11: SNP, InDel, “no extension,” and “#.” “No extension” represents the regions of no similarity, while their flanking sequences are quite similar between 93-11 and Nipponbare. There are 796 “no extension” in the database. “#” represents large chunks of rearranged, inverted, or inserted sequences. There is a total of 15,762 “#” in the database, and most of them are larger than 100 bp. Both “#” and “no extension” represent large-scale differences between these two genomic sequences.
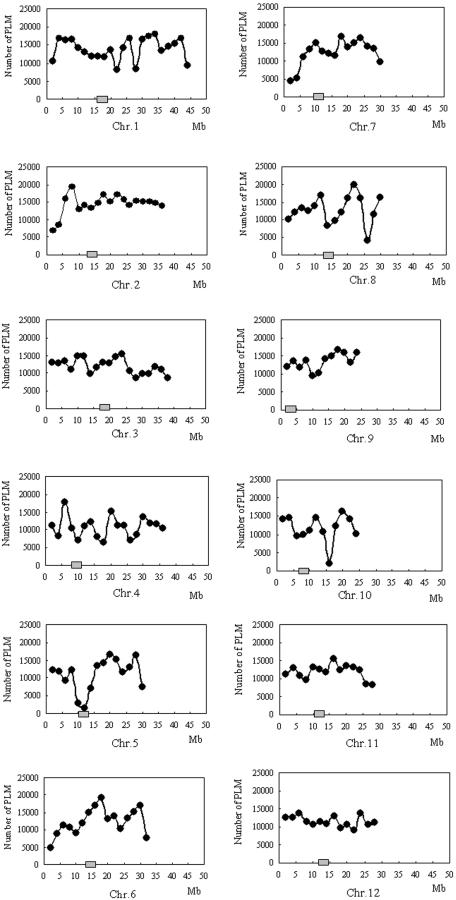
InDel represents insertion-deletion differences between two genome sequences, where one genome has an insertion of a number of nucleotides relative to the others (Jander et al., 2002). As InDel differences of larger than 100 bp are less reliable, MUMmer3.0 attributes these differences to “#.” The differences of less than 100 bp are attributed to InDel. Except for the “no extension” and “#,” our database contains 2,182,582 DNA polymorphisms, including 1,703,176 SNPs and 479,406 InDels. The frequencies of SNP and InDel polymorphisms between Nipponbare and 93-11 are 0.71% and 0.20%, respectively. The SNP frequency is comparable with that reported (0.68%–0.70%) between indica and japonica cultivars (Nasu et al., 2002). On average, the DNA polymorphism densities along the whole rice genome are one SNP every 268 bp and one InDel every 953 bp. Each Nipponbare BAC clone has an average of 652 polymorphisms. Of all 479,406 InDels, insertions in 93-11 and Nipponbare account for 47.5% and 52.5%, respectively, of all InDels, so there is no bias between Nipponbare and 93-11. The distribution of DNA polymorphisms along chromosomes was also surveyed (Fig. 1). They do not evenly distribute along each chromosome. The polymorphism densities are lower than adjacent regions in centromere of chromosomes 4, 5, 8, and 10. However, the centromere of chromosome 7 is a polymorphism-rich region.
Figure 1.
Distribution of DNA polymorphisms between Nipponbare and 93-11 of rice along each chromosome. The horizontal scale indicates the physical distance from the distal end of the short arm (on the IRGSP marker-based physical map; http://rgp.dna.affrc.go.jp/IRGSP/download.html, May 2003). The vertical scale indicates the number of polymorphisms (SNPs and InDels) at each region (2 Mb) between Nipponbare and 93-11. The gray box in each figure shows the location of the centromere.
The difference of InDel polymorphisms between Nipponbare/GLA4 and Nipponbare/93-11 was compared. Currently, there is only the 2.3-Mb GLA4 genome sequence available. We compared these sequences with the Nipponbare genome sequence and anchored them onto the Nipponbare genome. Then, we selected 318 InDels (from 1 bp to 82 bp) from our database in this region and compared them with InDels of Nipponbare/GLA4. A total of 228 (72%) InDels with the same size and 7 (2%) InDels with a different size were also found in the same locus in Nipponbare/GLA4. However, there was no InDel polymorphism between Nipponbare and GLA4 for 83 (26%) InDels found between Nipponbare and 93-11. This suggested that 74% of the InDel polymorphisms in our database could also be used for Nipponbare/GLA4 based on the InDel distribution survey described above.
The association of existing SSR markers with InDel polymorphisms in the database was analyzed. We have selected 213 SSR markers that dispersed along rice 12 chromosomes (McCouch et al., 2002). A total of 110 (52%) SSR markers were also found to be associated with the InDels in our database, and 26 (12%) SSR markers have no polymorphism between Nipponbare and 93-11. The loci of the remaining 77 markers (36%) could not be found in our database.
Experimental Validation of DNA Polymorphisms in the Database
The accuracy of both SNPs and InDels in the database was experimentally analyzed. The genome of Nipponbare was sequenced at 99.99% accuracy with each region sequenced for about 10× coverage (http://rgp.dna.affrc.go.jp). However, the genome sequence of 93-11 used in this work was a draft sequence. The accuracy of SNPs in our database was analyzed by resequencing some regions of the 93-11 genome. A total of 109 predicted SNPs in our database were chosen. PCR amplification and sequence analysis indicated that 107 SNPs are accurate, which can be converted into molecular markers. However, there are no polymorphisms for the remaining two predicted SNPs in our database because of sequence errors of 93-11 genome. Based on this data, the SNP accuracy of our database is about 98.2%.
The InDel accuracy of our database was also experimentally validated. A total of 108 primer pairs targeting some InDel polymorphisms in the database were designed. These primer pairs dispersed along all 12 chromosomes. The expected PCR products were about 120 to 250 bp, and the size differences of PCR products between japonica and indica were about 25 to 50 bp. PCR analysis indicated that 97 (90%) primer pairs gave reliable amplification using genomic DNA of Nipponbare and 93-11 as templates, and PCR products could be separated in agarose gel. There were no PCR products for three primer pairs, and there were no polymorphisms between Nipponbare and 93-11 for the remaining eight primer pairs. Based on these data, 90% of the InDel polymorphisms in this database could be transferred into InDel markers.
We used the 97 InDel markers developed above to further analyze polymorphisms between other japonica and indica cultivars. A total of four rice cultivars, including two japonica cultivars, 9522 and Zhong-hua 11 (ZH11), and two indica cultivars, GLA4 and Long-te-pu B (LTPB), were used. PCR analysis indicated that 68% to 89% of the 97 InDel markers also had polymorphisms for any other combinations of japonica and indica cultivars shown in Table II. Of all 97 InDel markers, 55 markers had polymorphisms for any combination of japonica (Nipponbare, ZH11, and 9522) and indica (93-11, LTPB, and GLA4) cultivars. The information for 50 of these markers was listed in Table III. These markers distributed along 12 rice chromosomes with each chromosome 3 to 5 marker.
Table II.
The number of InDel markers that could be used for different indica (93-11, GLA4, LTPB)/japonica (Nipponbare, 9522, ZH11) cultivars from 97 InDel markers developed for Nipponbare/93-11
| 93-11 | GLA4 | LTPB | |
|---|---|---|---|
| Nipponbare | 97 | 77 (79a) | 75 (77) |
| 9522 | 84 (87) | 70 (72) | 66 (68) |
| ZH11 | 86 (89) | 69 (71) | 70 (72) |
Percentage of InDel markers that could be used for this specific combination.
Table III.
The detail information of 50 InDel markers developed in this work for any combinations of japonica (Nipponbare, ZH11, and 9522) and indica (93-11, GLA4, and LTPB)
| Marker Name | Chromosome | BAC Accession | Positiona | Forward Primer (5′ 3′) | Reverse Primer (5′ 3′) | Expected PCR Product Size
|
|
|---|---|---|---|---|---|---|---|
| Nipponbare | 93-11 | ||||||
| R1M7 | 1 | AP002482 | 10,700 | ATTCCTGGTTCTACATTACTTA | CGCCTCACTAGAATATCGGA | 191 | 154 |
| R1M20 | 1 | AP004222 | 18,910 | TTGGAACAGGGAAGAAGC | AGGACATAGTTGTAATGGGTAG | 263 | 221 |
| R1M30 | 1 | AP002843 | 25,420 | AAGGGGCCCTAATTTATCTAG | TGTTTACTTTGTTCTTGGACTG | 246 | 197 |
| R1M37 | 1 | AP003453 | 30,500 | ATAGTTCGCCATCGTCAT | ACACGCCATAGCAAGGAA | 159 | 212 |
| R1M47 | 1 | AP003442 | 37,470 | AATAGAATTACTGATGAAACCTTA | GCCCGTTACCGCTTATGT | 159 | 108 |
| R2M10 | 2 | AP005394 | 6,610 | CCCAGTCTGCTGCCATCT | GAATGTATTTCAGTTCCAGTAAG | 134 | 182 |
| R2M24 | 2 | AP005414 | 11,340 | GGGCAACAACGGCTCTG | AGGGAATAAGGCGATACGG | 162 | 131 |
| R2M26 | 2 | AP005696 | 15,320 | GCAGCAAAGTGCGGAGTA | CAGGTGAATTGCCAATTT | 143 | 181 |
| R2M37 | 2 | AP004124 | 23,920 | ACTGTTACCCAAACGCTA | ACGTGCACCTACTACAGAAA | 212 | 147 |
| R2M50 | 2 | AP004888 | 30,870 | CCTGAAGGAAATGATAGCAATAG | GTTTTGTATGCTCTTCACTTGTC | 212 | 254 |
| R3M10 | 3 | AC105732 | 6,370 | CCGAGTACCATTGCTTTC | CTGCCATAGTTACTGCTCTGTT | 190 | 227 |
| R3M23 | 3 | AC099323 | 16,140 | TGCTTACAAGGGTCCAAT | GGAGGTGCCTACCAAGAG | 149 | 185 |
| R3M30 | 3 | AC091234 | 20,180 | AGGCTAAGTGAAGAAATAATAAG | CTCCGTATTCATTACTGGTTG | 175 | 199 |
| R3M37 | 3 | AC133930 | 27,130 | GCATTGAATTGTACTCTTATTATAT | ACGAATCAAAAGGAGACTAAAAT | 186 | 242 |
| R3M53 | 3 | AC091123 | 33,240 | ACACTGGCTACGGCAAAG | TTTGTTCGGGAATAATGATGC | 204 | 169 |
| R4M13 | 4 | AL606597 | 8,150 | TACACGGTAGACATCCAACA | ATGATTTAACCGTAGATTGG | 169 | 201 |
| R4M17 | 4 | AL731585 | 11,770 | AGTGCTCGGTTTTGTTTTC | GTCAGATATAATTGATGGATGTA | 169 | 220 |
| R4M30 | 4 | AL662979 | 18,220 | GCTTCTCCTGGTTGTATGC | AAAATAGGGAGGCAGATAGAC | 173 | 133 |
| R4M43 | 4 | AL662938 | 24,650 | CTTGAACCTGAGTGAGTGG | CGATGAAAATGATGTCTA | 160 | 194 |
| R4M50 | 4 | AL606639 | 29,080 | TTTTGTGAAACTTGACCCTC | GCGTCCATGTCTTTATTGTG | 132 | 165 |
| R5M13 | 5 | AC132493 | 5,820 | GAGAAAGAGTGGAAGGAG | AGTATCGTCAGGAGGGTC | 175 | 207 |
| R5M20 | 5 | AC137622 | 13,390 | CTCGCTGTTTACTGACTGG | TTTGATGTACTGCCTGCTCT | 175 | 214 |
| R5M30 | 5 | AC134930 | 20,980 | CTCAATTTCACCCATCCC | CGCTCCGTCTCCAACCTC | 224 | 178 |
| R5M43 | 5 | AC121365 | 25,960 | AGCGTGACTTGAGTTCCA | ATGACTTTCCCACCGTAT | 109 | 78 |
| R6M14 | 6 | AP004725 | 7,710 | AAATGTCCATGTGTTTGCTTC | CATGTGTGGAATGTGGTTG | 251 | 217 |
| R6M30 | 6 | AP005929 | 18,640 | CACAAGCCGTAGCAGAGC | TCACGAAAAAGACCCCAAG | 181 | 147 |
| R6M44 | 6 | AP005386 | 26,080 | TTAGGAATAAAGGCTGGATA | TTACCGTTAATAGGTGGAA | 156 | 122 |
| R7M7 | 7 | AP005719 | 6,350 | ACCTTCCCTCCCCTTTTGAT | AACTTGGTCTTCCTGTTTTATTG | 200 | 133 |
| R7M20 | 7 | AP004346 | 11,680 | GTTTTGTGCATTCCTTTAC | TTTATGACATTTTGACCG | 200 | 266 |
| R7M30 | 7 | AP005185 | 19,550 | ATGTCGCCTACGAGTTTTC | TTCATGTG ACCATTTGTGC | 187 | 219 |
| R7M37 | 7 | AP003747 | 23,720 | CAGCCCTAAATCTAAATACCC | ACGTTGAGACAGGCGAGC | 173 | 137 |
| R8M10 | 8 | AP004589 | 6,290 | ACCAAACAAGCCCTAGAATT | TGAGAAAGATGGCAGGACGC | 122 | 174 |
| R8M23 | 8 | AP005500 | 13,820 | CCTATTCACTCTACCGACAT | GTTTAGTTCCCATTGCTTT | 121 | 157 |
| R8M33 | 8 | AP003881 | 21,320 | CGAAAGAGGAGAGGGGTAGT | CGAAAACGAGAAACAAATA | 205 | 167 |
| R8M46 | 8 | AP004587 | 28,770 | CAGCAGAGTCCAGAGAAGAT | GCATAAGATGGCGAGTGA | 113 | 83 |
| R9M10 | 9 | AP005592 | 4,770 | CTTTGGATTCAGGGGGA | AACTTGAAACGGAGGCAG | 135 | 178 |
| R9M20 | 9 | AP005879 | 9,290 | ACTGCTTTGATGGCTTGTG | CTCCCCAAACTGAATCC | 142 | 182 |
| R9M30 | 9 | AP005397 | 14,940 | CTCACCTACCTAAAACCCAAC | CCACCCAAATCTGATACTG | 153 | 185 |
| R9M42 | 9 | AC108757 | 19,450 | CTATAAGACCAAAACGAAAACT | GAAAACCATTGTGTCACTGTA | 164 | 212 |
| R10M10 | 10 | AC098566 | 5,150 | GAATACAACCCCCTAAAAAC | ATGGACCGTTGAGGAGAC | 170 | 132 |
| R10M17 | 10 | AC090486 | 9,040 | TGAACAATAAACCACAGAAGCA | CCCTTTATTCCCTCCTTTG | 152 | 183 |
| R10M30 | 10 | AC027038 | 16,960 | CCCTAAAAATAGAGCAACCT | ACCCATAATACTACCAATCAAC | 152 | 133 |
| R10M40 | 10 | AC091122 | 19,470 | GTCCCTAGGCCATCTCTTG | GCGAATAGGGGTGGACAG | 166 | 133 |
| R11M17 | 11 | AC137589 | 14,370 | TGAGACGTTTGGGAGCAT | CGATCAGCAGCAACAGGT | 183 | 131 |
| R11M23 | 11 | AC134053 | 19,180 | AAGGTTGACAAGGACAGAAG | TCGCAGGAATGGATAAAA | 212 | 254 |
| R11M40 | 11 | AC125780 | 23,540 | AAGAAAAATATCTATTGAGGAGTG | GGAGGACCATAAATGACGG | 178 | 137 |
| R12M10 | 12 | AL954158 | 4,300 | ATCATTTCAGCCTGTGCC | AGCTTAATAGGGGGGACG | 214 | 261 |
| R12M27 | 12 | AL713927 | 17,370 | ATTTCATTGCCATCAGTT | GTAATCTTCTATCCGTTCA | 155 | 188 |
| R12M33 | 12 | AL731888 | 20,080 | TTGATGATAGTATTTGCTGATG | AGATAGTGTCGGCGGTGG | 208 | 250 |
| R12M43 | 12 | BX000457 | 26,290 | CCGCCGAGAAGAAACAAAG | CCCAAGAACAGGATTACA | 193 | 163 |
The marker location was calculated based on the IRGSP marker-based physical map, May 2003 (http://rgp.dna.affrc.go.jp/IRGSP/download.html).
DISCUSSION
The establishment of extensive genomic colinearity between the 127,551 contigs of 93-11 and reference genome Nipponbare was the critical step in constructing the DNA polymorphism database. We anchored contigs of 93-11 onto BAC clones of Nipponbare based on the BLASTN search. In most cases, contig sequence of 93-11 had high sequence similarity with only one region in the BAC clone of Nipponbare, and the colinearity can be identified without any problems. However, some contigs of 93-11 can align to several regions in the Nipponbare genome. Repetitive sequence in contig sequence is the major cause for such alignment. In this case, we just anchored these contigs onto the regions with the highest similarity based on the BLASTN search. In order to further improve the accuracy of our database, stringent selection criteria were applied to anchor the contig sequence of 93-11 to Nipponbare BAC clones. The first criterion was the threshold e-value of the BLASTN search, and the next criterion was the identity. After these selections, a total of 74,632 contigs were filtered out, and most of them are quite small. There were only 52,919 contigs left; however, they accounted for 68% of the total contig sequence and made up of 56% of the rice genome. Our database contains 1,703,176 SNPs and 479,406 InDels corresponding to one SNP every 268 bp and one InDel every 953 bp along the rice genome. Comparing it with the Arabidopsis polymorphisms database, the density of SNP and InDel in rice polymorphism is approximately 10 times denser (Jander et al., 2002). Thus, this database should meet the need of fine-scale genetic mapping in map-based cloning of rice genes.
The format of the rice DNA polymorphism database constructed in this work is essentially the same as that of Arabidopsis, the first and only genome-wide DNA polymorphism database of higher plants currently available (Jander et al., 2002). However, some improvements have been made. The Arabidopsis polymorphism database was constructed using MUMmer1.0. This program classified the differences between two genomes into SNPs and InDels. In the released Arabidopsis polymorphism database, InDels larger than 100 bp were omitted due to a high level of false positives mainly caused by repetitive sequence (README of Large InDels, http://www.arabidopsis.org/Cereon/index.jsp). In this work, a recent version of the program (MUMmer3.0) was used. MUMmer3.0 assigned the regions of no similarity between two cultivars (both regions cannot be aligned instead of one region in InDel polymorphisms), while their flanking sequences were quite similar to “no extension,” and large chunks of rearranged, inverted, or inserted sequence to “#.” This should improve the reliability of DNA polymorphisms in the database. On the other hand, the further investigation of “no extension” and “#” might provide insight into genome evolution. Another improvement is the “Identity” added in the rice database. The value of “Identity” was used not only to filter contigs with low similarity to Nipponbare BAC clones, but also as a reference to design primer pairs to develop molecular markers based on the polymorphisms in the database. In the rice polymorphism database, the information of 93-11 contig sequences, including “Direction,” “Contig_Name,” and “Contig_Length,” were also added. We did not assign names to SNPs and InDels so that this database can be updated when improvements are made in the future.
The draft sequence of 93-11 was used in this work. It is likely to include some errors (Han and Xue, 2003). While experimentally validating SNPs in our database, we also found sequence errors of draft sequence. However, 98.2% of SNPs can be converted into molecular markers when a total of 109 SNPs in the database are chosen for validation. Thus, we believe that the SNPs in our database can be used to develop molecular markers.
For most laboratories, InDel polymorphisms, including SSRs, might be the first choice to be used to develop molecular markers. Thus, a detailed analysis of the InDel polymorphisms in the database was performed, including InDel frequency, the survey of existing SSR markers, and the InDel survey between Nipponbare/93-11 and Nipponbare/GLA4. We also experimentally validated the InDel polymorphism between 93-11 and Nipponbare and found that about 90% of them could be transferred to InDel markers. All these suggested that the DNA polymorphism database could be quite useful for genetic mapping between Nipponbare and 93-11. We also used the 97 InDel markers to genotype other rice cultivars (Table II) and found that 68% to 89% of them can also be used for any combinations of japonica (Nipponbare, 9522, and ZH11) and indica (93-11, GLA4, and LTPB) cultivars. This suggested that this DNA polymorphism database is not only useful for genetic mapping of Nipponbare and 93-11, but also applicable for other combinations of japonica and indica cultivars. In order to facilitate rice research, this database can be downloaded and distributed without any limitations.
As a result of InDel polymorphism validation, we established a set of InDel markers that could be used for any combinations of japonica (Nipponbare, 9522, and ZH11) and indica (93-11, GLA4, and LTPB) cultivars. These markers distribute along all 12 chromosomes with each chromosome 3 to 5 marker. As the PCR products of these markers were about 120 to 200 bp and size differences of PCR products for each InDel marker were about 20 to 40 bp, they can be analyzed on agarose gels. Thus, these markers are easy to use and inexpensive. They might be used for some other combinations of japonica and indica cultivars.
MATERIALS AND METHODS
The genomic sequences of japonica cv Nipponbare and indica cv GLA4 and 93-11 were used for sequence comparisons in this work. They were downloaded from http://rgp.dna.affrc.go.jp/IRGSP/index.html for Nipponbare, http://btn.genomics.org.cn/rice/ for 93-11, and http://www.ncgr.ac.cn/english/ep2-i.htm for GLA4. As of September 9, 2003, 3,348 P1-derived artificial chromosome /BAC clones with a total size of 457 Mb of Nipponbare genomic sequence were downloaded. The genomic sequences of 93-11 were sequenced using a whole-genome shotgun approach and 127,551 contigs, totaling 352 Mb, were downloaded. The genomic sequence of GLA4 we downloaded contained 2.3 Mb of three contiguous segments of chromosome 4.
Polymorphism Detection between Rice Species
In order to detect DNA polymorphisms between rice (Oryza sativa) cv Nipponbare and 93-11, the contig sequences of 93-11 were first used as queries to BLASTN (stand-alone BLAST, ftp://ftp.ncbi.nih.gov/blast/executables) Nipponbare genomic sequences. Contigs of 93-11 were then anchored onto BAC clones of the Nipponbare genomic sequence with the highest score of BLASTN. Then, a simple Visual Basic 6.0 script was developed to automatically group the contigs to their corresponding BACs. MUMmer3.0 (ftp://ftp.tigr.org/pub/software/MUMmer/; Delcher et al., 2002) was used to align the contig sequences and BAC clones. Finally, a set of Visual Basic scripts was developed to calculate the overall sequence similarity between contigs and BAC clones and assign the data to “Identity” while transforming the result of MUMmer3.0 to our database. After contigs with low similarity with BAC clones were removed from the database, the DNA polymorphism data were sorted based on the location of BAC clones in chromosomes.
The DNA polymorphisms between GLA4 and Nipponbare were detected by the same method described above. In order to compare DNA polymorphisms between Nipponbare/93-11 and Nipponbare/GLA4, a total of 318 InDels (from 1 bp to 82 bp) from Nipponbare/GLA4 were selected and used to compare InDel polymorphisms between Nipponbare and 93-11. Also, a total of 227 SSR markers distributed along the 12 chromosomes (http://www.dna-res.kazusa.or.jp/9/6/05/spl_figure1/fig1.pdf; www.gramene.org) were chosen to survey the association of these SSRs with InDels in the database.
Software Used or Developed in This Study
A set of simple Visual Basic 6.0 scripts was developed to construct the rice DNA polymorphism database. “Datacollect” was used to anchor the contigs of 93-11 onto BAC clones with the highest score value of BLAST. “Contigcombine” was then used to group all contigs anchored to the same BAC clone together. After MUMmer3.0 analysis, “Mumtransform” transformed the result of MUMmer3.0 to our database. Meanwhile, it calculated the overall similarity between contigs of 93-11 and BAC clones and assigned the data to “Identity” in the database. “Bacorder” was applied to sort DNA polymorphisms in the database based on their physical locations in the chromosomes. Restriction sites of SNP polymorphisms were analyzed by the Remap application provided in the EMBOSS software package (Rice et al., 2000).
Experimental Validation of DNA Polymorphisms
Three japonica cultivars (Nipponbare, 9522, and ZH11), and three indica cultivars (93-11, GLA4, and LTPB) were used for InDel polymorphism validation. Genomic DNA was extracted from leaf issues as described (Murray and Thompson, 1980). A total of 108 primer pairs were designed targeting InDel polymorphisms in the database using PRIMER 5.0. These primer pairs evenly distributed along 12 rice chromosomes. The expected PCR products of these primer pairs were about 25 to 50 bp difference for Nipponbare and 93-11. PCR was performed in a 30-μL volume containing 20 ng of genomic DNA, 0.5 μm of each primer, 200 μm dNTP, 10 mm Tris-Cl, 1.5 mm MgCl2, 50 mm KCl2, and 1 unit of Taq polymerase. An MJ Research PTC-100TM was used for amplification with the following PCR profile: 94°C for 5 min, followed by 45 cycles of 94°C for 1 min, 52°C for 1 min, 72°C for 30 s, and a final extension at 72°C for 5 min. PCR products were analyzed in 3.5% agarose gel in Tris-acetate EDTA buffer (Sambrook and Russell, 2001).
The accuracy of SNPs in the database was analyzed by resequencing some regions of 93-11 genome. A total of 22 primer pairs distributed along the 12 rice chromosomes were designed based on the 93-11 genomic sequence. PCR was performed by the standard procedures described above. TaKaRa LA Taq (code no. DRR200A) was used instead of standard Taq polymerase to reduce the errors in amplification. PCR products were sequenced on Applied Biosystems (Foster City, CA) ABI Prism 377 sequencers.
Downloadable Information Resource
The rice DNA polymorphism database constructed in this work can be downloaded on our laboratory Web site (http://shenghuan.shnu.edu.cn/ricemarker) without any limitations.
Supplementary Material
Acknowledgments
We thank Prof. Xia Huijun from Wuhan University for providing rice seed.
This work was supported by the Shu-Guang Program of the Shanghai Education and Development Board (grant to Z.-N.Y.), by the Shanghai Education Committee (grant to Z.-N.Y.), and by the Shanghai Science and Technology Committee (grant no. 03DJ14015 to Z.-N.Y.).
The online version of this article contains Web-only data.
References
- Albar L, Ndjiondjop MN, Esshak Z, Berger A, Pinel A, Jones M, Fargette D, Ghesquiere A (2003) Fine genetic mapping of a gene required for rice yellow mottle virus cell-to-cell movement. Theor Appl Genet 107: 371–378 [DOI] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair MW, Garris AJ, Iyer AS, Chapman B, Kresovich S, McCouch SR (2003) High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.). Theor Appl Genet 107: 62–73 [DOI] [PubMed] [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, et al (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR (1997) Development of a microsatellite framework map providing genome wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95: 553–567 [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL (2002) Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30: 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Han B, Xue YB (2003) Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol 6: 134–138 [DOI] [PubMed] [Google Scholar]
- Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SP, Bhave SG, Chowdari KV, Apte GS, Dhonukshe BL, Lalitha K, Ranjekar PK, Gupta VS (2001) Use of DNA markers in prediction of hybrid performance and heterosis for a three-line hybrid system in rice. Biochem Genet 39: 179–200 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin SY, et al (1994) A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet 8: 365–372 [DOI] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, et al (2003) Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol 35: 89–99 [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraju J, Kathirvel M, Kumar RR, Siddiq EA, Hasnain SE (2002) Genetic analysis of traditional and evolved Basmati and non-Basmati rice varieties by using fluorescence-based ISSR-PCR and SSR markers. Proc Natl Acad Sci USA 99: 5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu S, Suzuki J, Ohta R, Hasegawa K, Yui R, Kitazawa N, Monna L, Minobe Y (2002) Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufigogon) and establishment of SNP markers. DNA Res 9: 163–171 [DOI] [PubMed] [Google Scholar]
- Ni JJ, Colowit MP, Mackill DJ (2002) Evaluation of genetic diversity in rice subspecies using microsatellite markers. Crop Sci 42: 601–607 [Google Scholar]
- The Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566–1569 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Saito A, Yano M, Kishimoto N, Nakagahra M, Yoshimura A, Saito K, Kuhara S, Ukai Y, Kawase M, Nagamine T, et al (1991) Linkage map of restriction fragment length polymorphism loci in rice. Jpn J Breed 41: 665–670 [Google Scholar]
- Sasaki T, Burr B (2000) International rice genome sequencing project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3: 138–141 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc Natl Acad Sci USA 98: 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100: 697–712 [Google Scholar]
- Wang YG, Xing QH, Deng QY, Liang FS, Yuan LP, Weng ML, Wang B (2003) Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor Appl Genet 107: 917–921 [DOI] [PubMed] [Google Scholar]
- Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J, et al (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14: 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K (2002) A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci USA 99: 7530–7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ren X, Weng Q, Zhu L, He G (2002) Molecular mapping and genetic analysis of a rice brown planthopper (Nilaparvata lugens Stal) resistance gene. Hereditas 136: 39–43 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. Indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Cheng Z, Chen M, Wang S, Feng Q, Huang Y, Li Y, Tang Y, Zhou B, et al (2002) A fine physical map of the rice chromosome 4. Genome Res 12: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.