Abstract
Pectic cell wall polysaccharides owe their high negative charge to the presence of d-galacturonate, a monosaccharide that appears to be present only in plants and some prokaryotes. UDP-d-galacturonate, the activated form of this sugar, is known to be formed by the 4-epimerization of UDP-d-glucuronate; however, no coding regions for the epimerase catalyzing this reaction have previously been described in plants. To better understand the mechanisms by which precursors for pectin synthesis are produced, we used a bioinformatics approach to identify and functionally express a UDP-d-glucuronate 4-epimerase (GAE1) from Arabidopsis. GAE1 is predicted to be a type II membrane protein that belongs to the family of short-chain dehydrogenases/reductases. The recombinant enzyme expressed in Pichia pastoris established a 1.3:1 equilibrium between UDP-d-galacturonate and UDP-d-glucuronate but did not epimerize UDP-d-Glc or UDP-d-Xyl. Enzyme assays on cell extracts localized total UDP-d-glucuronate 4-epimerase and recombinant GAE1 activity exclusively to the microsomal fractions of Arabidopsis and Pichia, respectively. GAE1 had a pH optimum of 7.6 and an apparent Km of 0.19 mm. The recombinant enzyme was strongly inhibited by UDP-d-Xyl but not by UDP, UDP-d-Glc, or UDP-d-Gal. Analysis of Arabidopsis plants transformed with a GAE1:GUS construct showed expression in all tissues. The Arabidopsis genome contains five GAE1 paralogs, all of which are transcribed and predicted to contain a membrane anchor. This suggests that all of these enzymes are targeted to an endomembrane system such as the Golgi where they may provide UDP-d-galacturonate to glycosyltransferases in pectin synthesis.
d-Galacturonate (GalUA) is a negatively charged monosaccharide that forms the backbone of pectic cell wall components either as an α-(1→4)-linked homopolymer in homogalacturonans (HGAs) and substituted galacturonans or as part of the repeat unit [→4)-α-d-GalpUA-(1→2)-α-l-Rhap-(1→] in rhamnogalacturonan-I (Bacic et al., 1988; Carpita and Gibeaut, 1993; Mohnen, 1999; Ridley et al., 2001). GalUA is also present in side chain A of the complex polysaccharide rhamnogalacturonan-II, which plays a unique role in borate-mediated cross-linking within the wall (O'Neill et al., 1996, 2001). Within the eukaryotic domain of organisms, GalUA appears to be confined to plants where it rivals Glc in overall abundance as a primary cell wall component. This suggests that GalUA is uniquely suited to fulfill the structural requirements of specific cell wall polysaccharides such as HGAs, which form cross-linked complexes with Ca2+ ions commonly referred to as the egg-box model (Grant et al., 1973).
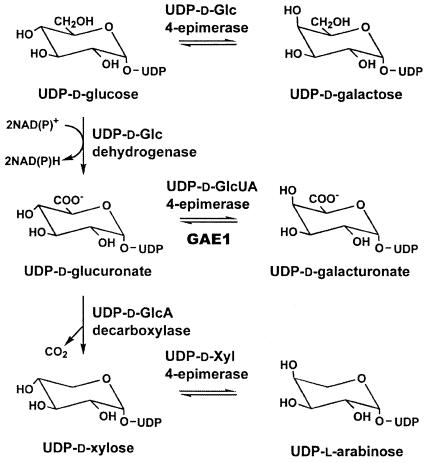
GalUA is synthesized as its UDP derivative by the 4-epimerization of UDP-GlcUA, a nucleotide sugar that is in turn formed either by the dehydrogenation of UDP-Glc (Tenhaken and Thulke, 1996) or by the so-called inositol oxygenation pathway (Loewus et al., 1973). Similar to the 4-epimerization of UDP-Glc and UDP-Xyl, the conversion of UDP-GlcUA to UDP-GalUA is believed to proceed via a transient 4-keto intermediate, which is generated by an enzyme-bound pyridine nucleotide cofactor (Maitra et al., 1974; Feingold and Avigad, 1980). Because of lack of stereospecificity, the resulting oxidation-reduction cycle leads to an equilibrium mixture of the two 4-epimers (Fig. 1).
Figure 1.
The de novo synthesis of UDP-GalUA and other UDP-sugars from UDP-Glc. Three types of UDP-sugar 4-epimerases are involved in the biosynthesis of UDP-Gal, UDP-GalUA, and UDP-Ara from UDP-Glc, UDP-GlcUA, and UDP-Xyl, respectively. All of the 4-epimerization reactions are freely reversible, whereas the dehydrogenation of UDP-Glc to UDP-GlcUA and the decarboxylation of UDP-GlcUA to UDP-Xyl are essentially irreversible.
UDP-GlcUA 4-epimerase activities have been detected in several prokaryotes (Smith et al., 1958; Ankel and Tischer, 1969; Gaunt et al., 1974) and plant species, including mung bean (Phaseolus aureus; Neufeld et al., 1958; Feingold et al., 1960; Mitcham et al., 1991), radish (Raphanus sativus; Neufeld, 1966; Liljebjelke et al., 1995), asparagus (Asparagus officinalis), and spinach (Spinacia oleracea; Neufeld et al., 1958); however, coding regions for these plant enzymes have not previously been described.
A bacterial UDP-GlcUA 4-epimerase (Cap1J) has recently been cloned from Streptococcus pneumoniae and functionally expressed in Escherichia coli (Muñoz et al., 1999). In agreement with the proposed catalytic mechanism, this enzyme belongs to the family of short-chain dehydrogenases/reductases, which catalyze various NAD(P)+-dependent reactions including many nucleotide sugar interconversion steps (Jörnvall et al., 1995). Homologous genes likely to encode UDP-GlcUA 4-epimerase activities have also been described for Sinorhizobium meliloti (Keating et al., 2002) and Klebsiella pneumoniae (Regué et al., 2004); however, functional expression of these two coding regions has not been reported. Based on sequence similarity to the Streptococcus enzyme, we used a bioinformatics approach to identify six likely homologs within the Arabidopsis genome, which were designated GAE1-GAE6 (Reiter and Vanzin, 2001). Here, we report the functional expression of isoform GAE1 in Pichia pastoris and demonstrate that it acts as a nucleotide sugar 4-epimerase specifically interconverting the UDP-GlcUA/UDP-GalUA pair. Unlike their bacterial counterparts, the plant GAEs are predicted to be type II membrane proteins, suggesting that they provide UDP-GalUA directly to Golgi-localized galacturonosyltransferases in pectin synthesis.
RESULTS
Arabidopsis Contains Six Coding Regions with Significant Sequence Similarity to UDP-GlcUA 4-Epimerase from S. pneumoniae
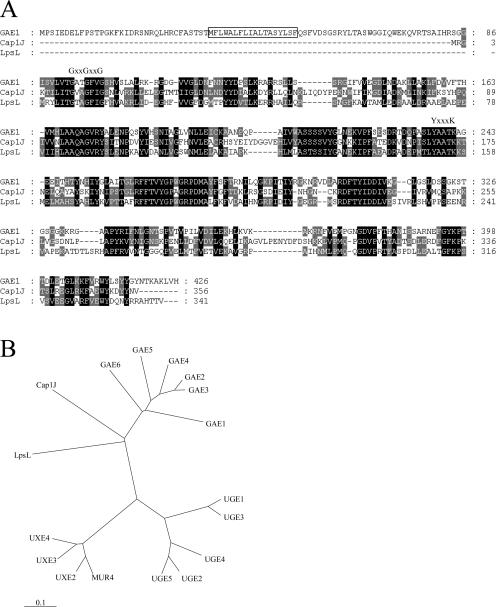
To identify candidate genes for UDP-GlcUA 4-epimerases in Arabidopsis, the amino acid sequence for the soluble UDP-GlcUA 4-epimerase Cap1J from S. pneumoniae (Muñoz et al., 1999) was used to search the Arabidopsis protein database. This search revealed six homologs corresponding to the Arabidopsis Genome Initiative identification numbers At4g30440, At1g02000, At4g00110, At2g45310, At4g12250, and At3g23820. These putative proteins were designated GAE1 through GAE6 for UDP-GlcUA 4-epimerase isoforms 1 to 6 (Fig. 2). All of these coding regions were predicted to be intronless and transcribed based on entries in dbEST. GAE1 and GAE6 were the by far most highly expressed isoforms with 29 and 51 expressed sequence tags (ESTs), respectively, whereas GAE2-GAE5 showed considerably lower expression levels (approximately 5–6 ESTs each). An evaluation of genomic and cDNA sequence databases revealed close homologs in virtually all angiosperms and gymnosperms for which cDNA or genome sequencing projects exist. Examples include potato (Solanum tuberosum), pea (Pisum sativum), rice (Oryza sativa), onion (Allium cepa), and pine (Pinus taeda). Interestingly, GAE-like sequences were also found in the moss Physcomitrella patens (see EST accessions BQ040396 and BJ164276) and the unicellular alga Chlamydomonas reinhardtii (e.g. EST accessions BI725633 and BI726335), whereas no obvious GAE homologs were present in eukaryotes other than plants, which correlates with the apparent absence of GalUA from animals, fungi, and protozoa. A phylogenetic comparison revealed a low but significant degree of sequence similarity of the GAEs to UDP-Glc 4-epimerases and UDP-Xyl 4-epimerases from Arabidopsis and other organisms (Fig. 2B). The GAE1 through GAE6 genes are predicted to encode proteins of 429 to 460 amino acids with deduced molecular masses of 47.4 to 50.6 kD. All putative proteins contain the amino acid sequence motif GxxGxxG that is involved in binding of the cofactor NAD(P)+ (Wierenga et al., 1986) and the YxxxK motif that is part of the catalytic domain of short-chain dehydrogenases/reductases (Jörnvall et al., 1995; Thoden et al., 1997; Fig. 2A). Alignment of the putative Arabidopsis proteins with Cap1J and related bacterial sequences revealed N-terminal extensions of about 90 amino acids in GAE1 through GAE6 (shown for GAE1 in Fig. 2A). Hydropathy profiles of the protein sequences and an evaluation by the Transmembrane Hidden Markov model algorithm (http://www.cbs.dtu.dk/services/TMHMM/) indicated that these N-terminal extensions contain a single transmembrane domain with type II topology, i.e. the catalytic domain is predicted to reside in the lumen of the endomembrane system. N-terminal transmembrane domains were also present in all five GAE homologs identified via TBLASTN searches in the rice genome and for homologs identified in a large variety of other plants based on EST sequence information (data not shown). This suggests that UDP-GlcUA 4-epimerases exist as membrane-anchored isoforms throughout the plant kingdom although the existence of soluble homologs cannot be ruled out.
Figure 2.
Comparison of GAE1 and other UDP-sugar 4-epimerases. A, Alignment of the amino acid sequences of GAE1 and UDP-GlcUA 4-epimerases from S. pneumoniae (Cap1J) and S. meliloti (LspL). Residues conserved between all three sequences are shaded in black, and residues conserved between two of the sequences are shaded in gray. The putative transmembrane domain at the N terminus of GAE1 is boxed, and the conserved GxxGxxG and YxxxK motifs involved in the binding of the NAD(P)+ cofactor and catalysis are indicated above the sequence alignment. B, Phylogenetic tree encompassing GAE1 to GAE6 from Arabidopsis, Cap1J from S. pneumoniae, LpsL from S. meliloti, and nucleotide sugar 4-epimerases from Arabidopsis that are known or believed to be involved in the interconversion of UDP-Glc (UGE1 to UGE5) or UDP-Xyl (MUR4, UXE2, UXE3, and UXE4).
Heterologous Expression of GAE1 in P. pastoris
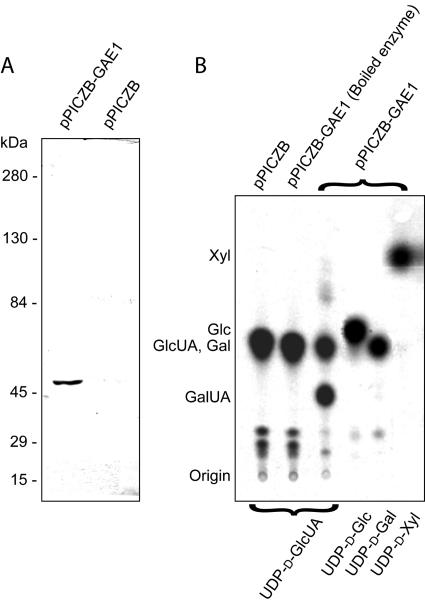
To test GAE1 for nucleotide sugar 4-epimerase activities, the full-length GAE1 coding region was cloned into the P. pastoris pPICZB expression vector, utilizing C-terminal myc epitope and polyhistidine tags. The construct was then transformed into P. pastoris strain KM71. Following a screen for transformants with high expression levels, crude cell-free extracts of P. pastoris expressing GAE1 were analyzed by SDS-PAGE and western blotting using a primary anti-myc antibody and an alkaline phosphatase-conjugated secondary antibody. Results from SDS-PAGE showed no significant difference in the protein profile between cells transformed with pPICZB-GAE1 and pPICZB (empty vector), which indicated a relatively low expression of the recombinant enzyme (data not shown). However, immunoblotting with a myc-specific antibody identified a protein with a molecular mass of approximately 50 kD (Fig. 3A), which is equal to the expected molecular mass of GAE1 plus the myc-epitope and polyhistidine tags (50.1 kD). This protein was not detectable in crude protein extracts from P. pastoris transformed with the empty vector (pPICZB; Fig. 3A).
Figure 3.
Heterologous expression of GAE1 in P. pastoris. A, Western blotting using an antiserum toward the myc epitope. Crude protein extracts from P. pastoris transformed with pPICZB-GAE1 and from an empty vector transformant (pPICZB) were separated by SDS-PAGE and transferred to a membrane prior to immunological detection. The antiserum recognizes a 50-kD protein in the crude extract from a pPICZB-GAE1 transformant, which fits the expected molecular mass of the recombinant myc epitope and polyhistidine tagged GAE1 (50.1 kD). This protein was not detectable in crude extracts from an empty vector transformant. B, Autoradiograph of a thin-layer chromatogram of the 14C-labeled reaction products from assays of UDP-sugar 4-epimerase activities using crude extracts from P. pastoris and UDP-d-[14C]sugar substrates. The types of protein extracts used are indicated at the top, and the nucleotide sugar substrates are indicated at the bottom. Nucleotide sugars were hydrolyzed to monosaccharides prior to TLC analysis. The only interconversion reaction observed was the reversible 4-epimerization of UDP-GlcUA to UDP-GalUA in the presence of recombinant GAE1 protein.
The Recombinant GAE1 Exhibits UDP-d-Glucuronate 4-Epimerase Activity
To assay GAE1 for UDP-sugar 4-epimerase activities, crude P. pastoris protein extracts solubilized with 4% (v/v) CHAPS were incubated with UDP-[14C]sugars, and reaction products were analyzed by thin-layer chromatography (TLC) after hydrolysis to monosaccharides. Assays of crude extracts containing the recombinant GAE1 with UDP-[14C]GlcUA as substrate established a mixture of two products with Rf values typical for GlcUA and GalUA, as determined by authentic standards (Fig. 3B). This indicated that GAE1 has UDP-GlcUA 4-epimerase activity and established an equilibrium between UDP-[14C]GlcUA and UDP-[14C]GalUA without formation of other detectable products. Determination of the GalUA to GlcUA ratio by phosphoimaging of thin-layer chromatograms showed that GAE1 establishes a 1.3:1 equilibrium between UDP-GalUA and UDP-GlcUA (Fig. 3B). Boiled samples and crude extracts from cells transformed with the empty vector (pPICZB) showed no interconversion activity (Fig. 3B). To determine the substrate specificity of GAE1, crude extracts containing recombinant GAE1 were incubated with UDP-[14C]Gal, UDP-[14C]Glc, and UDP-[14C]Xyl. These assays did not produce any detectable products, suggesting that the enzyme is specific for the UDP-GlcUA/UDP-GalUA pair. Positive controls with a plant UDP-Glc 4-epimerase and the MUR4-encoded UDP-Xyl 4-epimerase (Burget et al., 2003) yielded UDP-Gal from UDP-Glc and UDP-Ara from UDP-Xyl as expected (data not shown).
To verify that the reaction products from the GAE1 assays with radiolabeled UDP-GlcUA represented nucleotide sugars rather than monosaccharides or sugar phosphates, the reaction products were purified using anion-exchange chromatography (Bonin and Reiter, 2000; Burget et al., 2003). Ionic strengths required to elute the three sugar types were determined by loading and eluting GalUA, α-GalUA-1-phosphate, and a mixture of unlabeled UDP-GalUA and radiolabeled UDP-GlcUA. GalUA eluted at 50 mm potassium phosphate, pH 3.0, at 4°C, whereas α-GalUA-1-phosphate eluted at 200 mm, and the UDP-GalUA and UDP-GlcUA mixture eluted at 300 mm of this buffer. Once these conditions were established, the reaction products from the assays with GAE1 and UDP-[14C]GlcUA were loaded onto an anion-exchange column and eluted with two column volumes each of 10, 50, 200, and 300 mm potassium phosphate, pH 3.0, at 4°C. More than 90% of the radioactivity eluted with 300 mm of this buffer, indicating that the reaction products represented nucleotide sugars.
GAE1 Expressed in P. pastoris and UDP-GlcUA 4-Epimerases in Arabidopsis Localize to Microsomal Fractions
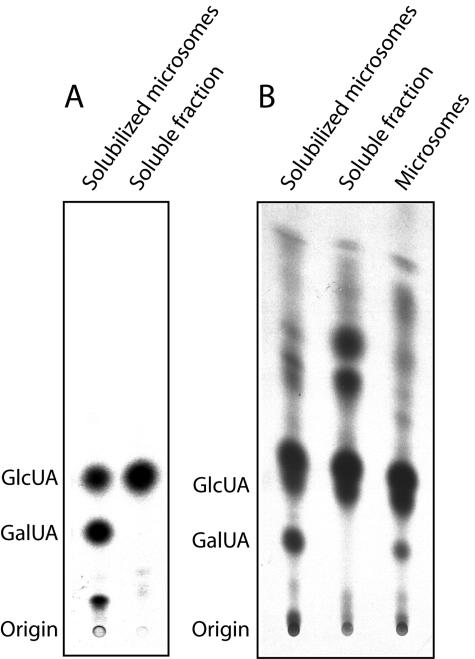
GAE1 and all of its paralogs in Arabidopsis are predicted to be type II membrane proteins. To determine if the recombinant GAE1 localizes to the membrane fractions of P. pastoris, we prepared microsomes from transgenic Pichia lines and assayed both microsomal and soluble fractions for UDP-GlcUA 4-epimerase activity using UDP-[14C]GlcUA as a substrate. Following hydrolysis, products were analyzed by TLC. These assays detected GAE1 activity in the solubilized microsomal fraction but not in the soluble protein fraction (Fig. 4A). To determine if UDP-GlcUA 4-epimerases in Arabidopsis are exclusively targeted to a membrane system, intact microsomes, detergent-solubilized microsomes, and the soluble protein fraction prepared from leaves of 3-week-old plants were assayed for interconversion activities. This experiment showed UDP-GlcUA 4-epimerase activity in solubilized microsomes and to a lesser extent in the intact microsomal fraction (Fig. 4B). Incubation of UDP-GlcUA with the soluble fraction did not yield any detectable GalUA. Instead, products with Rf values typical of Xyl and Ara were formed, which presumably reflects the action of cytoplamic UDP-Xyl synthase and UDP-Xyl 4-epimerase activities (Fig. 4B).
Figure 4.
Localization of recombinant GAE1 and Arabidopsis UDP-GlcUA 4-epimerase activities to microsomal fractions. A, Localization of GAE1 to the membrane fraction of P. pastoris. No UDP-GlcUA 4-epimerase activity could be detected in the soluble protein fraction of P. pastoris expressing GAE1. B, UDP-GlcUA 4-epimerase activity in Arabidopsis is exclusively found in the microsomal fractions.
Enzyme Stability, Cofactors, and Inhibitors of the Recombinant GAE1
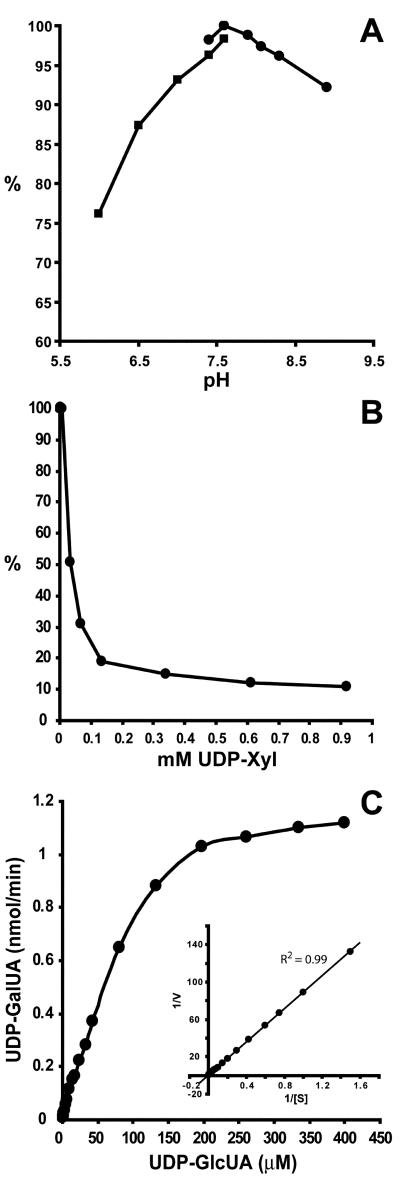
To characterize the recombinant GAE1 in more detail, we initially determined the stability and optimal assay conditions for the enzyme. Storage of crude extracts containing the recombinant GAE1 at 4°C for 16 h reduced GAE1 activity to 30%, and storage at 25°C for 16 h reduced activity to 14% of its original value (data not shown). Freezing and thawing of the crude extract in the absence of cryoprotectants abolished enzymatic activity entirely. In comparison, enzyme stored at −20°C or −80°C in 50% (v/v) glycerol for 7 d retained approximately 80% of its activity. To examine the influence of pH on GAE1 activity, protein preparations were assayed in 50 mm Tris-HCl, pH 7.4 to 8.9, and 50 mm potassium phosphate, pH 6.0 to 7.6. These experiments revealed enzymatic activity over a broad pH range with an optimum at pH 7.6 (Fig. 5A).
Figure 5.
Characterization of the recombinant GAE1. A, pH profile obtained by assaying GAE1 in 50 mm potassium phosphate, pH 6.0 to 7.6, and 50 mm Tris-HCl, pH 7.4 to 8.9. B, Effect of UDP-Xyl on GAE1 activity. C, The effect of UDP-GlcUA concentration on the rate of UDP-GalUA formation. Inset, Lineweaver-Burk plot of the data.
To analyze the effect of oxidized and reduced forms of pyridine nucleotide cofactors, NAD+, NADH, NADP+, and NADPH were added to GAE1 assays and the effects compared to assays conducted in the absence of these compounds. Addition of up to 3 mm NAD+, NADH, NADP+, or NADPH did not affect the interconversion activity, which suggests that GAE1 expressed in P. pastoris already contains tightly bound NAD+ or NADP+ to initiate the interconversion reaction. To test the effects of nucleotides, nucleotide sugars, and monosaccharides, up to 2 mm UDP, UDP-Gal, UDP-Glc, and UDP-Xyl, and up to 5 mm Xyl, Glc, Gal, GlcUA, GalUA, Rha, Ara, Man, or Fuc were included in the reaction mixture. Of these compounds, only UDP-Xyl was found to significantly inhibit GAE1 activity (Fig. 5B). This suggests that the concentration of UDP-Xyl may regulate GAE1 activity in vivo. Addition of EDTA up to a final concentration of 10 mm did not influence GAE1 activity, indicating that the enzyme is not dependent on divalent cations.
Substrate Binding by the Recombinant GAE1
To determine kinetic parameters of GAE1, the rates of product formation were determined as a function of substrate concentration. Computerized nonlinear regression fit and double reciprocal plot (Lineweaver-Burk plot) were constructed using GraphPad Prism Version 3.03, and the equation of the best fit was determined. The apparent Km of GAE1 was 0.19 mm for both plots (R2 = 0.98–0.99; Fig. 5C).
Expression Pattern of GAE1 and Its Homologs in Arabidopsis
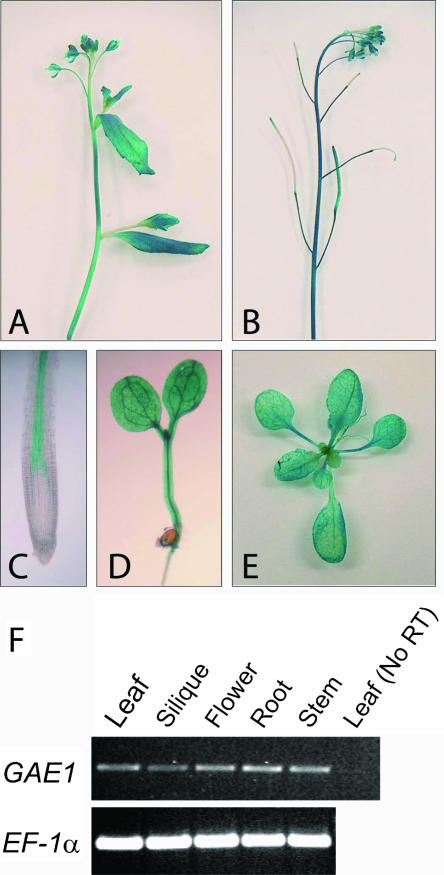
The expression pattern of GAE1 was determined by transforming a GAE1:GUS fusion construct into Arabidopsis and analyzing the transgenic plants for β-glucuronidase (GUS) activity. Results from this experiment showed that GAE1:GUS is strongly expressed in all major tissues of Arabidopsis (Fig. 6) indicating that GAE1 serves as a major housekeeping enzyme. In roots, the expression of GAE1:GUS appeared to be restricted to the stele (Fig. 6C). Semiquantitative reverse transcription (RT)-PCR results confirmed the ubiquitous expression of GAE1 (Fig. 6F). Expression patterns of the other GAE isoforms were evaluated via the massive parallel signature sequencing (MPSS) database for Arabidopsis (http://mpss.udel.edu/at/). Analysis of these data indicated strong expression of GAE1 and GAE6 in all major organs (leaves, roots, inflorescences, and siliques), whereas GAE2 through GAE4 showed moderate expression throughout the plant. GAE5 was moderately expressed in leaves and siliques but not in inflorescences or roots. The overall expression levels taken from the MPSS database were in good agreement with the number of GAE-derived cDNAs in dbEST.
Figure 6.
Expression analysis of GAE1 in Arabidopsis. A to E, Staining of plants transformed with a GAE1:GUS construct for GUS activity. A, Young influorescence. B, Inflorescence with siliques. C, Primary root tip. D, Young seedling. E, Rosette. F, Semiquantitative RT-PCR analysis of GAE1 expression; RNA was isolated from various organs and used for RT-PCR. RNA from rosette leaves was used in a control reaction without reverse transcription.
DISCUSSION
To identify candidate genes for UDP-GlcUA 4-epimerases in the Arabidopsis genome, the amino acid sequence of UDP-GlcUA 4-epimerase from S. pneumoniae (Muñoz et al., 1999) was used to search the Arabidopsis protein database for homologs, which led to the identification of six coding regions designated GAE1 to GAE6. All of these proteins are predicted to belong to the NAD(P)+-dependent family of short-chain dehydrogenases/reductases, which include various dehydrogenases, epimerases, decarboxylases, and dehydratases in nucleotide sugar interconversion reactions (Jörnvall et al., 1995). In contrast to UDP-GlcUA 4-epimerases from bacterial sources, all of the GAE proteins in Arabidopsis and rice are predicted to be type II membrane proteins, and EST data from many other plant species indicate the presence of N-terminal transmembrane domains. This is in line with the observation that all UDP-GlcUA 4-epimerase activity in Arabidopsis is associated with the microsome fraction (Fig. 4B) and that this activity has also been found in membrane fractions from mung bean seedlings (Neufeld et al., 1958; Feingold et al., 1960; Mitcham et al., 1991), radish roots (Neufeld, 1966; Mitcham et al., 1991; Liljebjelke et al., 1995), asparagus shoots, and spinach leaves (Neufeld et al., 1958). Feingold et al. (1960) reported the presence of both soluble and membrane-bound UDP-GlcUA 4-epimerase activities from mung bean seedlings, which may reflect the presence of cytoplasmically targeted isoform(s) or proteolytic events during enzyme purification, which may separate the catalytic domain from the transmembrane domain.
Coding regions for membrane-bound nucleotide sugar interconversion enzymes in Arabidopsis have previously been described for UDP-Xyl 4-epimerase (Burget et al., 2003) and UDP-GlcUA decarboxylases (Harper and Bar-Peled, 2002); however, soluble isoforms for these enzymes also exist (Harper and Bar-Peled, 2002; R. Verma and W.-D. Reiter, unpublished data). Because the MUR4-encoded UDP-Xyl 4-epimerase from Arabidopsis has recently been localized to the Golgi (Burget et al., 2003), it is tempting to speculate that the GAE proteins are also targeted to the Golgi where they could provide UDP-GalUA to galacturonosyltransferases in cell wall synthesis. The structures of MUR4 and GAE1 are surprisingly similar to each other with transmembrane domains of 15 and 18 amino acids, respectively, and N-terminal extensions of 36 amino acids in MUR4 and 34 amino acids in GAE1. This suggests that distinct ancestral nucleotide sugar interconversion enzymes have become fused to similar signal-anchor sequences during plant evolution. The absence of soluble GAE isoforms at least in Arabidopsis (and presumably also in rice) may reflect the exclusive use of UDP-GalUA for the synthesis of pectic cell wall material, obviating the need for a cytoplasmic pool of this nucleotide sugar.
In this study, we found that recombinant GAE1 catalyzed the interconversion of UDP-GlcUA to UDP-GalUA but did not act on UDP-Glc, UDP-Gal, or UDP-Xyl. This strongly suggests that GAE1 shows a high specificity for the UDP-GlcUA/UDP-GalUA pair. The recombinant GAE1 has a pH optimum of 7.6 and an apparent Km of 0.19 mm for UDP-GlcUA (Fig. 5, A and C). These values are similar to the recombinant Cap1J protein from S. pneumoniae, which has a pH optimum of 7.5 and a Km for UDP-GlcUA of 0.24 mm (Muñoz et al., 1999). The equilibrium constant of the GAE1-catalyzed reaction was determined to be 1.3 in the direction of UDP-GalUA, which is in line with an equilibrium constant of 1.1 for a UDP-GlcUA 4-epimerase preparation from mung bean (Feingold et al., 1960) and a value of 1.3 for the recombinant enzyme from S. pneumoniae (Muñoz et al., 1999).
GAE1 activity was strongly inhibited by UDP-Xyl but not by UDP, UDP-Gal, or UDP-Glc. This suggests that UDP-Xyl is a specific and physiologically relevant inhibitor of GAE1, which may serve to regulate cell wall synthesis on the precursor level. For instance, accumulation of UDP-Xyl may reflect low glycosyltransferase activities in the synthesis of Xyl-containing hemicelluloses (xylans and xyloglucans), so inhibition of GAE1 by UDP-Xyl would reduce the availability of UDP-GalUA to galacturonosyltransferases in pectin synthesis. The net result would be a coordinate regulation of the synthesis of pectic and hemicellulosic cell wall components.
The expression pattern of GAE1 was analyzed with a promoter:GUS construct, indicating that GAE1 was expressed in all organs consistent with a function as a housekeeping gene involved in the synthesis of an essential cell wall precursor (Fig. 6, A–E). This expression pattern was furthermore confirmed by RT-PCR analysis (Fig. 6F) and data from the MPSS project. The expression of GAE1 in all tissues raises some questions about the function of GAE2 to GAE6. Our recent observation that GAE6 expressed in P. pastoris has UDP-GlcUA 4-epimerase activity (M. Mølhøj, R. Verma, and W.-D. Reiter, unpublished data) suggests that all of the GAE genes encode proteins with this enzymatic activity. The substantial overlap in GAE expression suggests genetic redundancy; however, some of the isoforms may be expressed in specific cell types or tissues, or be localized to different compartments within the endomembrane system. This could have interesting consequences for the spatial organization of nucleotide sugar interconversion enzymes and their interaction with glycosyltransferases in cell wall synthesis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants were grown at 23°C and 60% to 70% humidity under continuous fluorescent light (60–70 μmol m−2 s−1). Arabidopsis of the ecotype Columbia was used to obtain protein, DNA, and RNA preparations.
Construction of Plasmids and Genetic Transformation of Escherichia coli, Pichia pastoris, and Arabidopsis
To generate an expression construct for GAE1, the full-length GAE1 gene was PCR amplified from genomic DNA using the oligonucleotides 5′-TCACAGGTACCAATAATGCCTTCAATAGAAGATGA-3′ and 5′-CTACATCTAGATGTACAAGCTTGGCTTTAGTATTG-3′ (KpnI and XbaI sites engineered into the primers are underlined). After cleavage of the PCR product with KpnI and XbaI, it was cloned into the Pichia pastoris vector pPICZB (Invitrogen, Carlsbad, CA) cleaved with the same two enzymes. This cloning strategy established a translational fusion between GAE1 and the C-terminal myc epitope and polyhistidine tags built into the vector. The resulting construct pPICZB-GAE1 was linearized with BstXI and transformed into P. pastoris strain KM71 by electroporation.
To analyze the expression pattern of GAE1, approximately 2.3 kb of nucleotide sequences upstream of the coding region were PCR amplified from genomic DNA using the oligonucleotides 5′-TATTGTCTGCAGAAGAAAACTAAACCGGGAAACTATTGATTACC-3′ and 5′-AGTAACCCATGGTCATAATTTAATTAAACTCTCTTTACAACAAAAATTC-3′ (PstI and NcoI sites are underlined) and cloned in frame with the uidA (GUS) reporter gene in the binary vector pCAMBIA1301 (CAMBIA, Canberra, Australia) giving pCAMBIA1301-GAE1.
PCR reactions were performed using PfuTurbo DNA polymerase (Stratagene, La Jolla, CA), and the sequences of all constructs were verified using the ABI Prism Big Dye Terminator cycle sequencing reaction kit (Perkin-Elmer Applied Biosystems, Foster City, CA) and an ABI Prism 377 DNA sequencer. Arabidopsis was transformed by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101 (pMP90).
Screening of P. pastoris Transformants
Twenty-four individual P. pastoris transformants were screened for expression of the recombinant GAE1 protein as follows: 25 mL of BMGY (buffered complex glycerol media) supplemented with 100 μg mL−1 zeocin in a 200-mL flask was inoculated with a single colony and grown at 28°C in a shaking incubator (260 rpm) for 24 h until an OD600 of approximately 2 was reached. Cells were harvested by centrifugation and resuspended in 50 mL of BMMY (buffered complex methanol media) at an OD600 of 1. Two-milliliter aliquots were removed every 24 h, and methanol was added to a final concentration of 0.5% (v/v) at these time points to maintain induction conditions for a total of 96 h. Cells were pelleted by centrifugation and stored at −80°C. Crude cell-free extracts were prepared by washing cells once in breaking buffer [50 mm Tris-HCl, pH 7.6, 1 mm EDTA, 2 mm dithiothreitol (DTT), 5% (v/v) glycerol] followed by sequential vortexing for 30 s and placing on ice for a total of eight cycles in 200 μL of breaking buffer containing 4% (w/v) CHAPS (Sigma, St. Louis), 1× Complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany), and an equal volume of acid-washed glass beads (425–600 μm; Sigma). After centrifugation for 10 min at 13,000 rpm in a microfuge, the cleared supernatants were transferred to fresh tubes and stored at −20°C.
Protein extracts were analyzed by immunodot blotting using an Immobilon-P transfer membrane (Millipore, Bedford, MA), a Bio-Dot apparatus (Bio-Rad Laboratories, Hercules, CA), and a 1:2,000 dilution of an alkaline phosphatase-conjugated anti-myc antibody (Invitrogen). The incubations were carried out in 50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 2% (v/v) Tween 20, and 5% (v/v) horse serum. Alkaline phosphatase was detected colorimetrically using 0.03% (w/v) 4-nitro blue tetrazolium chloride and 0.02% (w/v) 5-bromo-4-chloro-3-indolyl-phosphate (p-toluidine salt) in AP buffer (0.1 m Tris, pH 9.5, 0.1 m NaCl, 5 mm MgCl2). A transformant expressing high level of recombinant GAE1 after 2 d of induction was chosen for large-scale expression (200 mL). Cells were stored at −80°C and used for the preparation of crude protein extracts containing recombinant GAE1.
SDS-PAGE and Western Analysis
SDS-PAGE was performed in 10% polyacrylamide gels, and proteins were stained with 0.1% (w/v) Coomassie Brilliant Blue R-250 in methanol:acetic acid:water (9:2:9, v/v/v) and destained in the same solution without the dye. For western blots, proteins were transferred onto Immobilon-P transfer membranes (Millipore) by semidry electroblotting. For immunological detection, membranes were incubated overnight at 4°C with a 1:3,000 dilution of rabbit anti-myc primary antibody (Invitrogen) followed by incubation with a 1:2,000 dilution of an alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Sigma) for 3 h at room temperature in 50 mm Tris-HCl, pH 7.4, 500 mm NaCl, 2% (v/v) Tween 20, and 5% (v/v) horse serum. Alkaline phosphatase was detected colorimetrically as described above.
Assay of GAE1 Activity
Crude P. pastoris extracts containing 20 μg of protein were mixed with 25 nCi of UDP-[14C]GlcUA (300 mCi mmol−1), UDP-[14C]Gal (300 mCi mmol−1), UDP-[14C]Glc (300 mCi mmol−1), or UDP-[14C]Xyl (238 mCi mmol−1; American Radiolabeled Chemicals, St. Louis) contained in 50 μL of 50 mm Tris-HCl, pH 7.6, and incubated at 25°C overnight. The effects of pyridine nucleotide cofactors, UDP, UDP-sugars, monosaccharides, and EDTA were analyzed by incubating 20 μg extracted protein with 50 mm Tris-HCl, pH 7.6, and UDP-[14C]GlcUA for 30 s at 25°C in the presence or absence of additives. Nucleotide sugars were hydrolyzed by the addition of trifluoroacetic acid to 1.6 m final concentration and incubation at 95°C for 30 min. Samples were dried under vacuum and resuspended in 15 μL of 80% (v/v) ethanol. Resultant monosaccharides were separated by TLC on silica-coated plates (Aldrich, Milwaukee, WI) in a 6:2:1 (v/v/v) mixture of 1-propanol, saturated ammonia solution, and water. Radioactivity was visualized by phosphor imaging (Bio-Rad Laboratories, Hercules, CA) and quantified using Molecular Analyst Software (Bio-Rad Laboratories). Authentic sugar standards run in parallel were stained with aniline-hydrogen phthalate (Fry, 1988). The recombinant UDP-Xyl 4-epimerase MUR4 (Burget et al., 2003) and a recombinant UDP-Glc 4-epimerase were used as positive controls for the corresponding 4-epimerase assays. All assays were repeated at least once. Measurements of protein concentrations were performed using a bicinchoninic acid kit (Sigma).
For kinetic evaluation, the recombinant GAE1 was assayed using UDP-[14C]GlcUA (300 mCi mmol−1) at substrate concentrations below 6 μm, and a mix of labeled and unlabeled UDP-GlcUA at higher substrate concentrations. Linearity with respect to protein concentration and time was first established, and the concentration of radiolabeled UDP-GlcUA was then independently varied while maintaining all other parameters (20 μg extracted protein in 50 mm Tris-HCl, pH 7.6, with an incubation time of 30 s).
Anion Exchange Chromatography
Three separate Superclean LC-SAX solid-phase columns (Supelco, Bellefonte, PA) were used to determine the ionic strengths required to elute 100 μg of GalUA, 100 μg of α-GalUA-1-phosphate (Sigma), and a mixture of 100 μg of unlabeled UDP-GalUA and 100 nCi of UDP-[14C]GlcUA, respectively. The labeled nucleotide-sugars were identified by liquid scintillation counting and the unlabeled nucleotide sugars, monosaccharides, and sugar phosphates were identified by total uronic acid assays of the eluted fractions (Filisetti-Cozzi and Carpita, 1991).
Preparation of Microsomes from Pichia and Arabidopsis and Assay for UDP-GlcUA 4-Epimerase Activity
Cells from 50 mL of a P. pastoris flask culture induced with methanol for 2 d were centrifuged at 2,500g for 5 min and resuspended in 10 mL homogenization buffer (0.33 m Suc, 50 mm Tris-HCl, pH 7.6, 1 mm MgCl2, 1 mm EDTA, 1 mm DTT, 1× Complete protease inhibitor cocktail). The suspension was centrifuged at 2,500g for 5 min. The pellet was resuspended in 5 mL of homogenization buffer, and 2 mL of acid-washed 425 to 600 μm glass beads were added. The mixture was vortexed eight times for 1 min each, with 2 min on ice between each vortexing. The supernatant was centrifuged at 2,500g for 2 min to remove beads, then at 14,000g for 20 min to remove cell debris, and then finally at 150,000g for 60 min to obtain the membrane fraction. The microsomal pellet was resuspended by mixing in 50 μL of resuspension buffer (50 mm Tris-HCl, pH 7.6, 1 mm EDTA, 2 mm DTT, 5% [v/v] glycerol, 1× Complete protease inhibitor cocktail). The supernatant of the 150,000g centrifugation step was used as the soluble protein preparation. The microsomal membranes were solubilized with 4% (w/v) CHAPS, incubated for 20 min on ice, and centrifuged at 50,000g for 20 min. Ten micrograms of protein from each of the soluble preparation and the solubilized microsomes were used to assay for GAE1 activity in 50 μL of 50 mm Tris-HCl, pH 7.6, 25 nCi of UDP-[14C]GlcUA and incubated at 25°C for 16 h.
As with P. pastoris, microsomes from wild-type Arabidopsis plants were obtained by differential centrifugation. All steps of the homogenization and isolation procedure were performed on ice or at 4°C. Microsomes were isolated from rosette leaves of 3-week-old plants. Fifteen grams fresh weight of leaf material were ground with a mortar and pestle in 15 mL of homogenization buffer (0.4 m Suc, 50 mm Tris-HCl, pH 7.6, 1 mm MgCl2, 1 mm EDTA, 1 mm DTT, 1× Complete protease inhibitor cocktail). The suspension was homogenized in a blender for 10 s and filtered through a nylon mesh cloth (30-μm mesh size). Microsomes were isolated by differential centrifugation in three centrifugation steps: at 5,000g for 10 min, 15,000g for 10 min, and 100,000g for 1 h. The pellet and supernatant of the final centrifugation step were used as the microsome and soluble protein preparations, respectively. The microsomal pellet was resuspended by mixing in resuspension buffer [50 mm Tris-HCl, pH 7.6, 1 mm EDTA, 10 mm KCl, 2 mm DTT, 5% (v/v) glycerol] at 0.2 mL buffer g−1 fresh weight. A fraction of the microsomal membranes was solubilized with 0.5% (v/v) CHAPS and incubated for 20 min on ice. The suspension was centrifuged at 100,000g for 30 min. Fifteen micrograms of protein from each of the soluble preparation, microsomes, and solubilized microsomes were used to assay for UDP-GlcUA 4-epimerase activity. Assays were conducted in 50 μL of 50 mm Tris-HCl, pH 7.6, 25 nCi of UDP-[14C]GlcUA, 2 mm NAD+ and incubated at 25°C for 2 h.
Histochemical GUS Assays
Arabidopsis plants transformed with pCAMBIA1301-GAE1 were selected on 0.8% (w/v) agar plates containing half-strength MS medium (Sigma), 3% (w/v) Suc, 50 μg mL−1 hygromycin B (Calbiochem, La Jolla, CA), and 500 μg mL−1 vancomycin (Sigma). The T2 generation was analyzed for GUS staining using a protocol adapted from Jefferson et al. (1987). To determine the expression pattern of GAE1:GUS, 12 individual transformants were analyzed.
Semiquantitative RT-PCR
To determine the presence of GAE1 mRNA in various organs, the GAE1 transcript was amplified by RT-PCR using total RNA from all major plant organs. Total RNA was isolated from shoot tissues of 3-week-old soil-grown plants and from roots of axenically grown plants using the RNeasy plant mini kit (Qiagen, Valencia, CA) following the manufacturer's protocol. Altogether 1.5 μg total RNA from each tissue was treated with one unit of RNase free DNase-I (Promega, Madison, WI) for 30 min at 37°C prior to RT-PCR. The GAE1 transcript was amplified from the DNase-I treated total RNA (200 ng) using the OneStep RT-PCR kit (Qiagen) and the oligonucleotides 5′-TTAATTAAATTATGCCTTCAATAGAAGATGAGC-3′ and 5′-TTAATGTACAAGCTTGGCTTTAGTATTGTATCC-3′ (RT at 50°C for 30 min followed by 28 PCR cycles: 94°C for 45 s, 52°C for 30 s, and 68° C for 100 s). To verify the success of the DNase-I treatment, total RNA from leaves was used as a control without RT. Equal loading of RNA was verified by RT-PCR amplification of an 808-bp fragment from the coding region for EF-1α (accession no. X16432), using the oligonucleotides 5′-ATGCCCCAGGACATCGTGATTTCAT-3′ and 5′-TTGGCGGCACCCTTAGCTGGATCA-3′ and employing the same RT-PCR conditions as described above. The amplification products were resolved by 1% agarose gel electrophoresis and visualized by ethidium bromide staining.
Accession Numbers
GenBank accession numbers or AGI identification numbers for coding regions included in Figure 2 are as follows: GAE1, At4g30440; GAE2, At1g02000; GAE3, At4g00110; GAE4, At2g45310; GAE5, At4g12250; GAE6, At3g23820; UGE1, At1g12780; UGE2, At4g23920; UGE3, At1g63180; UGE4, At1g64440; UGE5, At4g10960; MUR4, At1g30620; UXE2, At2g34850; UXE3, At5g44480; UXE4, At4g20460; Cap1J, CAB05928; and LpsL, CAA10917. Note that our numbering convention for the putative UDP-Xyl 4-epimerases UXE2, UXE3, and UXE4 differs from that published by Seifert (2004). To avoid inconsistencies in the usage of these gene names, Dr. Seifert has kindly agreed to adopt the above numbering system (G. Seifert, personal communication).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY661562.
Acknowledgments
We thank the Center for the Application of Molecular Biology to International Agriculture (CAMBIA) for plant transformation vectors and Georg Seifert for his help in resolving a nomenclature issue.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–95ER20203) and by a fellowship from the Danish Agricultural and Veterinary Research Council (grant no. SJVF 23000237 to M.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043745.
References
- Ankel H, Tischer RG (1969) UDP-d-glucuronate 4-epimerase in blue-green algae. Biochim Biophys Acta 178: 415–419 [DOI] [PubMed] [Google Scholar]
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 14. Academic Press, New York, pp 297–371
- Bonin CP, Reiter W-D (2000) A bifunctional epimerase-reductase acts downstream of the MUR1 gene product and completes the de novo synthesis of GDP-l-fucose in Arabidopsis. Plant J 21: 445–454 [DOI] [PubMed] [Google Scholar]
- Burget EG, Verma R, Mølhøj M, Reiter W-D (2003) The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Feingold DS, Avigad G (1980) Sugar nucleotide transformations in plants. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 3. Academic Press, New York, pp 101–170
- Feingold DS, Neufeld EF, Hassid WZ (1960) The 4-epimerization and decarboxylation of uridine diphosphate d-glucuronic acid by extracts from Phaseolus aureus seedlings. J Biol Chem 235: 910–913 [PubMed] [Google Scholar]
- Filisetti-Cozzi TMCC, Carpita NC (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197: 157–162 [DOI] [PubMed] [Google Scholar]
- Fry SC (1988) The Growing Plant Cell Wall: Chemical and Metabolic Analysis. John Wiley and Sons, New York
- Gaunt MA, Maitra US, Ankel H (1974) Uridine diphosphate galacturonate 4-epimerase from the blue-green alga Anabaena flos-aquae. J Biol Chem 249: 2366–2372 [PubMed] [Google Scholar]
- Grant GT, Morris ER, Rees DA, Smith PJC, Thom D (1973) Biological interactions between polysaccharides and divalent cations: the egg-box model. FEBS Lett 32: 195–198 [Google Scholar]
- Harper AD, Bar-Peled M (2002) Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130: 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34: 6003–6013 [DOI] [PubMed] [Google Scholar]
- Keating DH, Willits MG, Long SR (2002) A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J Bacteriol 184: 6681–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljebjelke K, Adolphson R, Baker K, Doong RL, Mohnen D (1995) Enzymatic synthesis and purification of uridine diphosphate [14C]galacturonic acid: a substrate for pectin biosynthesis. Anal Biochem 225: 296–304 [DOI] [PubMed] [Google Scholar]
- Loewus F, Chen MS, Loewus MW (1973) The myo-inositol oxidation pathway to cell wall polysaccharides. In F Loewus, ed, Biogenesis of Plant Cell Wall Polysaccharides. Academic Press, New York, pp 1–27
- Maitra US, Gaunt MA, Ankel H (1974) The mechanism of uridine diphosphate-sugar-4-epimerase reactions. Isotope discrimination with 4-tritiated substrates. J Biol Chem 249: 3075–3078 [PubMed] [Google Scholar]
- Mitcham EJ, Gross KC, Wasserman BP (1991) Synthesis of uridinediphospho-[U-14C]-d-galacturonic acid by enzyme particulate fractions and purification via high performance liquid chromatography. Phytochem Anal 2: 112–115 [Google Scholar]
- Mohnen D (1999) Biosynthesis of pectins and galactomannans. In BM Pinto, ed, Comprehensive Natural Products Chemistry, Vol 3, Carbohydrates and Their Derivatives Including Tannins, Cellulose, and Related Lignins. Elsevier, Oxford, pp 497–527
- Muñoz R, López R, de Frutos M, García E (1999) First molecular characterization of a uridine diphosphate galacturonate 4-epimerase: an enzyme required for capsular biosynthesis in Streptococcus pneumoniae type 1. Mol Microbiol 31: 703–713 [DOI] [PubMed] [Google Scholar]
- Neufeld EF (1966) UDP-d-galacturonic acid 4-epimerase from radish roots. Methods Enzymol 8: 276–277 [Google Scholar]
- Neufeld EF, Feingold DS, Hassid WZ (1958) Enzymatic conversion of uridine diphosphate d-glucuronic acid to uridine diphosphate galacturonic acid, uridine diphosphate xylose, and uridine diphosphate arabinose. J Am Chem Soc 80: 4430–4431 [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill AG, Albersheim P (1996) Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. J Biol Chem 271: 22923–22930 [DOI] [PubMed] [Google Scholar]
- Regué M, Hita B, Piqué N, Izquierdo L, Merino S, Fresno S, Benedí VJ, Tomás JM (2004) A gene, uge, is essential for Klebsiella pneumoniae virulence. Infect Immun 72: 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W-D, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95–113 [PubMed] [Google Scholar]
- Ridley BL, O'Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Seifert GJ (2004) Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol 7: 277–284 [DOI] [PubMed] [Google Scholar]
- Smith EE, Mills GT, Bernheimer HP, Austrian R (1958) The presence of a uronic acid epimerase in a strain of pneumococcus type I. Biochim Biophys Acta 29: 640–641 [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Thulke O (1996) Cloning of an enzyme that synthesizes a key nucleotide-sugar precursor of hemicellulose biosynthesis from soybean: UDP-glucose dehydrogenase. Plant Physiol 112: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Hegeman AD, Wesenberg G, Chapeau MC, Frey PA, Holden HM (1997) Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 36: 6294–6304 [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Terpstra P, Hol WG (1986) Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol 187: 101–107 [DOI] [PubMed] [Google Scholar]