Abstract
A long-term goal of Arabidopsis research is to define the minimal gene set needed to produce a viable plant with a normal phenotype under diverse conditions. This will require both forward and reverse genetics along with novel strategies to characterize multigene families and redundant biochemical pathways. Here we describe an initial dataset of 250 EMB genes required for normal embryo development in Arabidopsis. This represents the first large-scale dataset of essential genes in a flowering plant. When compared with 550 genes with other knockout phenotypes, EMB genes are enriched for basal cellular functions, deficient in transcription factors and signaling components, have fewer paralogs, and are more likely to have counterparts among essential genes of yeast (Saccharomyces cerevisiae) and worm (Caenorhabditis elegans). EMB genes also represent a valuable source of plant-specific proteins with unknown functions required for growth and development. Analyzing such unknowns is a central objective of genomics efforts worldwide. We focus here on 34 confirmed EMB genes with unknown functions, demonstrate that expression of these genes is not embryo-specific, validate a strategy for identifying interacting proteins through complementation with epitope-tagged proteins, and discuss the value of EMB genes in identifying novel proteins associated with important plant processes. Based on sequence comparison with essential genes in other model eukaryotes, we identify 244 candidate EMB genes without paralogs that represent promising targets for reverse genetics. These candidates should facilitate the recovery of additional genes required for seed development.
The systematic identification of genes with essential functions has been described for several prokaryotes, including Mycoplasma (Hutchison et al., 1999), Staphylococcus (Ji et al., 2001), Haemophilus (Akerley et al., 2002), and Bacillus (Kobayashi et al., 2003), and two model eukaryotes, Saccharomyces (Giaever et al., 2002) and Caenorhabditis (Kamath et al., 2003). Significant advances have also been reported in the identification of essential genes associated with human diseases (Jimenez-Sanchez et al., 2001). These studies have provided insights into the minimal gene set required for basic cell functions in different organisms. Missing from this work has been a comparable dataset of essential plant genes. Here we characterize an initial collection of 800 genes that give a loss-of-function phenotype in Arabidopsis. Two hundred and fifty of these genes have a phenotype in the embryo. Sixty percent of these EMB identities were derived from a forward genetic screen of T-DNA insertion lines generated at Syngenta (McElver et al., 2001) and are published here for the first time. The dataset of 550 genes with other phenotypes was assembled through extensive literature searches and requests for input from the community (Meinke et al., 2003). We assess here the proposed functions and apparent redundancy of these genes, contrast our datasets with essential genes identified in yeast (Saccharomyces cerevisiae) and worm (Caenorhabditis elegans), and focus on 34 EMB genes that encode proteins with unknown functions. We propose that establishing a dataset of genes with a loss-of-function phenotype in Arabidopsis provides a foundation for comparative studies with essential genes in other organisms, a strategy for identifying unknowns with important cellular functions, and a mechanism for determining which genes are required to produce a viable plant with a normal phenotype.
In flowering plants, essential genes may be required for gametogenesis, seed development, or post-embryonic growth and development. Determining what constitutes an essential gene is limited by the absence of a complete collection of null alleles, the variable phenotypes exhibited by loss-of-function mutants, and the ability to rescue some mutants by altering the growth conditions. Arabidopsis genes that are required for viability under normal conditions and cannot be passed to subsequent generations when disrupted are often considered to be essential. Problems arise when considering genes such as LEC1, where homozygous mutant seeds cannot survive desiccation but give rise to viable plants if germinated precociously (Meinke, 1992), and EMB genes with late terminal phenotypes, where mutant embryos may produce callus (Franzmann et al., 1989) and germinate to form abnormal seedlings (Vernon and Meinke, 1995). If we define essential as required to complete the life cycle under normal conditions, then many genes with knockout phenotypes affecting flower development would fall into the essential class despite the absence of reduced viability. We have chosen instead to focus on a more comprehensive dataset of genes that we call indispensable because they give a loss-of-function phenotype of any kind, as detected through visual inspection, cellular characterization, or biochemical analysis under standard or specialized growth conditions. We have divided these indispensable genes into two major groups: those with a phenotype in the embryo, in part because we have an ongoing interest in this developmental pathway, and those with some other phenotype. Most genes in the first group are required for viability and may therefore be considered essential in a traditional sense. These genes are most comparable to essential genes of microorganisms. Those in the second group are often not required for viability but are needed for a normal phenotype. This classification system enables comparisons to be made with datasets from C. elegans, where genes with RNAi phenotypes have been divided into three classes: nonviable, growth defective, and viable with postembryonic phenotypes (Kamath et al., 2003). Because redundant genes are often not identified through forward genetic screens, our dataset of indispensable genes represents a subset of all genes needed to maintain plant functions. Complementary methods including the analysis of multiple mutants and suppression of multigene families will be required to determine which functions are overseen by duplicated genes, redundant cell processes, and parallel developmental pathways.
The focus of Arabidopsis research initiatives worldwide has shifted in recent years from genome sequencing (Meinke et al., 1998; Arabidopsis Genome Initiative, 2000) to functional analysis of the estimated 27,000 genes (Ausubel and Benfey, 2002; Wortman et al., 2003). Forward and reverse genetic screens of large populations of insertion mutants continue to be an important component of this research (Parinov et al., 1999; Sessions et al., 2002; Alonso et al., 2003). We have previously used T-DNA insertional mutagenesis to reveal large numbers of genes required for seed development in Arabidopsis (McElver et al., 2001) and constructed a sequence-based map of genes with mutant phenotypes (Meinke et al., 2003). Embryo defectives represent the largest single group of mutants examined in Arabidopsis. Many of these mutants have been characterized in some detail without knowledge of the molecular function of the disrupted gene. Other mutants have been described at length in more than 60 research publications from at least 40 different laboratories. We present here a collection of 250 EMB genes required for normal embryo development. Although this dataset is not complete and may not approach saturation for at least another 5 years, the number of genes available at present is sufficient to warrant analysis. The comparative approach described here should also facilitate the identification of additional genes with important roles in flowering plants.
Despite recent advances in the functional genomics of model organisms, many genes remain uncharacterized and are currently annotated to encode proteins with unknown functions. These genes are both intriguing and problematic, for while they may ultimately reveal the existence of novel cellular processes and biochemical pathways, they also present significant challenges with respect to classification and experimental characterization. Several different strategies have been pursued in the analysis of these unknowns: (1) structural and informatics-based approaches designed to identify informative protein motifs and establish families of related sequences; (2) cellular approaches intended to localize proteins to specific compartments; (3) molecular approaches designed to recover interacting protein partners and identify known genes with similar patterns of expression; and (4) genetic approaches aimed at determining the consequences of disrupting normal gene function. While considerable progress has been reported in the analysis of genes with unknown functions in yeast (Marcotte et al., 1999; Michaud and Snyder, 2002; Zhou et al., 2002; Vazquez et al., 2003) and selected multicellular eukaryotes (Giot et al., 2003; Grant and Wilkinson, 2003), advances with plant systems including Arabidopsis have been more limited. Most genes with unknown functions in plants also lack orthologs in other model organisms. We describe here a collection of Arabidopsis genes with unknown functions that share one defining feature in common, their requirement for normal embryo development. We propose that by focusing on unknowns that give a mutant phenotype when disrupted by a loss-of-function mutation, significant progress can be made toward assigning functions to those genes with indispensable yet uncharacterized functions.
We have long focused on genes that give a knockout phenotype in the seed (Meinke and Sussex, 1979) and have summarized information on cloned genes and their mutant alleles in a public database (Tzafrir et al., 2003) accessible through the Internet (www.seedgenes.org). Included among the 211 EMB genes described in a recent database update were 56 genes with cellular functions classified as unknown. We decided to examine these genes in more detail as a representative and informative sample of the Arabidopsis unknowns. Our approach was fourfold: (1) develop an operational system for defining what constitutes an unknown function; (2) determine which putative EMB genes with unknown functions could be confirmed through the recovery of duplicate mutant alleles; (3) characterize the diversity of confirmed genes identified and examine their patterns of expression; and (4) explore the feasibility of using epitope-tagged proteins to complement mutant phenotypes and facilitate the identification of interacting proteins.
We divided our collection of unknowns into two major groups depending on the availability of defined Pfam motifs (Bateman et al., 2002) and informative BLASTP matches (Altschul et al., 1997). The unknown class lacks these matches whereas the uncertain class has informative motifs or matches but lacks sufficient confidence or additional information needed to associate a given protein with a specific cellular function. In contrast to knockouts in genes with defined functions known to be essential in other organisms, knockouts in genes with unknown functions provide no supporting evidence that they are essential and that the correct gene has been identified. We therefore decided to focus on those EMB genes whose identity could be confirmed. Many of these genes have apparent orthologs in other plants but lack significant matches in the sequenced genomes of non-plant model organisms. Determining the functions of these unknowns will therefore require genetic analysis in a model plant such as Arabidopsis.
RESULTS AND DISCUSSION
Diversity of EMB Genes Identified
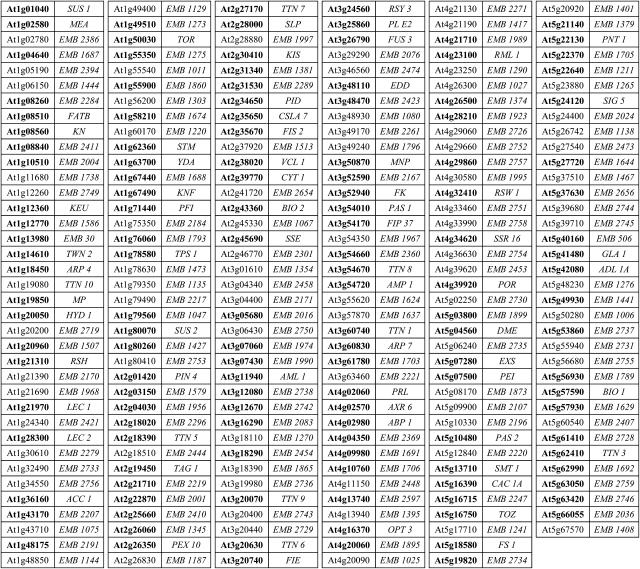
We present in Figure 1 a current list of EMB genes required for normal embryo development. Approximately 70% of these identities were obtained through forward genetics involving the Syngenta collection of seed mutants (McElver et al., 2001) and have not been published before. Confidence levels associated with gene identities are not uniform throughout the collection. Some identities have been confirmed through molecular complementation or the isolation of duplicate alleles. These represent about 60% of the total. Others are based on characterization of a single mutant allele and remain unconfirmed. Even when sequence information is available for both sides of the T-DNA insert, as with most Syngenta mutants, disruption of an adjacent gene may be responsible for the phenotype (Meierhoff et al., 2003). Some adjustments to the list of EMB genes presented here may therefore be required pending the identification of additional mutant alleles through reverse genetics.
Figure 1.
List of 220 EMB genes of Arabidopsis. Gene identities confirmed through duplicate alleles or molecular complementation are highlighted in boldface. Missing from this list are 14 confidential or questionable identities from the community and 16 identities from the Syngenta collection that are undergoing further review.
Several genes identified here are positioned adjacent to other genes with interesting knockout phenotypes. These include EMB2394 and AXR1 (Leyser et al., 1993), KEU and UVR2 (Landry et al., 1997), EMB2279 and MUR4 (Burget et al., 2003), SUS2 and TMM (Nadeau and Sack, 2002), EMB2742 and HUA1 (Li et al., 2001), EMB1967 and AP3 (Jack et al., 1992), AXR6 and LD (Lee et al., 1994), and EMB1401 and TSL (Roe et al., 1993). One cluster of six genes on chromosome 1 includes five with a knockout phenotype: FATB, PDE166, ABC1, NPQ1, and KN. The overall distribution of EMB genes, however, appears to be random. With respect to predicted sizes, EMB genes range from less than 1 kb (EMB2191 and EMB2184) to more than 10 kb (TOR, EMB2284, EMB2410, and EMB2016). Some of the smallest genes encode ribosomal proteins. Unusually large proteins include SUS2 (RNA splicing factor), ACC1 (acetyl-CoA carboxylase), TOR (protein kinase), EMB1507 (spliceosome-associated RNA helicase), EMB2016 (unknown function), EMB2410 (unknown function), and EMB2284 (subunit of DNA polymerase epsilon). The average size of EMB genes is larger than what is found genome-wide, consistent with the model that large genes provide good targets for random mutagenesis.
Full-length cDNA sequences are available for about 80% of the EMB genes in Figure 1. Another 12% have at least one matching Arabidopsis expressed sequence tag (EST) entry in GenBank. Many EMB genes are not embryo-specific in their pattern of expression, consistent with a requirement for their basal functions throughout the life cycle. Based on TargetP predictions (Emanuelsson et al., 2000), 32% of the gene products represented in Figure 1 are directed to chloroplasts and 13% are targeted to mitochondria. Proteins with locations supported by large-scale proteomics experiments (Heazlewood et al., 2004; www.mitoz.bcs.uwa.edu.au) include 17 recovered from chloroplasts, 5 from mitochondria, and 2 from both (SLP and EMB1956). Functional chloroplasts are known to be required for embryo development in Arabidopsis (Uwer et al., 1998; Despres et al., 2001; Apuya et al., 2002). We show here that mitochondrial functions are required as well. Disruption of nuclear genes encoding chloroplast and mitochondrial proteins can also result in a variety of defects later in development. Our dataset of 550 genes with other mutant phenotypes includes 40 with protein products recovered from chloroplasts and 15 from mitochondria. Some of these mutants may represent weak alleles of genes required throughout growth and development. We propose that a complete loss of critical chloroplast and mitochondrial functions in Arabidopsis results in embryo lethality, and in some cases gametophytic lethality, whereas partial loss of these functions or complete loss of peripheral functions results in defects after germination.
Diversity of emb Mutants Available
Embryo-defective mutants differ in their terminal phenotypes, extent of abnormal development, allele strength, nature of the underlying mutation, size and color of aborted seeds and embryos, efficiency of transmission through male and female gametes, capacity to produce mutant seedlings, and level of phenotypic analysis. We have summarized a considerable amount of information on more than 400 mutants in the SeedGenes database (Tzafrir et al., 2003). Embryo phenotypes are remarkably consistent from seed to seed in some mutants and exhibit considerable variation in others. Multiple alleles are available for more than half of the genes in Figure 1. In some cases, weak alleles have been found elsewhere through screens at later stages of the life cycle. Examples include sus1/caf/sin1/dcl1 alleles with defects in floral and ovule development (Jacobsen et al., 1999; Golden et al., 2002), cyt1/soz/vtc1 alleles resistant to ozone and deficient in vitamin C (Conklin et al., 1999), axr6 alleles resistant to auxin (Hobbie et al., 2000), and rsw1 (Arioli et al., 1998) and rml1 (Vernoux et al., 2000) alleles defective in root development. Such overlaps will become even more common in the future as additional genes with mutant phenotypes are identified. Some embryo-defective mutants may in turn represent weak alleles of genes required for gametogenesis, or knockouts of genes whose products are compensated in part by contributions from surrounding maternal or paternal tissues (Springer et al., 2000).
Functional Classification of Genes with Mutant Phenotypes
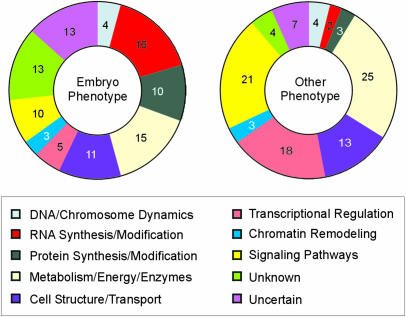
The 800 indispensable genes described here perform a wide range of cellular functions required for growth and development. The Gene Ontology (GO) Consortium (2001) system provides a standardized method for associating genes with molecular functions, biological processes, and cellular components. To date, 97% of these 800 genes have been associated with a GO molecular function, 88% with a biological process, and 89% with a cellular component. Molecular functions of 16% of the genes with assignments made are designated as unknown. Common functional assignments include catalytic (16%), transferase (12%), hydrolase (9%), transporter (8%), transcription factor (8%), and nucleotide binding (7%) activities. Process assignments include metabolism (11%), transport (7%), morphogenesis (6%), transcription (5%), and response to endogenous (6%) and abiotic (5%) stimuli. One current limitation to the GO system is the inability to combine information on molecular function and biological process into broad categories that allow comparisons with other published datasets. We therefore manually assigned each of the 800 genes to 1 of 10 functional categories that we devised based on published work on several organisms. We then compared results obtained for 250 EMB genes and 550 genes with other mutant phenotypes. Functional classifications are summarized in Figure 2 and detailed in Supplemental Table I (which may be viewed at http://www.plantphysiol.org).
Figure 2.
Functional diversity of indispensable genes of Arabidopsis. Numbers are percentages of two datasets: 250 genes with a mutant phenotype in the embryo and 550 genes with some other phenotype. Genes were manually assigned to functional classes based on current annotation and publications as described in “Materials and Methods.”
Genes required for basal functions associated with DNA, RNA, and protein synthesis, which should become critical during embryo development, are more prevalent among the embryo phenotype class (30% versus 9%), whereas transcription factors and components of signal transduction pathways are more common among the other phenotype class (39% versus 15%). These results reflect the complexity of molecular mechanisms that regulate postembryonic morphogenesis and responses to environmental signals in plants. Although many genes associated with basic metabolic functions give a knockout phenotype in the embryo, large numbers of genes with specialized metabolic functions have also been identified through post-embryonic screens for mutant phenotypes. Genes with unknown functions are scarce among the other phenotype class, perhaps because establishing gene function is often a prerequisite for publication.
Pentatricopeptide-repeat (PPR) proteins, which are thought to function in organelle RNA processing in plants (Small and Peeters, 2000), are especially common among embryo defectives. Seventeen have already been found among 250 EMB genes (6.8%) compared with 430 genome-wide (1.6%). Six predicted tRNA synthetases and 11 ribosomal proteins are products of EMB genes. Four of the ribosomal proteins have been found in chloroplasts (www.mitoz.bcs.uwa.edu.au), including one (SSR16) predicted to be targeted to mitochondria (Tsugeki et al., 1996). Seven ribosomal proteins are among the 550 genes with other mutant phenotypes. With respect to signaling pathways, 75% of the 22 protein kinases and 9 phosphatases in the list of 800 genes have postembryonic knockout phenotypes. The two datasets of indispensable genes therefore have distinctive functional profiles.
Comparisons with Published Datasets of Essential Genes
One reason for establishing a list of genes with mutant phenotypes in Arabidopsis is to enable comparisons with datasets from other model organisms. We chose to focus on yeast, worm, and human genomes for the comparisons summarized in Table I. Approximately 25% to 40% (205–301) of the 800 Arabidopsis genes have a significant match in at least one of these genomes. EMB genes have a higher percentage of matches among essential (nonviable) genes of yeast and worm than among an equivalent number of nonessentials, indicative of shared functions required for cell maintenance. EMB genes are also more likely than Arabidopsis genes with other phenotypes to have a counterpart among essential genes of yeast and worm. Examining these essential gene datasets may therefore facilitate the future identification of additional EMB genes. Less overlap exists between these EMBs and human disease genes, consistent with specialization of factors responsible for heritable human diseases, particularly those with post-embryonic phenotypes.
Table I.
Indispensable genes of Arabidopsis with a significant match in another model eukaryote
| Organism Datasetb | Entries | Genes with a Matcha
|
|
|---|---|---|---|
| Embryo | Other | ||
| Yeast genome | 5,889 | 90 (36%) | 115 (21%) |
| Worm genome | 21,711 | 96 (38%) | 157 (29%) |
| Human genome | 37,490 | 116 (46%) | 185 (34%) |
| Yeast nonviable | 1,009 | 54 (22%) | 34 (6%) |
| Yeast other genes | 4,880 | 58 (23%) | 107 (20%) |
| Worm nonviable | 1,158 | 57 (23%) | 57 (10%) |
| Worm other phenotype | 524 | 16 (6%) | 18 (3%) |
| Worm viable, no phenotype | 20,029 | 65 (26%) | 141 (26%) |
| Human recessive disease | 305 | 14 (6%) | 30 (6%) |
| Human other disease | 528 | 6 (2%) | 38 (7%) |
| Human other genes | 36,657 | 114 (46%) | 175 (32%) |
Number (percentage) of Arabidopsis genes with a significant BLASTP match (<e−30). Datasets: 250 genes with an embryo phenotype; 550 genes with other phenotypes. Subclass numbers (e.g. nonviable; other) do not add up to genome totals because a single Arabidopsis gene can have a match in >1 subclass.
See “Materials and Methods” for nature and sources of datasets utilized.
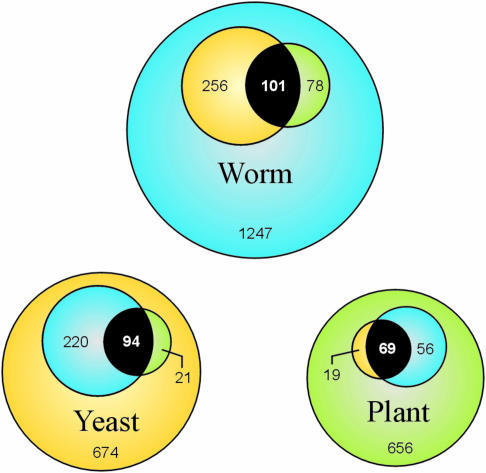
Overlaps between datasets of genes with mutant phenotypes in yeast, worm, and Arabidopsis are shown in Figure 3. Identities of shared genes are presented in Supplemental Table II. Yeast and worm have the most matches, reflecting in part the completeness of their datasets. Of the 69 Arabidopsis genes with a significant match in both yeast and worm, 60% are EMB genes and 35% are predicted to perform a basal cellular function represented by the first three categories in Figure 2. Shared Arabidopsis genes are enriched for components of translation (15% versus 4% in the combined list of 800), protein kinases (10% versus 3%), and chromatin remodeling factors (9% versus 3%), but devoid of transcription factors and proteins with completely unknown functions. Searching for Arabidopsis orthologs of yeast and worm genes with important functions defined by mutant phenotypes may therefore result in a biased sample of candidate genes deficient in certain specialized functions. Fifty-five percent of the 656 Arabidopsis genes with no significant match (e−30 cutoff) to yeast or worm essential genes appear to be plant-specific based on BLASTP searches. More than half of these genes are predicted to encode transcriptional regulators or proteins of unknown function.
Figure 3.
Overlapping sets of essential and indispensable genes in model eukaryotes. BLASTP comparisons were performed using <e−30 as a significant match. Circle sizes are proportional to numbers of genes: 1,682 C. elegans genes with an RNAi phenotype (Kamath et al., 2003); 1,009 essential genes of S. cerevisiae (Giaever et al., 2002); and 800 indispensable genes of Arabidopsis. Numbers represent matches (e.g. 357 worm genes with an RNAi phenotype have at least 1 match among yeast essentials and 1,247 have no match in either the yeast or Arabidopsis datasets). Numbers in white represent genes with matches in all 3 datasets. Differences in reciprocal comparisons (e.g. 115 yeast genes with an Arabidopsis match; 88 Arabidopsis genes with a yeast match) reflect different levels of sequence redundancy and sizes of datasets.
Sequence Redundancy of EMB Genes
Genes with an RNAi phenotype are on average less redundant than other genes in C. elegans (Kamath et al., 2003). We found a similar pattern with EMB genes of Arabidopsis: 55% are unique compared with 34% genome-wide (Table II) and fewer EMB genes (16% versus 34%) are members of large families (≥4 members). These results are consistent with the assumption that redundant genes are less likely to exhibit a mutant phenotype than unique genes, provided the redundant genes perform overlapping functions. Surprisingly, Arabidopsis genes with other mutant phenotypes exhibit a level of redundancy similar to that of the whole genome. This suggests that many duplicated genes, particularly those associated with specialized rather than basal cellular functions, exhibit a loss-of-function phenotype because putative paralogs are not functionally redundant and have instead evolved nonoverlapping roles or patterns of expression. The intermediate redundancy observed with genes adjacent to EMBs on the chromosome may indicate that segmental duplication events in Arabidopsis (Vision et al., 2000; Simillion et al., 2002) have resulted in pockets of paralog-depleted sequences that often contain genes with mutant phenotypes. The amount of overlap between genes with mutant phenotypes in different plant species remains to be determined. We suspect that different sets of genes will be left unprotected by functional redundancy and that distinctive collections of genes will ultimately be identified.
Table II.
Sequence redundancy of indispensable genes in Arabidopsisa
| Arabidopsis Dataset | Entries | Percentage with Number of Paralogs
|
||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ≥ 4 | ||
| Embryo phenotype | 250 | 54.8 | 21.2 | 6.4 | 2.0 | 15.6 |
| Adjacent to EMBb | 487 | 42.7 | 18.3 | 7.4 | 5.3 | 26.3 |
| Other phenotype | 550 | 37.3 | 15.6 | 7.8 | 6.9 | 32.4 |
| Total genome | 27,375 | 34.2 | 17.4 | 9.0 | 5.8 | 33.6 |
BLASTP comparisons were performed using <e−30 and a match covering ≥80% of the protein length as a cutoff. Embryo phenotype and adjacent gene distributions both differ significantly from those expected genome-wide (P < 0.001).
Genes immediately above and below a given EMB gene on the physical map, excluding pseudogenes and other ambiguities.
If functional redundancy is responsible for the absence of a seed phenotype in some gene knockouts, then double mutants disrupted in both redundant genes should result in a visible or enhanced phenotype. At least 10 examples of such gene pairs have already been described: SEC/SPY (Hartweck et al., 2002), CUC1/CUC2 (Aida et al., 1997), PNH/AGO1 (Lynn et al., 1999), ACX3/ACX4 (Rylott et al., 2003), SPDS1/SPDS2 (Takahashi et al., 2003), POL/PLL1 (Song and Clark, 2003), PP2A-Beta/PP2A-Gamma (Zhou et al., 2003), ASK1/ASK2 (Liu et al., 2004), ZIG/VTI12 (Surpin et al., 2003), and TTN3/AtSMC2 (Muralla et al., 2003; Siddiqui et al., 2003). Additional details on these redundant genes are presented in Supplemental Table III. In most cases, partners identify each other as their closest match in the genome, as expected for genes with redundant functions. However, the amino acid identities differ widely, from 26% for SEC/SPY to 75% for ASK1/ASK2. Alignments to the next most similar gene in Arabidopsis also differ (e.g. 72% identity, ASK1/At4g34210; 24% identity, TTN3/At2g27170). It may therefore be difficult to identify future candidates for additional gene pairs based on BLASTP results alone. The presence of a close paralog does not necessarily protect against a loss-of-function phenotype. We have identified at least 458 examples of genes with mutant phenotypes, including 113 with an embryo phenotype, which have paralogs, some with more than 80% amino acid sequence identity. These are likely to represent genes with critical differences in spatial and temporal patterns of expression.
EMB Genes with Unknown Functions
Table III summarizes information on 34 EMB genes of Arabidopsis that are predicted to encode proteins of unknown function. These genes are divided into two classes (unknown and uncertain) and four different ranks that reflect available information on possible function: (1) no significant BLASTP match to an informative protein (e−10 cutoff value) or Pfam motif (gathering threshold) other than domains of unknown function; (2) questionable motif assignment or annotation comment from another source; (3) match to an informative protein (e−10 cutoff marginal; e−30 significant) but not a Pfam motif; and (4) informative Pfam motif but unresolved cellular function. The uncertain class includes genes assigned to the third and fourth ranks. All gene identities have been confirmed through molecular complementation or recovery of duplicate mutant alleles. Three genes (EMB506, RSY3, and TOZ) are described in recent publications (Albert et al., 1999; Apuya et al., 2002; Boniotti and Griffith, 2002).
Table III.
Overview of 34 confirmed EMB genes with unknown functions
| Locus | Symbol | Function Ranka | Plant ESTb
|
Non-Plant Modelc
|
||
|---|---|---|---|---|---|---|
| Identity | Organism | Identity | Organism | |||
| % | % | |||||
| Unknown Class: | ||||||
| At1g49510 | EMB 1273 | 1 | 63 | Orange | – | – |
| At2g25660 | EMB 2410 | 1 | 81 | Maize | – | – |
| At2g03150 | EMB 1579 | 2 | 60 | Grape | – | – |
| At2g31340 | EMB 1381 | 1 | 72 | Potato | – | – |
| At3g05680 | EMB 2016 | 2 | 50 | Sorghum | – | – |
| At3g07060 | EMB 1974 | 1 | 43 | Medicago | – | – |
| At3g20070 | TTN 9 | 1 | 52 | Cotton | – | – |
| At3g48470 | EMB 2423 | 2 | 86 | Brassica | 26 | Mouse |
| At3g61780 | EMB 1703 | 1 | 71 | Tomato | – | – |
| At4g13740 | EMB 2597 | 1 | – | – | – | – |
| At4g20060 | EMB 1895 | 2 | 44 | Medicago | – | – |
| At4g28210 | EMB 1923 | 1 | 77 | Brassica | – | – |
| At5g53860 | EMB 2737 | 1 | 70 | Potato | – | – |
| At5g57930 | EMB 1629 | 1 | 65 | Rice | – | – |
| At5g62990 | EMB 1692 | 1 | 75 | Grape | – | – |
| Uncertain Class: | ||||||
| At1g10510 | EMB 2004 | 4 | 78 | Medicago | 34 | Human |
| At1g48175 | EMB 2191 | 4 | 71 | Onion | – | – |
| At1g58210 | EMB 1674 | 3 | 56 | Brassica | – | – |
| At1g67440 | EMB 1688 | 3 | 76 | Soybean | 36 | Synechocystis |
| At1g76060 | EMB 1793 | 4 | 85 | Brassica | – | – |
| At2g21710 | EMB 2219 | 4 | 70 | Lettuce | – | – |
| At3g07430 | EMB 1990 | 4 | 63 | Brassica | – | – |
| At3g12080 | EMB 2738 | 4 | 85 | Ipomoea | 46 | Synechocystis |
| At3g24560 | RSY 3 | 4 | 51 | Grape | – | – |
| At4g26500 | EMB 1374 | 4 | 81 | Brassica | 50 | Synechocystis |
| At5g16750 | TOZ | 4 | 75 | Poplar | 35 | Human |
| At5g21140 | EMB 1379 | 3 | 67 | Poplar | – | – |
| At5g22370 | EMB 1705 | 4 | 81 | Wheat | 60 | Mouse |
| At5g22640 | EMB 1211 | 4 | 80 | Grape | – | – |
| At5g40160 | EMB 506 | 4 | 73 | Barley | – | – |
| At5g49930 | EMB 1441 | 4 | 69 | Lettuce | 37 | Mouse |
| At5g56930 | EMB 1789 | 4 | 66 | Soybean | – | – |
| At5g63420 | EMB 2746 | 4 | 86 | Aspen | 36 | Synechocystis |
| At5g66055 | EMB 2036 | 4 | 73 | Soybean | – | – |
Genes were divided into two classes: those without Pfam motifs or informative BLASTP matches (Ranks 1 and 2) and those with informative motifs or matches but uncertain functions (Ranks 3 and 4).
TBLASTN searches were performed against the EST database at NCBI (www.ncbi.nlm.nih.gov). The top significant match (<e−30) is listed.
BLASTP searches were performed against the nonredundant database at NCBI (www.ncbi.nlm.nih.gov). Top significant matches (<e−30) were identified from an initial list of model organisms that also included D. melanogaster, C. elegans, D. discoideum, S. cerevisiae, S. pombe, and E. coli. Dash indicates no significant match.
Table IV lists examples of Pfam motifs found among 19 proteins in the uncertain class. Some of these motifs (WD-40, ATP binding, and ankyrin repeat) are not informative with respect to precise cellular function. Others such as cytidine deaminase and Fe-S metabolism are not supported by significant matches. Examples of potentially informative BLASTP matches but uncertain functional classification include At1g58210 and a kinase-interacting protein of petunia (Skirpan et al., 2001), At1g10510 and a human caspase recruitment domain (CARD15) protein associated with Crohn disease and Blau syndrome (Miceli-Richard et al., 2001), and At5g49930 and a human colon cancer antigen of unknown function (Carbonnelle et al., 1999). Although the boundary between genes with known functions predicted from sequence comparisons and those assigned to the uncertain class based on the presence of informative motifs is subjective, we found this classification system to be helpful in identifying genes with the least amount of information available on predicted cellular functions in plants. Ninety percent of the genes in the unknown class and 25% of those in the uncertain class in Table III were annotated (www.tigr.org) to code for hypothetical or expressed proteins. Text searches of annotation comments are therefore useful but not always accurate predictors of the level of information available.
Table IV.
Pfam motifs in EMB proteins with uncertain functions
| Locus | Informative Pfam Motif | Informative Protein Match |
|---|---|---|
| At1g10510 | Leu-rich repeat | Yes |
| At1g48175 | Cytidine deaminase | Marginal |
| At1g76060 | Complex 1 LYR | No |
| At2g21710 | mTERF | No |
| At3g07430 | YGGT | No |
| At3g12080 | GTPase | Yes |
| At3g24560 | ATP binding | No |
| At4g26500 | Fe-S metabolism | Marginal |
| At5g16750 | WD-40 repeat | Yes |
| At5g22370 | ATP binding | Yes |
| At5g22640 | MORN repeat | No |
| At5g40160 | Ankyrin repeat | Marginal |
| At5g49930 | Zinc knuckle (CCHC) | Yes |
| At5g56930 | Zinc finger (CCCH) | No |
| At5g63420 | Lactamase B | Yes |
| At5g66055 | Ankyrin repeat | Marginal |
Determining the cellular locations of proteins with unknown functions can provide valuable clues to possible functions. TargetP predictions (Emanuelsson et al., 2000) for 34 EMB proteins with unknown functions are consistent with patterns observed for the entire EMB collection but differ somewhat from those obtained with PSORT (Nakai, 2000). Based on top scores alone, half of these proteins appear to be localized to different compartments. Similar problems have been noted with the mitochondrial proteome (Heazlewood et al., 2004), where only half of the proteins recovered were predicted to be targeted to the mitochondrion. We are therefore reluctant to propose models of gene function for selected unknowns based to any significant extent on predicted protein localizations. The production of epitope-tagged EMB proteins described here, when combined with fluorescence tagging of proteins with unknown functions (Tian et al., 2004), should generate more definitive information on protein localization in the future.
Approximately half of the genes listed in Table III have no paralogs based on sequence comparisons, about a quarter have one paralog, and the rest have two or more paralogs. These results are similar to those obtained for the entire collection of EMB genes and reflect a reduced level of redundancy in comparison to the genome as a whole. To determine the distribution of related sequences in other plants, we searched the EST database at GenBank using TBLASTN and listed the most significant match in Table III. All but one of the 34 genes have a significant match among plant ESTs (e−30 cutoff value) and 94% of the top matches have predicted amino acid sequences that are more than 50% identical to the Arabidopsis unknown. Eighteen species are represented among the top EST matches in Table III. Some of the best matches are from cDNA libraries prepared from developing flowers, seeds, and fruits. We then used BLASTP to search GenBank for significant matches among representative organisms with well-characterized genomes: human, mouse, Drosophila, Caenorhabditis, Schizosaccharomyces, Saccharomyces, and Synechocystis. Ninety-three percent of genes in the unknown class and 58% of those in the uncertain class did not have a significant match (e−30) in this dataset. This supports our conclusion that the unknown class of EMBs is enriched for genes with counterparts in flowering plants but not elsewhere. The functions of many of these genes will therefore not emerge from work on yeast, worm, or fly genomes, but rather from continued analysis of Arabidopsis and other model plants.
Identification of Duplicate Alleles
We uncovered duplicate alleles for 13 of the 34 genes listed in Table III through forward genetics by screening T-DNA insertion lines for defects in seed development, cloning sequences flanking insertion sites in tagged mutants, and crossing mutants that appeared to be disrupted in the same gene (McElver et al., 2001). We then sought to identify second alleles for other genes in our collection by searching the Salk database of T-DNA insertion lines (Alonso et al., 2003), planting lines that contained promising insertions expected to result in a null phenotype, screening these plants for the presence of defective seeds, crossing confirmed heterozygotes with the putative Syngenta allele, and confirming insert locations through sequencing of PCR products generated using T-DNA border and gene-specific primers (see Supplemental Table IV).
No promising Salk lines were identified for six genes on the original list of unknowns: At1g21390, At2g37920, At3g20400, At5g06240, At5g27540, and At5g55940. These genes are excluded from Table III, along with three genes where the Salk lines failed to produce any plants with the expected insertion (At1g55540, At1g56200, and At4g21190) and one where analysis is ongoing (At1g19080). Eleven other genes were excluded because Salk lines gave contradictory results (At1g13120, At1g22090, At4g14590, and At5g40600) or produced plants with confirmed insertions but no seed phenotype (At1g04590, At1g71190, At2g34780, At3g14225, At4g30090, At5g06910, and At5g49820). Another gene (At5g08170) was removed when it was shown to encode an enzyme in polyamine biosynthesis (Janowitz et al., 2003).
Confirmed Salk alleles were obtained for 22 of the genes listed in Table III. All combined, the identities of 31 EMB genes with unknown functions were confirmed here through the analysis of 25 Salk mutants and 46 Syngenta mutants. Genetic, molecular, and phenotypic data for these 71 embryo-defective mutants are summarized in Supplemental Table V. Additional details are found at www.seedgenes.org. One important distinction between the Salk and Syngenta mutants is that flanking sequences for most Syngenta lines were obtained from both sides of the insert (Rank A), whereas sequence information for the Salk lines is available for just one side (Rank B). We have therefore not determined the size of any deletions associated with insertion sites in the Salk alleles. As expected, terminal phenotypes of confirmed allelic mutants were in most cases similar. For 23 of the 31 confirmed genes, mutant alleles were assigned to the same terminal seed phenotype class (3 preglobular, 10 globular, 1 transition, and 9 cotyledon). In another 5 cases, the phenotype classes were overlapping (globular and transition; transition and cotyledon) and results obtained were generally consistent with confirmed locations of insertions. In the remaining cases (At1g76060, At3g48470, and At5g22370), seed phenotypes of mutant alleles were more difficult to reconcile with the confirmed insertion sites. These results demonstrate some of the challenges faced when attempting to predict the severity of mutant phenotypes based on the known locations of T-DNA insertions within a defined coding region. Only a few genes had very early (preglobular) phenotypes whereas more had later (cotyledon) terminal phenotypes. For genes with no paralogs and mutants with insertion sites consistent with a null phenotype, survival of mutant embryos to the cotyledon stage is consistent with a gene function that is not required for basic cell viability.
Expression of Genes with Unknown Functions
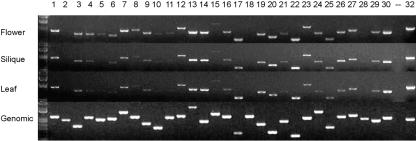
Expression data for genes with unknown functions can provide critical validation of gene models, direct evidence of transcription at different stages, and clues to molecular functions when compared to expression patterns genome-wide. Full-length cDNA sequences are available for 74% of the 34 genes in Table III. Four genes in the unknown class have no matching EST from Arabidopsis and one of these (At4g13740) has no match among other plant ESTs. In order to obtain further evidence of expression in reproductive and nonreproductive tissues, we designed gene-specific primers flanking introns for 30 genes with confirmed identities from the Syngenta collection and performed RT-PCR with RNA isolated from inflorescences, immature siliques, and leaves. Results are summarized in Figure 4 and Supplemental Table VI. Products of the expected sizes were obtained for 29 of the 30 genes from all three RNA samples. Six genes appear to be expressed at low levels in both reproductive and vegetative tissues: At2g25660, At3g05680, At4g13740, At4g20060, At5g62990, and At5g49930. Most of the remaining genes are transcribed at more significant levels and without apparent tissue specificity. Our failure to detect expression of At1g58210 in any of the samples examined was surprising in light of the availability of a full-length cDNA but consistent with low levels of transcription reflected in public MPSS (Meyers et al., 2002) and microarray (Finkelstein et al., 2002) datasets (see http://mpss.udel.edu/at/java.html and http://ssbdjc2.nottingham.ac.uk/narrays/experimentbrowse.pl). Expression of the remaining 29 genes in both reproductive and vegetative tissues is consistent with general functions throughout the life cycle rather than specialized functions limited to seed development.
Figure 4.
RT-PCR expression patterns for 30 EMB genes with confirmed identities but unknown cellular functions. Locus numbers for products loaded in lanes 1 to 30 are provided in Supplemental Table VI. Lane 32 contains an actin control (At3g18780). PCR products from genomic DNA (bottom section) differed in size from RT-PCR products.
Molecular Complementation with Tagged Proteins
We attempted molecular complementation of three embryo-defective mutants with epitope tagged proteins in order to explore the feasibility of combining the confirmation of gene identities with the analysis of interacting protein complexes. Because many cellular processes are carried out by complexes of interacting proteins rather than by individual gene products working in isolation, understanding the nature of these interactions and identifying known protein partners are valuable steps in characterizing proteins with unknown functions. We decided to focus on His (6×His) and tandem affinity purification (TAP) tags (Rigaut et al., 1999) for molecular complementation. The TAP tag is a fusion cassette composed of 183 amino acids that includes a calmodulin-binding domain, protease cleavage site, and proteinA site for IgG binding. TAP tagging and analysis of protein complexes by mass spectrometry have recently been performed on a large scale in yeast (Gavin et al., 2002). Successful experiments exploiting the TAP system have been reported with mammalian cells (Bouwmeester et al., 2004), protozoan parasites (Aphasizhev et al., 2003), and plants (Rivas et al., 2002; Rohila et al., 2004). In a recent study, RNA interference was combined with TAP tagging to reduce the amount of endogenous protein incorporated into protein complexes (Forler et al., 2003).
Three genes were chosen here for initial studies, two with unknown functions (At3g20070 and At5g57930) and another (At3g01610) with sequence similarity to an intriguing AAA-ATPase (cdc48) involved in spindle disassembly and cell division control (Cao et al., 2003). Putative wild-type genes under the control of their own promoters and carrying the appropriate 6×His and TAP tags were introduced into a T-DNA construct carrying the aacC1 gene conferring plant resistance to gentamycin and then transformed into Arabidopsis plants by Agrobacterium infiltration. The treated T0 plants were allowed to produce T1 seeds, which were subsequently harvested in bulk and germinated on gentamycin to select for the presence of the transgene. Resistant plants were then crossed with plants heterozygous for the loss-of-function mutation and the progeny seeds germinated on Basta and gentamycin to select for plants carrying both the mutation and the tagged transgene. Seeds produced by F1 plants that survived double selection were scored for mutant phenotypes by dissecting immature siliques.
Results of three complementation experiments involving 6×His tags and another with a TAP tag are summarized in Table V. With full complementation, siliques of transformed EMB/emb plants carrying a single unlinked copy of the transgene were expected to contain 6.25% aborted seeds instead of 25% after self-pollination. This result was obtained with emb1629-1 complemented with the His-tagged protein, confirming that the correct gene had been identified and that the His tag did not interfere with protein function. In contrast, partial complementation appeared to take place with His-tagged TTN9. Some seeds were completely rescued, because the percentage of mutant seeds was significantly reduced from 25%, some were partially rescued and gave rise to atypical mutant phenotypes, and some did not get beyond the normal stage of arrest, resulting in more aborted seeds than expected with complementation. Three plants were found in which the transgene appeared to have been inactivated, resulting in a complete loss of complementation. These results suggest that the His tag interferes somewhat with TTN9 function and that the transgene is susceptible to suppression. A different result was encountered with His-tagged emb1354 plants. The percentage of mutant seeds in these plants was significantly reduced from the expected 6.25% but was also higher than the 1.5625% (1/64) expected for plants containing two unlinked transgenes. In this case, we conclude that full complementation has occurred and that two linked copies of the transgene are responsible for the intermediate percentage of mutant seeds observed.
Table V.
Complementation of mutant seeds with epitope-tagged proteins
| Complemented Allele | Protein Tag | Seeds Screened | Mutant (Typical) | Mutant (Atypical) |
|---|---|---|---|---|
| % | % | |||
| emb 1629-1 | His | 1,497 | 7.1a | 0.3 |
| 1,540 | 5.4a | 0.3 | ||
| emb 1629-1 | TAP | 2,261 | 5.5a | 16.7 |
| 3,053 | 5.3a | 19.8 | ||
| ttn 9 | His | 1,960 | 10.7b | 3.5 |
| 1,845 | 10.6b | 2.5 | ||
| 1,399 | 9.1b | 1.5 | ||
| emb 1354 | His | 6,288 | 3.8b | 0.7 |
| 4,067 | 3.5b | 0.5 | ||
| 1,589 | 3.3b | 0.3 | ||
| 1,097 | 3.6b | 0.5 |
Differs significantly from 25.0% but not from 6.25% at P = 0.001.
Differs significantly from both 6.25% and 25.0% at P = 0.001.
A single experiment involving complementation with a TAP-tagged protein has been completed to date. As summarized in Table V, partial complementation of the emb1629 seed phenotype was observed. Siliques segregating for both the transgene and the knockout allele contained 5% to 6% mutant seeds arrested early in development, as expected for complementation, along with another 17% to 20% pale-green seeds with minor alterations in cotyledon morphology late in development. Embryonic lethality was corrected and plants homozygous for the knockout allele were obtained. The altered pigmentation observed in rescued seeds appears to result from limited interference of the TAP tag with normal protein function. These results demonstrate the value of testing the functionality of TAP-tagged proteins in plants lacking the normal gene product before embarking on intensive screens for interacting proteins.
Approaching Saturation for EMB Genes
Forward genetics will not enable the identification of every essential gene through random mutagenesis. Determining the full spectrum of mutant phenotypes will be a major challenge. Knocking out small genes will be especially difficult. Based on estimates of 500 to 1,000 EMB genes in Arabidopsis (Franzmann et al., 1995; McElver et al., 2001), the 250 genes described here represent about 25% to 50% of the total. One strategy for uncovering additional EMB genes might be to focus on reverse genetics of specific pathways or protein complexes for which at least one knockout is known to result in embryo defects. For example, because mutants disrupted in two different steps of biotin synthesis are embryo lethals (Patton et al., 1998), disruption of the remaining steps should also result in embryo abortion if those genes are not functionally redundant. Similarly, when disruption of a single component of a large protein complex results in a seed phenotype, as with det/fus/cop mutants defective in the COP9 signalosome (Wei and Deng, 2003), then remaining members of the complex might be promising candidates for reverse genetic analysis. Alternatively, a comprehensive screen of tRNA synthetase or ribosomal protein knockouts might reveal additional EMB genes based on their high frequency in existing collections. Comparative studies with embryo-defective mutants identified in other plant species could also be explored if more EMB genes were known outside of Arabidopsis. An interesting example is a Syngenta mutant (emb1275) disrupted in the apparent ortholog (At1g55350) of the DEK1 gene of maize (Zea mays), which encodes a calpain-domain Cys proteinase required for embryonic pattern formation and cell fate specification (Becraft et al., 2002; Lid et al., 2002; Wang et al., 2003).
We have adopted instead a bioinformatics-based approach to identify promising candidates for additional EMB genes based on sequence comparisons with essential genes found in other eukaryotes. Results are summarized in Table VI and Supplemental Table VII. Arabidopsis contains more than 2,300 genes with a significant match to a yeast or worm essential gene. Some of these Arabidopsis genes are known to be indispensable (73 embryo phenotype and 64 other phenotype). Of the 636 instances where the top match is not known to give a knockout phenotype, 299 contain 1 to 3 paralogs and 93 contain ≥4 paralogs. This leaves 244 unique genes of Arabidopsis not yet identified through forward genetics that represent the top match to a known essential gene in another model eukaryote. These genes are promising candidates for reverse genetic analysis. Toward this end, we have identified potential T-DNA insertions in approximately 75% of these genes in the Salk collection (Alonso et al., 2003) of insertion lines. A variety of predicted functions are included among these genes. Because the kanamycin resistance marker in Salk lines may become suppressed, we cannot readily pursue the genetic strategy used to identify tagged mutants in the Feldmann (Castle et al., 1993) and Syngenta (McElver et al., 2001) collections. We have therefore chosen to focus initially on candidate EMB genes with at least two independent insertions predicted to fall within the coding region of a candidate gene. Putative knockouts in these 75 genes are being screened for a seed phenotype. This strategy will be expanded to include additional knockouts from other collections of insertion lines if the initial screen is successful.
Table VI.
Candidate EMB genes in Arabidopsis based on sequence similarity to essential genes in other model eukaryotes
| Result of Querying Arabidopsis Genome | Query Dataset of Essential Genesa
|
||
|---|---|---|---|
| Yeast | Worm | Total | |
| Matched Arabidopsis genes found | 1,542 | 1,870 | 2,362 |
| Not a known indispensable geneb | 1,454 | 1,756 | 2,225 |
| Top match is not a known indispensable | 394 | 421 | 636 |
| Unique gene without paralogsc | 165 | 139 | 244 |
BLASTP comparisons were performed using <e−30 as a significant match. Datasets: 1,009 Saccharomyces (Giaever et al., 2002) and 1,158 Caenorhabditis (Kamath et al., 2003) nonviable genes. Total refers to a nonoverlapping set from yeast and worm combined, with duplicate matches removed.
Number of Arabidopsis genes that represent a significant match to a yeast or worm essential gene but do not correspond to a known Arabidopsis gene with a mutant phenotype.
Top Arabidopsis match to a yeast or worm essential; lacks a paralog. None of these corresponds to a known EMB gene.
The potential therefore exists over the next 5 years to approach saturation for genes with a wide range of critical functions during seed development. This should provide the foundation needed to saturate for other classes of mutants in Arabidopsis and ultimately define a minimal set of genes that cannot be eliminated without altering the normal phenotype of a plant. When a comparable dataset becomes available for another model plant such as maize, rice, or Medicago, it should be possible to determine whether genome duplication and deletion events in these plants have exposed similar types of genes to the deleterious effects of recessive mutations.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
More than 120,000 T-DNA insertion lines of Arabidopsis were generated at Syngenta (Research Triangle Park, NC) and screened for mutants with a seed phenotype as part of a large-scale project designed to identify plant genes with essential functions (Budziszewski et al., 2001; McElver et al., 2001). This population of insertion lines is distinct from the Syngenta collection designed for reverse genetics at Torrey Mesa Research Institute (Sessions et al., 2002). Some lines also carry the quartet mutation that prevents separation of pollen tetrads (Preuss et al., 1994). Seed stocks for the Syngenta mutants and their Salk alleles, both in the Columbia ecotype, are available through the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University. Plants were grown in 7.5-cm pots containing a mixture of vermiculite (Strong-Lite coarse; medium for transplants; Sun-Gro Horticulture, Pine Bluff, AR), potting soil (Scotts Redi-Earth, Plug and Seedling mix, Scotts-Sierra Horticultural Products, Marysville, OH), and sterilized sand (12:3:1 v/v/v), placed in a growth room at 24 ± 2°C under fluorescent lights on 16-h-light/8-h-dark cycles, and watered daily from below with a nutrient solution (0.35 g L−1) containing Excel 15-5-15 fertilizer (Scotts Miracle-Gro, Port Washington, NY). Mutants were maintained as heterozygotes that produced 25% defective seeds upon selfing.
Sequence Datasets
The following protein datasets were used for BLASTP: Arabidopsis, ATH1_pep_20030417.Z from ftp://ftp.Arabidopsis.org/home/tair/Sequences/blast_datasets; Saccharomyces, orf_trans.200307309.fasta.gz from ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/genomic_sequence/orf_protein/archive; Caenorhabditis, ftp://ftp.wormbase.org/pub/wormbase/wormpep106.tar.gz; and human, protein. fa.gz (April 29, 2003) from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/protein/. A single protein sequence was chosen for genes with multiple products predicted from differential splicing. Published lists of essential genes (Jimenez-Sanchez et al., 2001; Giaever et al., 2002; Kamath et al., 2003) were updated to eliminate those not represented in current protein datasets. This resulted in lists of 1,009 nonviable genes of Saccharomyces cerevisiae; 1,158 nonviable, 524 growth and postembryonic phenotype, and 1,682 total RNAi phenotype genes of Caenorhabditis elegans; and 305 autosomal recessive and 528 other disease genes of humans. The C. elegans viable phenotype classes were included in Table I and Figure 3 to facilitate comparison with Arabidopsis genes with other phenotypes but were excluded from Table VI to focus on essential genes most likely to have counterparts among EMB genes. The 250 EMB genes include 211 examples listed at www.seedgenes.org (February 2004), another 19 from Syngenta to be added to SeedGenes by September 2004, 11 awaiting future release from the community, and 9 undergoing further review. Arabidopsis genes with other knockout phenotypes include those already on the sequence-based map (Meinke et al., 2003) along with recent additions from meetings and publications.
Functional Classification
A classification system patterned after that used for C. elegans (Fraser et al., 2000) was used to assign Arabidopsis essential genes to functional categories. The following guidelines were used to make functional assignments. DNA/chromosome dynamics includes DNA synthesis and repair along with proteins associated with chromosome structure. Chromatin remodeling proteins were placed in a separate category to underscore their importance in plant growth and development. Cell structure and transport includes components of the cytoskeleton and membrane systems and proteins involved in transport of ions and molecules. Signaling includes protein kinases and phosphatases, protein degradation pathways, and proteins involved in hormone synthesis, light perception, and signal transduction.
Sequence Comparisons
The BLASTP (Altschul et al., 1997) program (Version 2.2.5, National Center for Biotechnology Information [NCBI]) was used for sequence database searches. The expectation threshold was set to −30 and simple sequence filtering was turned off. The following searches were performed: Arabidopsis versus yeast, worm and human; yeast versus worm and human; and worm versus human. Results were parsed into a relational database for subsequent data mining. Alignments for Arabidopsis paralogs were required to extend over 80% or more of the protein length. To determine which indispensable genes of Arabidopsis were plant specific, a subset of the GenBank “nr” protein set was filtered to include metazoa, fungi, bacteria, and archae (ftp.ncbi.nih.gov/pub/taxonomy) and then examined for the presence of significant matches.
Reverse Genetics of Salk Insertion Lines
Putative Salk alleles of Syngenta mutants were identified by searching http://signal.salk.edu and requesting seeds from ABRC for lines with open reading frame insertions likely to generate a null phenotype. We typically planted 30 seeds per Salk line, screened immature siliques from the resulting plants for defective seeds, and harvested leaf tissue to confirm the presence of the insertion. When Salk lines segregated for a seed phenotype, heterozygotes were crossed with the corresponding Syngenta lines to test for allelism. Genomic DNA was extracted using a modified CTAB protocol (Lukowitz et al., 2000). DNA was isolated from heterozygotes to confirm the insertion by sequencing PCR products generated with gene-specific and T-DNA left border (LB1) primers. Five heterozygous and three wild-type plants were often examined to confirm cosegregation of the mutant phenotype and T-DNA. For lines without a phenotype, DNA was analyzed from pooled plants to determine whether the expected insertion was present.
Gene-specific primers for each Salk line were designed to flank the T-DNA insertion (see Supplemental Table IV). Primers were 21 to 27 bp long with 50% to 70% GC content. The PCR parameters used were: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 55°C for 2 min, 72°C for 3 min, and a final elongation step of 72°C for 5 min. Reactions were performed with a Biometra (Goettingen, Germany) Uno II thermocyler. PCR products were separated in agarose gels, stained with ethidium bromide, and visualized with a Kodak (New Haven, CT) EDAS 290 camera. Products amplified using a gene-specific primer in combination with Salk LB1 primer were gel purified (Qiagen, Valencia, CA), sequenced (OSU Recombinant DNA/Protein Resource Facility), and insertion points verified.
RT-PCR Analysis of Gene Expression
For confirmation of gene expression, cauline leaves, young flowers, and siliques with embryos up to the transition stage were harvested from plants grown in soil and flash frozen in liquid nitrogen. Tissues were stored at −80°C without thawing until RNA extraction. An RNase-free mortar and pestle was used to homogenize frozen tissue (1.5 g) in liquid nitrogen. Total RNA was prepared from powdered tissues using the RNeasy Plant Mini kit (Qiagen), treated with RNase-free DNase I (Takara Bio, Kyoto), quantified with a Shimadzu (Kyoto) UV-160 spectrophotometer, and visualized on a 1.0% formaldehyde agarose gel. For the 2-step RT-PCR reaction, 5 μg total RNA was reverse transcribed using the Superscript First-Strand Synthesis system (Invitrogen) followed by RNaseH treatment. A 1-μL aliquot of reverse transcribed reaction was used as template for PCR with REDTaq DNA polymerase (Sigma-Aldrich, St. Louis). PCR parameters were: 94°C 1 min, followed by 30 cycles of 94°C 1 min, 55°C 1 min, 72°C 2 min, and a final elongation step at 72°C for 10 min. Primers used for RT-PCR are listed in Supplemental Table VIII.
Complementation with Tagged Proteins
Wild-type genomic sequences corresponding to the coding regions of three genes (At5g57930, At3g01610, and At3g20070) and their promoters were PCR-amplified from Arabidopsis (Col-0) DNA. Forward primers (fAt5g57930, 5′-CCTGAACACAGCATGGAGTTTG-3′; fAt3g01610, 5′-TGCTGAATTGGATTGGATGG-3′; fAt3g20070, 5′-GTCAGGTTCAATCCTCGAATG-3′) were designed to anneal 850 to 970 bp upstream of the predicted start codons. Reverse primers were extended to add a sequence encoding six His and a stop codon (6×His, 5′-TAAGTGATGGTGATGGTGATG-3′); these were derived from the 3′-ends of the genes (rAtg557930, 5′-6×His-AACAACCATCTCAGCTTCTTTCAC-3′; rAt3g01610, 5′-6×His-CTCGAGGGTAAAAGATG GCC-3′; rAt3g20070, 5′-6×His-ACAATTGAGCTTAATGACTCGAG-3′). Primer pairs fAt5g57930/rAtg557930, fAt3g01610/rAt3g01610, and fAt3g20070/rAt3g20070 were utilized to amplify DNA fragments of 2,171; 3,918; and 2,664 bp, respectively.
For complementation with 6×His-tagged proteins, the PCR product was recovered after gel electrophoresis using QIAquick Gel Extraction kit (Qiagen) and then cloned in Escherichia coli using TOPO XL PCR Cloning kit (Invitrogen) following the manufacturer's protocols. Tagging of At5g57930 with TAP (Rigaut et al., 1999) was done as follows: (1) DNA fragments with the sequence encoding the TAP-tag were obtained by PCR using the primer pair TAP1/TAP2 (5′-CATCACCATCACCATCACATGGAAAAGAGAAGATGGAAAA-3′ and 5′-TCAGGTTGACTTCCCCGC-3′) and plasmid pBS1479 (Puig et al., 2001) as template; (2) another PCR was performed amplifying the gene of interest already tagged with 6 × His from plasmid DNA with primers fAt5g57930 and 6HTAP (5′-TTTTCCATCTTCTCTTTTCCATGTGATGGTGATGGTGATG-3′); (3) the two amplification products were isolated after gel electrophoresis and combined in an overlap PCR reaction with primers fAt5g57930 and TAP2; and (4) the amplification product was recovered after gel electrophoresis and cloned in E. coli as described.
Cloned DNA fragments containing the promoters and tagged genes of interest were isolated from plasmid DNA by double restriction digests and subcloned in E. coli using the binary vector pPZP121 (Hajdukiewicz et al., 1994), which contains the cmr gene for bacterial chloramphenicol resistance and the aacC1 gene conferring plant resistance to gentamycin. The plasmids were transferred into Agrobacterium strain GV3101 and desired colonies selected on Luria-Bertani agar containing 60 mg L−1 chloramphenicol and 50 mg L−1 gentamycin. Transformation of wild-type Arabidopsis (Col-0) plants was performed with the floral dip method (Clough and Bent, 1998). Seeds from Agrobacterium-treated plants were bulk harvested, air-dried and refrigerated. They were then surface sterilized, resuspended in 8 mL 0.8% agar and dispersed on Murashige and Skoog-Glc agar plates containing 100 mg L−1 gentamycin. Plants resistant to gentamycin were transferred to soil and maintained as described. Transgenic plants were used as the male parent in crosses with BASTA-resistant plants heterozygous for the knockout mutation of interest. Progeny seeds were surface sterilized and plated on Murashige and Skoog-Glc agar containing 100 mg L−1 gentamycin and 50 mg L−1 BASTA to select for plants containing both the knockout allele and the transgene.
Supplementary Material
Acknowledgments
We thank Amy Fesler, Clay Holley, Christi Schultz, Sandrine Casanova, Laura Meinke, Thomas Showalter, Ryan Jenlink, and Scott Meinke for assistance with the SeedGenes project and for updating the list of genes with mutant phenotypes, Suparna Mundodi at TAIR for GO annotations, Patricia Ayoubi for assistance with human genome datasets, and the ABRC for providing seed stocks for Salk insertion lines.
This work was supported by the NSF Developmental Mechanisms and Arabidopsis 2010 Programs and the S.R. Noble Foundation (Ardmore, OK).
The online version of this article contains Web-only data.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ (2002) A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA 99: 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Despres B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M (1999) The EMB 506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J 17: 169–179 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L (2003) Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 22: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Goldberg RB, Harada JH (2002) RASPBERRY3 gene encodes a novel protein important for embryo development. Plant Physiol 129: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Ausubel F, Benfey P (2002) Arabidopsis functional genomics. Plant Physiol 129: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y (2002) The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225 [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Griffith ME (2002) “Cross-talk” between cell division cycle and development in plants. Plant Cell 14: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al (2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, et al (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Verma R, Molhoj M, Reiter WD (2003) The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Nakajima R, Meyer HH, Zheng Y (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115: 355–367 [DOI] [PubMed] [Google Scholar]
- Carbonnelle D, Liehr T, Jacquot C, Masson D, Lustenberger P, Denis MG, Roussakis C (1999) Assignment of the serologically defined colon cancer antigen 1 gene (SDCCAG1) to human chromosome band 14q22 by in situ hybridization. Cytogenet Cell Genet 86: 248–249 [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW (1993) Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet 241: 504–514 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96: 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres B, Delseny M, Devic M (2001) Partial complementation of embryo defective mutations: a general strategy to elucidate gene function. Plant J 27: 149–159 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Finkelstein D, Ewing R, Gollub J, Sterky F, Cherry JM, Somerville S (2002) Microarray data quality analysis: lessons from the AFGC project. Plant Mol Biol 48: 119–131 [DOI] [PubMed] [Google Scholar]
- Forler D, Koecher T, Rode M, Gentzel M, Izaurralde E, Wilm M (2003) An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol 21: 89–92 [DOI] [PubMed] [Google Scholar]
- Franzmann L, Patton DA, Meinke DW (1989) In vitro morphogenesis of arrested embryos from lethal mutants of Arabidopsis thaliana. Theor Appl Genet 77: 609–616 [DOI] [PubMed] [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW (1995) Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J 7: 341–350 [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J, et al (2000) Functional genomic analysis of C. elegans chromosome 1 by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res 11: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Wilkinson HA (2003) Functional genomic maps in Caenorhabditis elegans. Curr Opin Cell Biol 15: 206–212 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE (2002) Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32 [DOI] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286: 2165–2169 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Janowitz T, Kneifel H, Piotrowski M (2003) Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett 544: 258–261 [DOI] [PubMed] [Google Scholar]
- Ji Y, Zhang B, Van Horn SF, Warren P, Woodnutt G, Burnham MK, Rosenberg M (2001) Identification of critical Staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293: 2266–2269 [DOI] [PubMed] [Google Scholar]
- Jimenez-Sanchez G, Childs B, Valle D (2001) Human disease genes. Nature 409: 853–855 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Hays JB, Walbot V, Last RL (1997) An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA 94: 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13: 2269–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99: 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D (2004) The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell 16: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D (1999) A combined algorithm for genome-wide prediction of protein function. Nature 402: 83–86 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M (1998) Arabidopsis thaliana: a model plant for genome analysis. Science 282: 662–682 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Meinke LK, Showalter TC, Schissel AM, Mueller LA, Tzafrir I (2003) A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol 131: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Sussex IM (1979) Embryo-lethal mutants of Arabidopsis thaliana: a model system for genetic analysis of plant embryo development. Dev Biol 72: 50–61 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP (2001) CARD15 mutations in Blau syndrome. Nat Genet 29: 19–20 [DOI] [PubMed] [Google Scholar]
- Michaud GA, Snyder M (2002) Proteomic approaches for the global analysis of proteins. Biotechniques 33: 1308–1316 [DOI] [PubMed] [Google Scholar]
- Muralla R, Rogers R, Meinke D (2003) Genetic analysis of condensin (SMC) mutants with a titan seed phenotype. In 14th International Conference on Arabidopsis Research, June 20–24, Madison, WI
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakai K (2000) Protein sorting signals and prediction of subcellular localization. Adv Protein Chem 54: 277–344 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 112: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rivas S, Romeis T, Jones JDG (2002) The Cf-9 disease resistance protein is present in an ∼420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC (1993) The TOUSLED gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75: 939–950 [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Cerny R, Fromm ME (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J 38: 172–181 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA (2003) Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130: 3283–3295 [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan AL, McCubbin AG, Ishimizu T, Wang X, Hu Y, Dowd PE, Ma H, Kao T (2001) Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, a receptor-like kinase of petunia. Plant Physiol 126: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Song SK, Clark SE (2003) POLTERGEIST (POL) and POLTERGEIST-LIKE (PLL1) are essential for the establishment of both shoot and root meristems in Arabidopsis. In 14th International Conference on Arabidopsis Research, June 20–24, Madison, WI
- Springer PS, Holding DR, Groover A, Yordan C, Martienssen RA (2000) The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G1 phase and is required maternally for early Arabidopsis development. Development 127: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Imai A, Matsuyama T, Hanzawa Y, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al (2003) Spermidine synthase genes are essential for survival of Arabidopsis thaliana. Plant Biology 2003, American Society of Plant Biologists, Annual Meeting, July 25–30, Honolulu, HI
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV (1996) A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J 10: 479–489 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes project. Nucleic Acids Res 31: 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T (1998) Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10: 1277–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Flammini A, Maritan A, Vespignani A (2003) Global protein function prediction from protein-protein interaction networks. Nat Biotechnol 21: 697–700 [DOI] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW (1995) Late embryo-defective mutants of Arabidopsis. Dev Biol 16: 311–320 [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inze D, et al (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Wang C, Barry JK, Min Z, Tordsen G, Rao AG, Olsen OA (2003) The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J Biol Chem 278: 34467–34474 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HW, Nussbaumer C, Chao Y, DeLong A (2003) Functional specificity of PP2A regulatory A subunits in Arabidopsis. In 14th International Conference on Arabidopsis Research, June 20–24, Madison, WI
- Zhou X, Kao MCJ, Wong WH (2002) Transitive functional annotation by shortest-path analysis of gene expression data. Proc Natl Acad Sci USA 99: 12783–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.