Abstract
NAD kinase (NADK; ATP:NAD 2′-phosphotransferase, EC 2.7.1.23), an enzyme found in both prokaryotes and eukaryotes, generates the important pyridine nucleotide NADP from substrates ATP and NAD. The role of NADKs in plants is poorly understood, and cDNAs encoding plant NADKs have not previously been described to our knowledge. We have cloned two cDNAs from Arabidopsis predicted to encode NADK isoforms, designated NADK1 and NADK2, respectively. Expressed as recombinant proteins in bacteria, both NADK1 and NADK2 were catalytically active, thereby confirming their identity as NADKs. Transcripts for both isoforms were detected in all tissues examined and throughout development. Although the predicted catalytic regions for NADK1 and NADK2 show sequence similarity to NADKs from other organisms, NADK2 possesses a large N-terminal extension that appears to be unique to plants. Using recombinant glutathione-S-transferase fusion proteins and calmodulin (CaM)-affinity chromatography, we delineated a Ca2+-dependent CaM-binding domain to a 45-residue region within the N-terminal extension of NADK2. Although recombinant NADK2 was not responsive to CaM in vitro, immunoblot analysis suggests that native NADK2 is a CaM-binding protein. In Arabidopsis crude extracts, CaM-dependent NADK activity was much greater than CaM-independent activity throughout development, particularly in young seedlings. A native CaM-dependent NADK was partially purified from Arabidopsis seedlings (KmNAD = 0.20 mM, KmMg2+−ATP = 0.17 mM). The enzyme was fully activated by conserved CaM (S0.5 = 2.2 nm) in the presence of calcium but displayed differential responsiveness to eight CaM-like Arabidopsis proteins. Possible roles for NADKs in plants are discussed in light of our observations.
NAD kinase (NADK; ATP:NAD 2′-phosphotransferase, EC 2.7.1.23) catalyzes the de novo synthesis of the important diphosphopyridine nucleotide NADP from the substrates NAD and ATP. NADKs have been found in all organisms examined to date, suggesting a fundamental role for the enzyme (McGuinness and Butler, 1985; Zielinski, 1998; Kawai et al., 2001a, 2001b; Lerner et al., 2001). While the importance of the pyridine nucleotides NAD(H) and NADP(H) in energy metabolism and reductive biosynthesis is well established, the role of NADK itself remains unclear. Furthermore, while the enzyme has been studied for decades in various organisms, it was only recently that cDNAs encoding NADKs were cloned from bacteria (Kawai et al., 2000, 2001a), yeast (Kawai et al., 2001b), and human (Lerner et al., 2001) sources. Until this study, a cDNA encoding a plant NADK had not been isolated, and the lack of molecular tools has hindered research progress on the roles of NADKs in plants. Nevertheless, there is considerable interest in plant NADKs mostly arising from the observation that plants possess both calmodulin (CaM)-independent and CaM-regulated NADK isoforms (Roberts and Harmon, 1992; Harding et al., 1997; Pou De Crescenzo et al., 2001). Indeed, NADK was the first CaM-regulated enzyme identified in plants (Muto and Miyachi, 1977; Anderson et al., 1980) and remains among the most studied at the biochemical level (Zielinski, 1998). CaM is a ubiquitous and critical Ca2+ sensor found in all eukaryotes where it regulates a diverse array of targets involved in cytoskeletal rearrangements, signaling, metabolism, transcription, and ion homeostasis (Snedden and Fromm, 2001; Yang and Poovaiah, 2003). In plants, a CaM-dependent NADK has been suggested to play a metabolic role (Jarrett et al., 1982) and to participate in Ca2+-mediated cellular defense against invading pathogens by helping to provide reductant (indirectly) for the NADPH-dependent oxidative burst (Harding et al., 1997). It is interesting to note that early reports suggested NADKs found in neutrophils (Williams and Jones, 1985) and sea urchin eggs (Epel et al., 1981; Iwasa and Mohri, 1983), both systems capable of generating a substantial oxidative burst, are activated by CaM. However, it remains unclear whether this modulation is direct or indirect (e.g. via CaM-dependent kinase), and recent studies with a human recombinant NADK did not reveal any effect of CaM on in vitro NADK activity (Lerner et al., 2001). Conversely, the modulation of plant NADK by CaM is well established (for review, see Zielinski, 1998). It is noteworthy that plants possess unique CaM targets not found in animals and have also evolved targets whose animal homologs are not CaM regulated (Reddy et al., 2002; Yang and Poovaiah, 2003). Moreover, plants possess not only the evolutionarily conserved form of CaM but also an extended family of CaM-like proteins (CMLs) not present in other organisms (McCormack and Braam, 2003). Physiological roles for many of these putative Ca2+ sensors remain to be established, but several show differential regulation of plant NADK (Liao et al., 1996; Lee et al., 1997). Furthermore, the potent Ca2+ mobilizing agents, cADP-Rib and nicotinic acid adenine dinucleotide (NAADP) are derived from NAD and NADP, respectively, and play important roles in intracellular Ca2+ signaling in both plants (Navazio et al., 2000) and animals (Patel et al., 2001). It remains an interesting and open question as to why plants have evolved a CaM-dependent means of generating NADP and how this process fits into other Ca2+-mediated signal transduction pathways. As a step toward answering this question, we have cloned cDNAs encoding two different NADKs from Arabidopsis, termed NADK1 and NADK2. Both isoforms show sequence similarity with NADKs from other organisms. In addition, NADK2 contains a unique N-terminal extension that includes a CaM-binding domain. We provide evidence that Arabidopsis possesses both CaM-dependent and -independent NADKs, and we discuss possible roles for NADKs in plants.
RESULTS
Isolation of cDNAs and Sequence Analyses
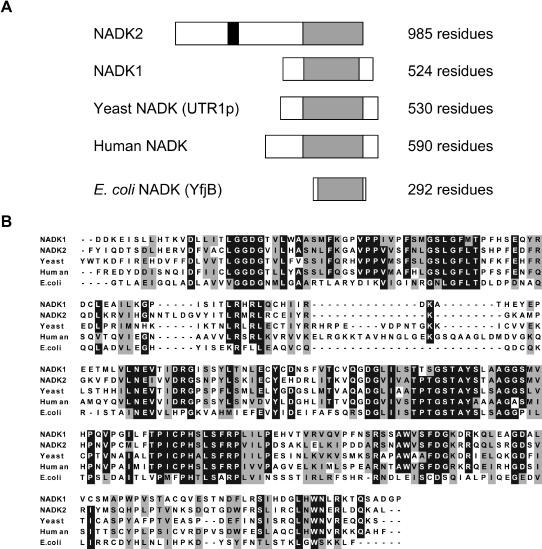
Although NADK activity has been studied in various plant species for several decades, no molecular data were available. Consequently, we set out to clone and characterize NADKs from the model plant Arabidopsis. Using sequences of the recently cloned human (Lerner et al., 2001), yeast (Kawai et al., 2001b), and bacteria (Kawai et al., 2001b) NADKs, database searches identified two putative NADKs in the Arabidopsis genome. We have named these genes NADK1 and NADK2, and they correspond to loci At3g21070 and At1g21640, respectively. Based upon DNA sequence (including full-length mRNA transcript data, AF337912) information derived from database searches, we designed specific oligonucleotide primers to isolate the full-length cDNA for NADK2 by PCR (see “Materials and Methods”). DNA sequencing and analysis (BLAST, National Center for Biotechnology Information) of the subcloned PCR product confirmed it as NADK2 with complete identity to the full-length sequence deposited in GenBank (AF337912). Similarly, DNA sequencing of an EST clone (received from Kasuza DNA Research Institute, Kisarazu, Japan) for NADK1 (AV547501; Asamizu et al., 2000) agreed with the predicted full-length cDNA (AY090937) deposited in GenBank. The protein sequence for NADK1 differed slightly from the most recent annotation from The Institute for Genomic Research (http://www.tigr.org) predicted for this locus (At3g1070) based upon genomic DNA that includes an additional six amino acids in the central part of the protein. Based upon our cDNA data, the predicted amino acid sequences for both NADK1 and NADK2 are presented (Supplemental Fig. 1, available at www.plantphysiol.org). The cDNAs are predicted to encode proteins of 524 residues and 985 residues corresponding to molecular masses of approximately 58.2 kD and 109.2 kD for NADK1 and NADK2, respectively. Figure 1A shows a schematic presentation of the structural organization for various NADKs (Pfam, PF01513; Bateman et al., 2000). Note the significant N-terminal extensions for both NADK1 and, in particular, NADK2, relative to other NADKs. Interestingly, database searches for proteins with sequence similarity to the N-terminal region of NADK2 identified putative homologs only among higher plants, including rice (Oryza sativa), maize (Zea mays), tomato (Lycopersicon esculentum), and soybean (Glycine max; data not shown), and it would thus appear that this large isoform is unique to, and conserved among, plants. Furthermore, the position of the experimentally delineated CaM-binding domain lies within this N-terminal extension of NADK2. Putative NADK1 homologs from various plant species, including rice, tomato, and soybean, were revealed by genomic and expressed sequence tag (EST) database searches (data not shown). With the exception of the conserved C-terminal region, no other identifiable structural motifs were predicted in either NADK1 or NADK2. A more detailed sequence alignment of the C-terminal regions, believed to include the catalytic domain (Kawai et al., 2001a), for various NADKs is presented in Figure 1B. The NADK motifs, XXX-XXGGDG-XL and GDXXX-TPTGSTAY (where X represents a hydrophobic residue) are also largely conserved in both NADK1 and NADK2 (Kawai et al., 2001a). It is noteworthy that the genomes of yeast (Kawai et al., 2001b; Outten and Culotta, 2003) and Arabidopsis (this work) predict multiple NADKs, whereas only a single NADK has been described in humans (Lerner et al., 2001).
Figure 1.
Sequence comparison of Arbidopsis NADKs NADK1 and NADK2 with NADKs from other species. A, Schematic representation of the structural organization of various NADKs: Arabidopsis NADK1, NADK2; yeast NADK (UTR1p; P21373); human NADK (NP_075394); E. coli NADK (YFjB; NP_417105). Gray bars indicate the predicted catalytic domain, white bars indicate the nonconserved regions, and the black bar denotes the CaM-binding domain of NADK2. B, Alignment of the conserved catalytic domains of the NADKs described in A; NADK1 (residues 271–534), NADK2 (residues 731–985), yeast (residues 191–451), human (residues 309–581), E. coli (residues 55–292). Black and gray boxes indicate identical or similar residues, respectively, conserved among at least 60% of the sequences, whereas dashes indicate gaps introduced to optimize the alignment.
Characterization of Recombinant and Native NADK Activity and Identification of NADK2 as CaM Binding
Although a number of reports in the literature have described NADK activity in different plant species (Muto and Miyachi, 1977; Dieter and Marme, 1984; Delumeau et al., 2000; Pou De Crescenzo et al., 2001), evidence of an NADK in Arabidopsis has not previously been reported. Consequently, we examined Arabidopsis extracts for NADK activity. We observed a substantial level of activity in crude extracts, most of which (>90%) was dependent upon Ca2+ and CaM for activity. Data describing the partial purification (about 400-fold) of a CaM-dependent NADK from Arabidopsis seedlings grown in liquid culture are presented in Table I. The CaM-affinity chromatography stage gave the greatest fold purification and resulted in activity that was completely Ca2+- and CaM-dependent. As previously reported for other plant NADKs (Jarrett et al., 1982; Roberts and Harmon, 1992; Delumeau et al., 2000), Arabidopsis native NADK was very labile, and much of the activity was lost during purification. Nevertheless, we obtained preparations with specific activities comparable to that observed in other reports on plant NADKs (Harmon et al., 1984; Delumeau et al., 1998).
Table I.
Purification of CaM-dependent NADK from Arabidopsis
| Step | Volume | Protein | Activity | Specific Activity | Purification | Yield |
|---|---|---|---|---|---|---|
| mL | mg | unitsa | units mg−1 protein | fold | % | |
| Clarified Extract | 110 | 253 | 141 | 0.56 | 1 | 100 |
| DEAE-Sephacel (unbound) | 25 | 200 | 115 | 0.58 | 1 | 82 |
| Butyl-Superose | 17 | 9.50 | 22.0 | 2.3 | 4 | 16 |
| CaM-Agarose | 4 | 0.04 | 8.5 | 213 | 380 | 6 |
Two-week-old seedlings (150 g) grown in liquid culture were used for purification. Data from a typical extraction are shown. Samples were assayed in the presence of 1 mm Ca2+ and 300 nm CaM. CaM independent activity was undetectable following DEAE chromatography.
One unit of activity is defined as 1 μmol NADP−1 h.
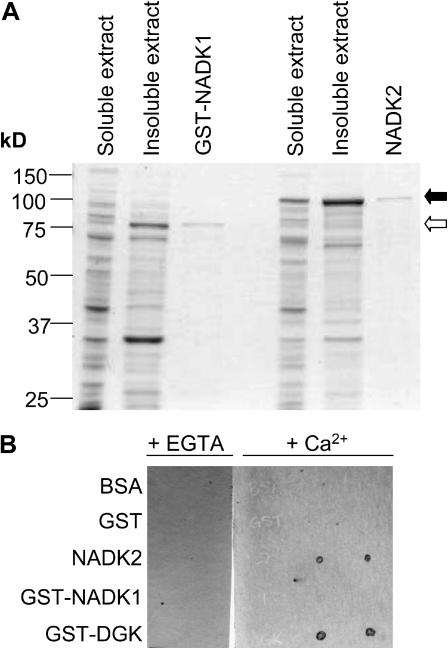
In order to compare the catalytic properties of the native and recombinant proteins, we expressed recombinant NADK1 and NADK2 in Escherichia coli (Fig. 2A). NADK1 was expressed as a full-length glutathione S-transferase (GST) fusion protein and purified by glutathione-affinity chromatography. The intact GST-fusion protein was used for further study of NADK1 due to the lability of enzyme activity during thrombin cleavage of the GST tag. NADK2 was expressed without any fusion tag and, due to its inherent ability to bind to CaM, was purified using CaM-affinity chromatography (Fig. 2A). We confirmed the ability of recombinant NADK2 to bind CaM by using fluorescently labeled CaM81 (CaM81-AF532) as a probe in a nondenaturing overlay assay (Fig. 2B). Recombinant NADK2 bound CaM81-AF532 in a Ca2+-dependent manner, whereas GST-NADK1 did not.
Figure 2.
Expression and purification of recombinant NADK1 and NADK2 and identification of NADK2 as a CaM-binding protein. A, NADK1 was expressed as a GST-fusion protein (GST-NADK1) and purified from the soluble extract of E. coli lysates using GSH-affinity chromatography. NADK2 was expressed as an untagged protein and purified from the soluble extract of E. coli lysates using CaM-affinity chromatography as described in “Materials and Methods.” Open and closed arrows indicate the migration positions in SDS-PAGE gels for GST-NADK1 and NADK2, respectively. A Coomassie Blue-stained gel showing 20.0 μg of soluble and insoluble bacterial extract and 1.0 μg of each purified recombinant protein from a typical purification is presented. B, An overlay spot-blot assay using fluorescently labeled recombinant CaM81 as a probe, in the presence of either 1 mm CaCl2 (+Ca2+) or 10 mm EGTA (+EGTA), was used to examine the CaM-binding abilities of NADK1 and NADK2. Purified, recombinant, nondenatured proteins (50 ng) were spotted (in duplicate) onto nitrocellulose, blocked in 25 mm Tris-HCl, pH 7.5, containing 5% (w/v) nonfat milk in the presence of either CaCl2 or EGTA and then incubated with 110 nm fluorescently labeled CaM81. After washing the blots, CaM-binding proteins were detected by fluorescent imaging. A GST-fusion protein containing the CaM-binding domain of a tomato diacylglycerol kinase (GST-DGK) was used as a positive control, whereas GST and bovine serum albumin (BSA) served as negative controls.
Table II summarizes the kinetic properties of the partially purified native NADK and the purified recombinant enzymes, GST-NADK1 and NADK2. All activities were linear with respect to time and protein concentration (data not shown) and displayed Michaelis-Menten saturation kinetics. The native and recombinant enzymes all displayed a similar broad and symmetrical pH activity profile with an optimum at about pH 8.0. Preparations of native enzyme showed greater specific activity (Vmax) and a lower Km for both substrates (NAD and ATP) than either of the recombinant NADKs. The native and recombinant enzymes were able to utilize either UTP or ATP with comparable efficiency as nucleoside triphosphate substrates (data not shown) but were unable to use tetrapolyphosphate or pyrophosphate as has been described for some bacterial NADKs (Kawai et al., 2000). Neither the native nor the recombinant enzymes were able to phosphorylate NADH under our assay conditions. In general, the catalytic properties of NADK1 and NADK2 were similar. Partially purified native NADK showed a sigmoidal response to physiological Ca2+ and CaM concentrations with S0.5 values of 0.8 μm and 2.2 nm, respectively. Neither Ca2+ nor CaM alone had any effect on catalytic activity, whereas activity was completely inhibited by the Ca2+ chelator EGTA or the CaM antagonist trifluoperazine (Table II). By contrast, although NADK2 bound to CaM in a Ca2+-dependent manner (Fig. 2, A and B), no effect of either Ca2+ or CaM was observed on its catalytic activity in vitro under a variety of assay conditions. NADK1 activity was inhibited by about 50% at 2 mm CaCl2 but only inhibited by 12% at 200 μm CaCl2.
Table II.
Kinetic properties of Arabidopsis native and recombinant NADKs
| Parameter | Native (CaM Dependent) | Rec. NADK1 | Rec. NADK2 |
|---|---|---|---|
| Optimum pH | 8.0 | 7.9 | 7.9 |
| VmaxMg2+·ATP (units mg−1 protein) | 213 ± 7.2 | 11.1 ± 0.61 | 14.3 ± 0.30 |
| KmNAD (mm) | 0.20 ± 0.011 | 0.52 ± 0.030 | 0.43 ± 0.017 |
| KmMg2+·ATP (mm) | 0.17 ± 0.014 | 0.73 ± 0.040 | 0.74 ± 0.012 |
| S0.5Ca2+ (μm) | 0.8 ± 0.07 | 12% inhibition at 0.2 mm Ca2+ | No effect |
| S0.5CaM (nm) | 2.2 ± 0.079 | No effect | No effect |
| I50TFP (μm)b | 57 ± 2.90 | No effect | No effect |
Partially purified CaM-dependent native NADK from 14-d-old seedlings grown in liquid culture or purified recombinant (Rec.) NADK expressed in E. coli were assayed as described in “Materials and Methods.” Data represent mean ± se.
aOne unit of activity is defined as 1 μmol NADP h−1 mg−1 protein.
TFP, Trifluoperazine.
Effect of Various CaM-Related Proteins on Native NADK Activity
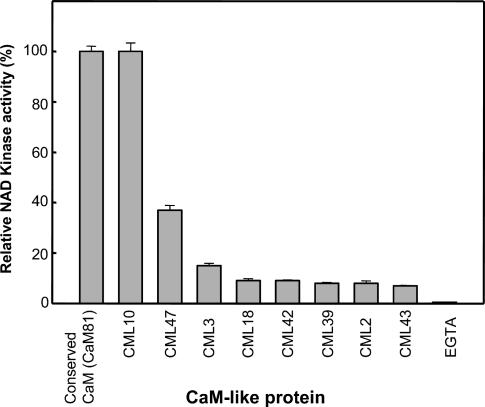
Previous studies on pea (Pisum sativum) NADK have demonstrated a differential response of the enzyme to various CMLs unique to plants (Lee et al., 1995, 1997; Liao et al., 1996). We examined the responsiveness of Arabidopsis native NADK to seven novel Arabidopsis CML recombinant proteins and a previously described protein, CML10 (originally termed CaBP-22; Ling and Zielinski, 1993). These CMLs were chosen because they represent a reasonable selection from across the various CML subfamilies found in Arabidopsis (McCormack and Braam, 2003). Activity was measured relative to that observed using a recombinant conserved CaM (petunia CaM81). Supplemental data shows a protein sequence alignment of the CML proteins tested compared to the conserved CaM81 (Supplemental Fig. 2). Most of the CML proteins were only able to stimulate NADK activity to a level of 5% to 35% of the maximum (Fig. 3) and were not able to competitively inhibit NADK activity even when present at 5-fold higher concentrations (data not shown). However, CML10, which possesses a unique C-terminal extension of 42 amino acids (Supplemental Fig. 2) relative to the conserved CaM was able to fully activate native NADK.
Figure 3.
Comparative responsiveness of Arabidopsis native NADK to various CML proteins. Activity of purified native NADK was examined in the presence of 1 mm EGTA or 2 mm Ca2+ and 100 nm of either the conserved CaM (CaM81) or 8 different Arabidopsis CMLs: CML10 (AAD12002), CML47 (CAB61974), CML3 (AAF02168), CML18 (AAF26959), CML42 (CAB79078), CML39 (AAF04446), CML2 (CAB78328), and CML43 (BAB09153). The CMLs were cloned, expressed as recombinant proteins in E. coli, and purified as described in “Materials and Methods.” Data represent mean ± se from triplicate assays of a typical experiment. A sequence alignment of various CML proteins can be found as supplemental data (Supplemental Fig. 2).
Developmental Profile of Native NADK Activity
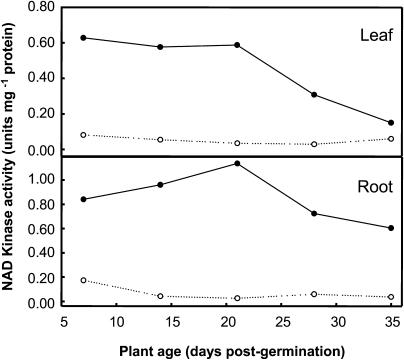
NADK assays of clarified crude Arabidopsis extracts revealed the predominance of CaM-dependent activity that followed a similar profile in both roots and leaves throughout the developmental period examined (Fig. 4). Only a low level of CaM-independent activity was observed at any stage of development or in any of the tissues examined. CaM-dependent NADK activity was already high by 7 d postgermination. By 35 d postgermination in mature plants, there was a substantial decline in CaM-dependent NADK activity in leaves and to a greater extent in roots.
Figure 4.
Developmental profile of Arabidopsis native NADK activity. CaM-dependent (black circles) and CaM-independent (white circles) activity was measured in crude, clarified extracts of leaf or root tissue as described in “Materials and Methods.” Data represent specific activities (mean ± se; error bars are smaller than symbols) from triplicate assays.
Transcript Expression Analysis
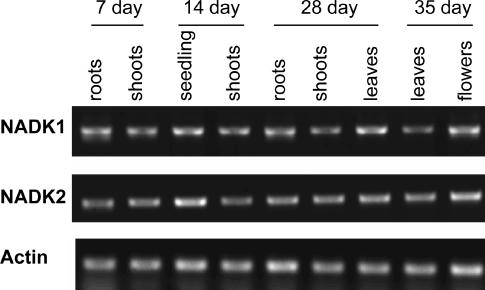
Reverse transcription (RT)-PCR (nonquantitative) was used to examine the expression profiles among various tissues for NADK1 and NADK2. Transcripts for both isoforms were detected in all tissues examined and at all stages of development (Fig. 5).
Figure 5.
Expression of NADK1 and NADK2 transcripts in various Arabidopsis tissues. Plant growth and RT-PCR conditions are described in “Materials and Methods.” For 7-d-old tissue, plants were grown in petri dishes on Murashige and Skoog media. For 14- and 28-d-old tissue, plants were grown in liquid culture and 35-d-old tissue was taken from soil grown plants. Control RT-PCR reactions lacking reverse transcriptase did not show any bands. Actin2 (U41998) was used as a reference control to ensure equal RNA quantification during RT and equal PCR efficiency.
Mapping of the CaM-Binding Domain of NADK2
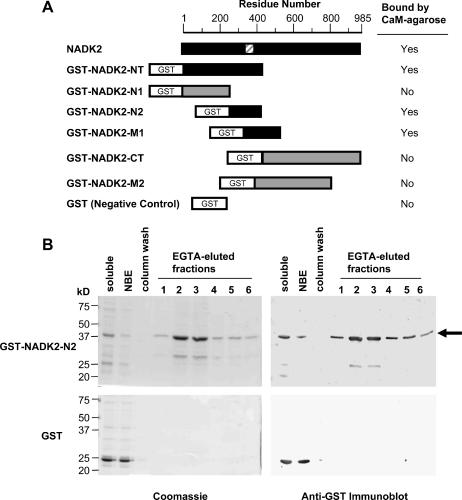
Recombinant NADK2 bound CaM in a Ca2+-dependent manner (see Fig. 2, A and B). We therefore set out to delineate the CaM-binding domain by testing the ability of different regions of the NADK2 protein, expressed in E. coli as GST-fusion proteins, to bind to CaM-agarose in the presence of Ca2+ (Fig. 6). Immunoblotting, using GST anti-serum, was used to confirm the soluble expression and elution of GST-fusion proteins from the CaM-affinity column using the Ca2+ chelator, EGTA. Figure 6B shows an example of a GST-fusion protein (GST-NADK2-N2) that bound strongly to CaM-agarose, whereas the negative control, GST, was unable to bind to CaM-agarose. Only GST-fusion proteins (or the untagged full-length protein) containing a region of 45 amino acids (residues 335–380) within the long N-terminal extension of NADK2 were able to bind to CaM-agarose. Consistent with our experimental analysis, this region contains a domain (residues 355–375) that is predicted by computer analysis (Yap et al., 2000) to bind CaM. Moreover, this predicted CaM-binding domain is conserved among higher plant NADK2s (Fig. 7).
Figure 6.
Mapping of the CaM-binding domain of NADK2. A, Different regions of NADK2 were expressed in E. coli as recombinant GST-fusion proteins and tested for their ability to bind to a CaM-agarose affinity column in a calcium-dependent manner. A schematic of the GST-fusion constructs tested shows their positions relative to the full-length NADK2. Black boxes denote recombinant proteins that were able to bind to CaM-agarose, whereas gray boxes indicate proteins that did not bind CaM. The striped box indicates the position of the delineated CaM-binding domain. B, A typical example of how GST-NADK2 fusion proteins described in A (data for GST-NADK2-N2 is shown) and CaM-affinity chromatography were used to delineate the CaM-binding domain of NADK2. Left and right panels show Coomassie-stained SDS-PAGE gels and anti-GST immunoblots, respectively, of the EGTA-elution profile from a CaM-affinity column for E. coli extracts containing either GST-NADK2-N2 fusion protein (top panels) or control GST (bottom panels). For Coomassie-stained gels (left panels), about 10 μg protein of bacterial soluble extract (soluble) or nonbinding eluate (NBE) was loaded for both the GST-NADK2-N2 and GST samples. EGTA-eluted fractions (lanes 1–6) were loaded on an equal volume (30 μL) basis to show the GST-fusion protein elution profile. The immunoblots (right panels) correspond to the Coomassie-stained gels with the exception that one-third the amount of sample was loaded per lane. A black arrow indicates the position of GST-NADK2-N2.
Figure 7.
Sequence comparison of the predicted CaM-binding domains for putative NADK2 homologs from various plant species. Black and gray boxes indicate identical or similar residues, respectively, conserved among at least 60% of the sequences. GenBank accession numbers for the sequences are follows: Arabidopsis NADK2 (AF337912), rice (AK065215), lettuce (BQ998926), soybean (AW620785), wheat (BE402274), and sorghum (BE364471).
Anti-NADK Antiserum Detects a 110-kD CaM-Binding Arabidopsis Protein
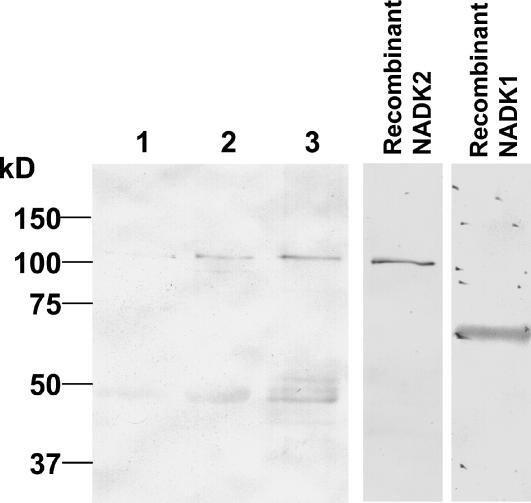
Immunoblots, using anti-NADK antiserum, detected an immunoreactive band with a similar electrophoretic mobility as the 110-kD recombinant NADK2 (Fig. 8). This immunoreactive protein was not detected in clarified crude extracts (or very weakly in some preparations) but was enriched by CaM-affinity chromatography, indicating that it is a CaM-binding protein. CaM-dependent NADK activity in fractions eluted from a CaM-affinity chromatography column correlated with the presence of this immunoreactive band. Although recombinant GST-NADK1 is recognized by our antiserum (Fig. 8), no immunoreactive band of a comparable size was detected in plant extracts. However, a diffuse, weakly immunoreactive protein that migrated with a predicted molecular mass of about 45 kD was observed. Whether this protein is a proteolytic product of NADK2, an immunologically related protein, or a nonspecific immunoreactive band remains unknown at this point.
Figure 8.
Anti-NADK antiserum detects a 110-kD CaM-binding Arabidopsis protein. Crude, clarified extracts from 20 g of total tissue from 14-d-old liquid culture grown Arabidopsis seedlings were passed over an anion-exchange column and the flow-through, containing the CaM-dependent NADK activity, was used for CaM-affinity chromatography as described under “Materials and Methods.” Protein samples were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted using anti-NADK antiserum: lane 1, 25 μg of protein from crude clarified extract; lane 2, 25 μg of protein from anion-exchange nonbinding eluate; and lane 3, 25 μg of protein from a CaM-affinity fraction showing highest NADK activity. The two panels on the right show immunoblots for purified, recombinant NADK2 (12.5 ng) and NADK1 (250 ng) with the GST-tag removed by proteolytic cleavage to allow for comparison of electrophoretic mobilities with the endogenous immunoreactive proteins. A typical immunoblot is presented where similar results were obtained from three independent experiments.
DISCUSSION
Research progress on plant NADKs has been hindered by a lack of molecular and genetic tools. This article describes the first report on the cloning and molecular characterization of NADKs from a higher plant. In addition, this is the first documentation, to our knowledge, of NADK enzyme kinetics and of NADK gene expression in Arabidopsis. We have identified, cloned, and characterized cDNAs encoding two distinct NADK isoforms, denoted NADK1 and NADK2, and demonstrated that transcripts for both genes are present in all tissues examined (Fig. 5). The sequence similarity between the predicted proteins for NADK1 and NADK2 is restricted to the C-terminal region that is conserved across kingdoms (Fig. 1). Based upon database analyses (sequence homology searches), the long N-terminal extension of NADK2, which includes a CaM-binding domain (Figs. 6 and 7), appears to be unique to plants. NADK2, at 985 residues and a predicted molecular mass of about 110 kD, is by far the largest NADK subunit described to date from any organism. Database analyses, based upon strong sequence conservation in the extended N-terminal region (including the CaM-binding domain), revealed widespread distribution of putative NADK2 homologs in other higher plants (e.g. canola, soybean, lettuce, sorghum, wheat, and others). However, apart from Arabidopsis, there is at present only full-length cDNA sequence data available for rice. A full-length rice mRNA (AK065215), encoding a putative NADK of 981 residues and a predicted molecular mass of about 109 kD, shows very strong sequence similarity (about 60% identity) to NADK2 suggesting that these large NADKs may be widely distributed among higher plants. NADK2 (and the putative rice homolog) are considerably larger than the 56-kD subunit size described for a purified native tomato CaM-binding NADK (Delumeau et al., 2000). Although no other pure plant CaM-binding NADKs have been described, several studies have observed proteins ranging from 34 to 110 kD in SDS-PAGE gels of highly enriched preparations (Simon et al., 1982; Roberts et al., 1990; Delumeau et al., 2000). It therefore remains unclear whether different species possess CaM-dependent NADKs of markedly different sizes or whether proteolytic or other posttranslational processing may have affected predictions of NADK size in earlier studies. Sequence-based identification of purified native NADKs and the isolation of cDNAs from various species are needed to address these possibilities. NADK1, with a predicted mass of roughly 60 kD, is similar to the subunit size predicted for a tomato CaM-dependent NADK (Delumeau et al., 2000), but we found no evidence of CaM binding to NADK1 or effect of CaM on its activity. Although CaM-independent NADKs have been described in a number of higher plant species (Simon et al., 1982, 1984; Gallais et al., 2001; Delumeau et al., 1998), none have been purified to homogeneity (nor encoding cDNAs isolated) and, thus, estimates of subunit size are not available for comparison to NADK1. Nevertheless, it is clear from database searches and sequence comparisons that, as with NADK2, homologs of NADK1 are widely distributed among higher plants (data not shown). For example, a full-length mRNA from rice (AK099730) is predicted to encode a protein of similar size and primary sequence to NADK1.
We detected both CaM-dependent and CaM-independent NADK activity in extracts from Arabidopsis throughout development. In crude extracts, most of the activity of the native enzyme was dependent on Ca2+ and CaM similar to reports on NADK activity in pea (Muto, 1983; Simon et al., 1984), tomato (Delumeau et al., 1998), oat (Pou De Crescenzo et al., 2001), and maize (Dieter and Marme, 1984). In this study, CaM-independent activity separated from CaM dependent activity at an early stage of purification, suggesting that two distinct enzymes are responsible for the different activities. Most efforts at purification of CaM-dependent NADK to homogeneity from plant tissues have been unsuccessful due to the highly unstable nature of the native enzyme (Jarrett et al., 1982; Roberts and Harmon, 1992; Delumeau et al., 2000). Indeed, the only group to describe purification of a plant NADK to homogeneity used a partially pure preparation for kinetic characterization due to the extreme lability of the homogenous sample (Delumeau et al., 2000). Although it remains unclear as to why native NADK is so labile, a strong sensitivity to oxidation has been described (Delumeau et al., 2000).
Although considerable variability between plant species has been reported for the Vmax of plant CaM-dependent NADKs, the specific activity of native Arabidopsis CaM-dependent NADK is within the range described for partially pure NADKs from plants such as pea (Harmon et al., 1984) and tomato (Delumeau et al., 1998). Similarly, the pH optimum (pH 8.0) for Arabidopsis NADKs (Table II) is comparable to that for human (Lerner et al., 2001), yeast (Kawai et al., 2001b), and E. coli (Kawai et al., 2001a) NADKs as well as other plant NADKs (Muto, 1983; Gallais et al., 2001). The Kmvalues for native CaM-dependent NADK, for both Mg2+-ATP (200 μm) and NAD (170 μm), are comparable to reports on CaM-dependent NADKs from pea (Muto, 1983), Avena sativa (Gallais et al., 2001), and tomato, with the exception that the tomato enzyme appears to have a much higher affinity for Mg2+-ATP (Delumeau et al., 1998). In general, Km values, for both substrates, are higher for nonplant NADKs with KmATP values as high as 2.5 mm reported for E. coli (Kawai et al., 2001a). This may reflect differences among organisms with respect to the cellular levels of NADK substrates. The high affinity of Arabidopsis NADK for CaM (S0.5 of 2.2 nm) is consistent with reports on other plant NADKs (Harmon et al., 1984; Lee et al., 1997; Delumeau et al., 2000) and other plant CaM-binding proteins (Snedden et al., 1996; Bouche et al., 2002; Zhang et al., 2002; Chandok et al., 2003). Similarly, the S0.5 value for Ca2+ (0.8 μm) suggests stimuli that elevate Ca2+ from resting levels in vivo may elicit a CaM-modulated NADK response. Specific activity and affinity for the substrate NAD were substantially higher for the native enzyme than for either NADK1 or NADK2. Considerable variability among recombinant NADK activity has previously been observed (Kawai et al., 2000, 2001a, 2001b; Lerner et al., 2001). Although we demonstrated that the N-terminal region of NADK2 contains a CaM-binding domain (Figs. 6 and 8), we did not observe any responsiveness of either NADK1 or NADK2 to CaM under various in vitro assay conditions. Improper tertiary folding in E. coli or the absence of posttranslational modifications or other regulatory components (such as additional, unidentified subunits) are possible explanations for the biochemical differences between the recombinant and native enzymes, but they remain speculative at this point. Our observations are reminiscent of a recent report on a CaM-binding catalase from tobacco in which the native enzyme was responsive to CaM, whereas the recombinant protein expressed in E. coli was not (Yang and Poovaiah, 2002). However, our immunoblot analysis (Fig. 8) using anti-NADK2 antiserum, showing an enrichment of an approximately 110-kD immunoreactive protein by CaM-affinity chromatography, suggests that native NADK2 is indeed a CaM-binding protein and is therefore likely the isoform responsible for CaM-dependent activity in Arabidopsis. Nevertheless, we cannot exclude the possibility that other, unrelated NADK(s) exist in Arabidopsis. It is conceivable that CaM may also act indirectly on native NADK by modulating the activity of additional CaM-binding proteins (e.g. a CaM kinase or phosphatase). Future experimental approaches using reverse genetic analyses, microsequencing of purified NADKs, and isolation of other interacting proteins will help determine the identities of all components responsible for CaM-dependent and -independent NADK activities in Arabidopsis.
It has been hypothesized that the large repertoire of CMLs in plants may serve to regulate specific targets or compete for CaM-binding proteins (Snedden and Fromm, 2001; Yang and Poovaiah, 2003). A diversity of Ca2+ sensors, including Ca2+-dependent protein kinases and calcineurin-like proteins, likely endow plants with a highly specific and complex Ca2+ signaling network (Luan et al., 2002). Consistent with earlier reports (Lee et al., 1995, 1997; Liao et al., 1996), we observed a differential response of native NADK activity to a variety of CaM-related proteins, including seven that had not previously been cloned or studied. Interestingly, CML10, which is about 64% identical to the conserved CaM but possesses a unique 42-residue C-terminal extension, was able to fully activate native NADK. Previous reports have shown that both C-terminal and N-terminal regions of CaM are important in achieving full activation of plant NADK (Liao et al., 1996; Lee et al., 1997), and, thus, it is perhaps surprising that we observed full activation of Arabidopsis NADK by CML10. Although the function of the C-terminal extension in CML10 is unknown, our results suggest that this extension acts independently of the hydrophobic regions important in the activation of NADK (Liao et al., 1996). By contrast, the more divergent CaM-related proteins we examined were only able to weakly activate native NADK above background levels. Given the predominance of CaM-dependent NADK activity throughout development in Arabidopsis (Fig. 4), future work should address which Ca2+-binding proteins interact with NADK in vivo.
We delineated the CaM-binding domain of NADK2 to a region of less than 45 residues within the N-terminal extension (Fig. 6). Interaction of fluorescently labeled CaM81 or CaM-agarose with this region of NADK2 was completely Ca2+-dependent. By comparison, despite strong expression of recombinant NADK1, we did not observe an interaction with fluorescent CaM81 in overlay assays or with CaM-agarose in vitro, and we conclude that NADK1 is not a CaM-binding protein. By contrast, we were able to purify recombinant NADK2 from bacterial extracts (Fig. 2A) with a single CaM-affinity step, demonstrating the specificity and affinity of the CaM-binding domain of NADK2. Although nondenatured recombinant NADK2 bound to fluorescently labeled CaM81, denatured NADK2 protein run on SDS-PAGE and used in CaM overlay assays was not able to bind CaM (data not shown), unlike many other plant CaM-binding proteins (Reddy et al., 2002; Yang and Poovaiah, 2003). This suggests that the CaM-binding domain, or associated region of the recombinant protein, is unable to refold after denaturation and may reflect a general lability comparable to that of the native enzyme. Sequence database analysis suggests that the predicted CaM-binding domain of NADK2 is conserved among putative homologs in other plants (Fig. 7), a finding consistent with the observation that CaM-modulated NADK activity has been observed among various dicotyledonous and monocotyledonous plant species (Jarrett et al., 1982; Dieter and Marme, 1984; Harding et al., 1997; Pou De Crescenzo et al., 2001). It is thus an intriguing question as to why plants possess a CaM-regulated means to generate NADP.
A number of different physiological roles have been proposed for NADKs in plants. Having multiple, differentially regulated NADKs may afford additional metabolic flexibility in coordinating cellular NAD to NADP ratios. An important consideration, however, and one which merits additional research, is the subcellular localization of NADK isoforms in plants given the importance of NADP in different organelles. There has been substantial debate as to where CaM-dependent and -independent isoforms are localized within plant cells, and the issue remains unresolved. CaM-dependent NADK activity has been observed in mitochondrial fractions (Dieter and Marme, 1984; Sauer and Robinson, 1985; Pou De Crescenzo et al., 2001), chloroplastic fractions (Muto et al., 1981; Jarrett et al., 1982; Simon et al., 1984), and the cytosolic fraction (Simon et al., 1982; Gallais et al., 2000; Pou De Crescenzo et al., 2001). By comparison, CaM-independent NADK activity in plant cells has been observed in the cytosolic fraction of cells (Gallais et al., 2000; Pou De Crescenzo et al., 2001) and in the chloroplast (Simon et al., 1984). The roles of these isoforms remain unclear, although it has been suggested that chloroplastic NADK might participate in the light-stimulated conversion of NAD to NADP (Jarrett et al., 1982). NADP(H) also plays an important role in plant mitochondria metabolism and, thus, how mitochondrial-associated NADK activity might impact upon cell energetics in plants needs to be examined. It should be noted that yeast possess an NADH kinase (Pos5p) that localized to the mitochondrion and plays a critical role in oxidative stress response (Outten and Culotta, 2003). The Pos5p kinase, although related to NADKs, utilizes NADH rather than NAD as a substrate and appears to play a distinct role from yeast NADKs (Outten and Culotta, 2003). We have recently isolated a cDNA encoding a CaM-independent putative Pos5p NADH kinase homolog from Arabidopsis, and its subcellular localization and physiological roles are under investigation (W. Turner, J. Waller, and W. Snedden, unpublished data). Thus, a complex picture of NADP(H) metabolism in plant cells is emerging. One of the most intriguing examples is the involvement of a CaM-modulated NADK in the NADPH-mediated oxidative burst associated with pathogen defense (Harding et al., 1997). Transgenic tobacco plants expressing a modified CaM, which hyperactivates endogenous NADK, showed increased levels of NADPH and a more rapid and enhanced oxidative burst than wild-type plants when challenged with a number of stimuli (Harding et al., 1997). Additional studies are needed that examine the contribution of NADKs to processes, such as the oxidative pentose phosphate pathway (Pugin et al., 1997), known to supply reductant for the NADPH oxidase during an oxidative burst. It is interesting to note that the only other biological systems reported to possess a CaM-dependent NADK, human neutrophil cells (Williams and Jones, 1985) and sea urchin eggs (Epel et al., 1981; Iwasa and Mohri, 1983), are capable of generating an NADPH-oxidase mediated oxidative burst.
In addition, there is mounting evidence that the NADP-derived metabolite, NAADP, is an important regulatory molecule in both plants and animals (Navazio et al., 2000; Churchill et al., 2002; Sanders et al., 2002; Rutter, 2003). NAADP, in a manner similar to inositol 1,4,5-trisphosphate and cyclic-ADP-Rib, is a potent mediator of Ca2+ release from intracellular stores (Patel et al., 2001; Rutter, 2003) and thus plays a vital role in Ca2+ signal transduction. It will be interesting to determine if a possible role of NADKs in plants is to contribute to NAADP biosynthesis by providing the substrate, NADP.
By cloning NADKs from a higher plant and identifying the CaM-dependent isoform in Arabidopsis as NADK2, this study makes an important advance in our understanding of this enzyme family and should help facilitate the development of genetic and molecular tools needed to provide a comprehensive picture of the role of plant NADKs. Emerging evidence in plants for an involvement in processes such as the oxidative burst and redox balance are exciting avenues for future work.
MATERIALS AND METHODS
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich Canada (Oakville, Canada). Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Plant Material
Arabidopsis (accession Col-0) was used for all experiments, unless stated otherwise, and general growth conditions consisted of a 3-d, 4°C stratification period in the dark, followed by a 16-h-light (120 μE), 8-h-dark photoperiod at 22°C. For partial purification of native NADK, plants were grown in 200 mL of sterile liquid culture using 0.5× Murashige and Skoog liquid media in 1-L flasks on an orbital shaker (60 rpm). Tissue was harvested by filtration over cheesecloth, powdered under liquid nitrogen, and stored at −80°C until use. Liquid cultures were also used for some NADK transcript analyses. In addition, Arabidopsis seedlings were grown on sterile media in petri dishes or, for analysis of mature plants, transferred to growth chambers under the conditions described above. Seeds were sown on petri dishes of sterile 0.5× Murashige and Skoog media containing 0.8% (w/v) agar, 1% (w/v) Suc, stratified, then transferred to soil following germination and grown in growth chambers as described above. Tissues were harvested at various stages of development, frozen in liquid nitrogen, and stored at −80°C until use.
Partial Purification of Native CaM-Dependent NADK
All steps were carried out at 0°C to 4°C. Arabidopsis tissue (150 g) from liquid culture 14-d-old seedlings was homogenized in buffer A (50 mm Tris-Cl, pH 7.5, 200 mm KCl, and 3 mm MgCl2), 1 mm EGTA, 0.5 mm EDTA, 2% (w/v) polyvinylpolypyrrolidone, 1 mm dithiothreitol (DTT), 1 mm phenyl methyl sulfonyl fluoride (PMSF), 1 mm benzamidine, 50 μm leupeptin, and 10 μm N-(N-(l-3-transcarboxyoxirane-2-carbonyl)-l-leucyl) agmatine (E-64) using a Polytron (Kinematica; Fisher Scientific, Toronto) homogenizer. The homogenate was filtered through two layers of Miracloth (Calbiochem, San Diego) and centrifuged at 14,000g for 15 min. Solid (NH4)2SO4 was added to the supernatant to give a final concentration of 50% (w/v) and the solution stirred for 20 min. Precipitated proteins were collected by centrifugation (14,000g, 15 min), resuspended in 100 mL of buffer B (50 mm Tris-Cl, pH 7.5, 100 mm KCl, 3 mm MgCl2, 1 mm EGTA, 0.5 mm EDTA, and 1 mm DTT) and applied to a column (60 mL) of DEAE-Sephacel. The nonbinding protein eluate, containing CaM-dependent NADK activity, was collected and brought to 3.5 m NaCl. The sample was clarified by centrifugation (14,000g, 10 min) and applied (3 mL min−1) to a column (30 mL) of butyl-Sepharose (Bio-Rad, Mississauga, Canada). The column was washed with buffer C (50 mm Tris-Cl, pH 7.5, 3.5 m NaCl, 3 mm MgCl2, and 1 mm DTT) until the A280 decreased to baseline and adsorbed proteins were eluted with a linear gradient (150 mL) of 0% to 100% buffer D (50 mm Tris-Cl, pH 7.5, 30% [v/v] ethanediol, 100 mm KCl, 3 mm MgCl2, and 1 mm DTT). NADK activity eluted as a single peak of activity at approximately 95% buffer D. Fractions (7 mL) were collected and those containing greater than 20% peak NADK activity were pooled, made 0.5 mm with respect to Ca2+, and applied to a column (5 mL) of CaM-agarose. The column was washed (1.5 mL min−1) sequentially with 50 mL of buffer E (50 mm Tris-Cl, pH 7.5, 50 mm KCl, 3 mm MgCl2, and 1 mm DTT), then 100 mL of buffer E (enriched in KCl to 400 mm), and finally10 mL of buffer E without supplemental KCl. NADK activity was eluted (1 mL min−1) with buffer E containing 1.5 mm EGTA in place of Ca2+. Fractions of 1 mL were collected and those containing NADK activity were pooled, concentrated 10-fold over a YM-30 membrane (Millipore, Bedford, MA), frozen in liquid nitrogen, and stored in aliquots at −80°C. For analysis of NADK activity in crude extracts, samples were prepared using the extraction buffer described above and tissue (100 mg) was ground using sterile sand in a chilled mortar and pestle. Crude homogenates were centrifuged at 14,000g for 5 min and the clarified extract used for NADK assays.
Enzyme Assays
NADK activity was assayed (180 μL final volume) in microtiter plates essentially as described (Harmon et al., 1984). The standard assay mixture, unless otherwise specified, contained 50 mm Tris-Cl (pH 7.9), 4 mm ATP, 4 mm NAD, 10 mm MgCl2, 1 mm CaCl2, and 300 nm recombinant petunia CaM81 (S70768, prepared as described in Fromm and Chua, 1992). It should be noted that petunia CaM81, which is 100% identical to Arabidopsis CaM7 (P59220) and about 90% identical to human CaM (NP_001734) at the peptide sequence level, is considered an evolutionarily conserved form of CaM (McCormack and Braam, 2003) and thus served as a standard reference for CaM-dependent NADK activity throughout our analyses. The reaction was initiated with up to 18 μL of protein extract and allowed to proceed for 30 min at 25°C. NADP formed was immediately detected by adding 20 μL of the NADK assay mixture to a cycling assay (250 μL final volume) containing 50 mm Tris-HCl (pH 7.9), 5 mm Glc-6P, 1 mm EGTA, 0.5 units of yeast Glc-6-P dehydrogenase, 1 mg mL−1 2,6-dichlorophenolindophenol (DCIP), and 1 mg mL−1 methyphenazinium methylsulfate (PMS). Reduction of DCIP was monitored at 600 nm using a SpectroMax Plus microplate reader (Molecular Devices, Sunnyvale, CA) and the amount of NADP quantified by comparison to a standard curve produced using analytical-grade NADP. One unit of NADK activity is defined as the amount of enzyme resulting in the production of 1 μmol of NADP h−1 at 25°C. Apparent Vmax and Km values were calculated from the Michaelis-Menten equation using a nonlinear least-squares regression algorithm (Brooks, 1992). For activation by CaM, S0.5 values were calculated from data fitted by nonlinear regression to the equation v = (Vmax)[Act]/S0.5 + [Act] using the aforementioned computer program. Kinetic parameters are the means of three or more independent determinations and are reproducible to within ±10% (se) of the mean value. Concentrations of free Mg2+ and Ca2+ were calculated based upon their respective binding to organophosphates, nucleotides, and/or Cl− using a computer program that automatically corrects for temperature, pH, and ionic strength (Brooks and Storey, 1992). Protein concentration was determined by the dye-binding method of Bradford (1976) as modified by Bollag et al. (1996) with bovine γ-globulin as the protein standard.
Cloning of NADK1 and NADK2 cDNAs
EST cDNA clones encoding two unique, putative NADKs (designated NADK1 and NADK2) were found in The Arabidopsis Information Resource database (http://Arabidopsis.org/) based upon their sequence similarity to yeast, mammalian, and bacterial NADKs. The ESTs included 5′ untranslated region sequence and were predicted to encode full-length proteins. The full-length cDNA clone encoding NADK1 (RZL33e02f, AV547501) was obtained from the Kasuza DNA Research Institute, Japan (Asamizu et al., 2000). The complete cDNA for NADK1 was amplified by PCR using the full-length cDNA as a template and high-fidelity Pfu DNA polymerase (MBI Fermentas, Burlington, Canada) according to the manufacturer's instructions. PCR primers were designed to add a BamHI site at the predicted start position (NADK1-F, 5′-GGATCCATGTCGTCGACCTACAAG-3′) and an EcoRI restriction site at the predicted termination position (NADK1-R, 5′-CGGATTTCCTAAGGTCCATCAGCAGAT-3′) to facilitate subcloning into pGEX-4T-3 (Amersham Biosciences, Piscataway, NJ) vector to generate a GST-fusion protein. In order to isolate a full-length cDNA encoding NADK2, PCR forward (NADK2-F, 5′-CTCATATGTTCCTATGCTTTTGCC-3′) and reverse (NADK2-R, 5′-CGGTCGACTCAGAGAGCCTTTTGATC-3′) primers were designed according to the GenBank cDNA sequence (AF337912) at the predicted start and termination positions, respectively. The forward and reverse NADK2 primers introduced NdeI and SalI sites, respectively, into NADK2 cDNA. Template cDNA for PCR reactions was prepared by reverse transcribing total mRNA isolated from 14-d-old total seedling tissue as described below under the heading “RT-PCR.” Amplified NADK2 cDNA was ligated into a SmaI-digested pBluescript SKII− vector (Stratagene, La Jolla, CA) and then subsequently excised and subcloned into an NdeI and SalI-digested pET 29a expression vector (Novagen, Madison, WI). All subcloned fragments were verified by DNA sequencing (Cortec DNA Service Laboratories, Kingston, Canada).
RT-PCR
For developmental RT-PCR analysis, total RNA was extracted from Arabidopsis tissues at different stages of development with the RNeasy plant mini kit (Qiagen, Valencia, CA) and 5 μg was treated with DNase (Amersham Biosciences) as per manufacturer's instructions to remove any contaminating genomic DNA. First strand cDNA was reverse-transcribed using SuperScript II RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA) in the presence of 1 unit RNase inhibitor (MBI Fermentas). To control for the possible presence of contaminating genomic DNA, parallel negative-control first strand cDNA synthesis reactions were run in which no reverse transcriptase was added. The Arabidopsis Actin2 gene (U41998), using primers flanking an intron as an additional control to detect genomic DNA contamination (forward primer, 5′-TCGGTGGTTCCATTCTTGCT; reverse primer, 5′-GCTTTTTAAGCCTTTGATCTTGAGAG-3′), was amplified as a control to ensure equal amounts of cDNA template were used in each PCR reaction. Internal primers were designed to amplify fragments of NADK1 (forward primer, 5′-GGATGGAGCACTTAGCAAAGTCTCCGCGGC-3′; reverse primer, 5′-CCCATAGCACTGTGCCATCCCCACCAAGAG-3′), and NADK2 (forward primer, 5′-GGAGGGGAGGACAGGTAACCCAAGAAGG-3′; reverse primer, 5′-CCAAGCTTAGTTTCAGAAACTCCCGACGC-3′). The PCR (30 cycles, saturating, nonquantitative) was performed using an annealing temperature of 58°C. PCR products were run on 1.2% agarose gels and images captured using a Typhoon 8600 (Amersham Pharmacia, Uppsala) phosphor imager.
Expression and Purification of Recombinant NADKs
For recombinant NADK expression, Escherichia coli strain BL21 Codon Plus (DE3) RIL (Stratagene) was used according to the manufacturer's instructions. Full-length NADK1 and NADK2 were expressed from the pGEX4-T3 and pET29a constructs, respectively, described above. Routinely, 1-L bacterial cultures were inoculated with 5 mL from an overnight starter culture, grown at 28°C with strong agitation until an A600 of 0.6 was reached, isopropyl-β-d-thiogalactopyranoside added to 1 mm, and grown for an additional 3 h. Bacteria were collected by centrifugation, resuspended (3 mL per 100 mL of bacterial culture) in extraction buffer (50 mm Tris-HCl, pH 7.5, 1.0 mm EDTA, 10% [w/v] glycerol, 1 mm DTT, 1 mm benzamidine, and 1 mm PMSF). Cells were lysed with a single pass through a SLM-Aminco French pressure cell (Fisher Scientific) at 140 MPa and the soluble and insoluble bacterial extracts separated by centrifugation at 10,000g, at 4oC, for 12 min. Recombinant GST-NADK1 was purified by GSH-affinity chromatography as per manufacturer's (Amersham Biosciences) directions. Column fractions containing purified GST-NADK1 were pooled and desalted using a Sephadex G-25 PD-10 (Amersham Biosciences) column that had been pre-equilibrated with 50 mm Tris-Cl (pH 7.5), 10% (w/v) glycerol, 1 mm DTT, and 1 mm PMSF. Samples were either assayed immediately for NADK activity or were frozen in liquid nitrogen and stored at −80°C. Recombinant NADK2, which is able to bind CaM-agarose, was purified using CaM-affinity chromatography essentially as described (Snedden and Blumwald, 2000). The soluble bacterial extract containing recombinant NADK2 was adjusted to a final concentration of 3.0 mm CaCl2 and loaded onto a CaM-agarose affinity column (300 μL) pre-equilibrated with CaM-binding buffer (50 mm Tris-HCl, pH 7.5, 1 mm CaCl2, 10% [w/v] glycerol, 1 mm DTT, and 1 mm PMSF). The column was washed with 20 column-volumes of washing buffer (50 mm Tris-HCl, pH 7.5, 1 mm CaCl2, 100 mm KCl, 10% [w/v] glycerol, 1 mm DTT, and 1 mm PMSF) in order to remove weakly adsorbed bacterial proteins. Adsorbed NADK2 was eluted with elution buffer (50 mm Tris-HCl, pH 7.5, 2 mm EGTA, 10% [w/v] glycerol, 1 mm DTT, and 1 mm PMSF), and fractions were either assayed immediately for enzyme activity or were frozen in liquid nitrogen and stored at −80°C. For mapping of the CaM-binding domain of NADK2, different regions of NADK2 cDNA were subcloned into pGEX-4T-3 vector. PCR primers for making these vector constructs are presented below in the order in which they appear in Figure 6: GST-NADK2-NT forward (5′-GGATCCATGTTCCTATGCTTTTGCCC-3′), GST-NADK2-NT reverse (5′-GTCGACTAGACTTTATCAGTCTGCTCATCAGG-3′), GST-NADK2-N1 forward (same as GST-NADK2-NT forward), GST-NADK2-N1 reverse (5′-GTCGACTATGACGAGTAAATAGGGTCC-3′), GST-NADK2-N2 forward (5′-GGATCCAATACGAAAGAGGATATTG-3′), GST-NADK2-N2 reverse (same as GST-NADK2-NT reverse), GST-NADK2-M1 forward (5′-GAATTCGCTCCTAAAGCTGAGCAGGTCGAGC-3′), GST-NADK2-M1 reverse (5′-CTCGAGCTAAACTTTGAGAACTTGTAGG-3′), GST-NADK2-CT forward (5′-GAATTCAGATCTGCTTCAAGCC-3′), GST-NADK2-CT reverse (5′-CGGTCGACTCAGAGAGCCTTTTGATC-3′), GST-NADK2-M2 forward (5′-GAATTCCGACCGATCACGAAAGAAATTCCAG-3′), and GST-NADK2-M2 reverse (5′-CTCGAGCTACGTGTTATTCCCATGGATGACTCG-3′). Recombinant GST-fusion proteins for the different regions of NADK2 were expressed in E. coli BL21 Codon Plus (DE3) RIL cells and tested for their ability to bind to CaM-agarose either directly from total soluble bacterial extracts, or after purification by GSH-affinity chromatography. Purity of recombinant NADK preparations were assessed by SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining or immunoblot analysis, for GST-fusion proteins, using rabbit anti-GST antiserum (Amersham Biosciences), and goat anti-rabbit alkaline phosphatase conjugated secondary antiserum (Sigma) as per manufacturer's directions.
Fluorescent Labeling of CaM81 and Use in Overlay Assays
One milligram of purified, recombinant CaM81, prepared as described above, was conjugated to the succinimidyl ester of Alexa Fluor 532 (Molecular Probes, Eugene, OR) according to manufacturer's directions. The fluorescently labeled CaM, termed CaM81-AF532, was used as a probe to examine the ability of GST-NADK1 and NADK2 to bind CaM using overlay assays. Purified recombinant proteins were spotted onto nitrocellulose membranes, allowed to dry, and then blocked overnight at 4°C in 25 mm Tris-HCl, pH 7.5, containing 5% (w/v) nonfat milk in the presence of either 1 mm CaCl2 or 10 mm EGTA. CaM81-AF532 was then added to a final concentration 110 nm and the membranes were incubated for 2 h at room temperature with gentle shaking. Membranes were washed three times for 5 min with the same buffer described above but containing 500 mm NaCl and lacking milk. CaM81-AF532 interacting proteins were detected by fluorescence imaging using a Typhoon 8600 phosphor imager (Uppsala) at excitation and emission wavelengths of 532 and 555 nm, respectively. A GST-fusion protein containing the CaM-binding domain of tomato diacylglyerol kinase (Snedden and Blumwald, 2000) was used as a positive control for CaM81-AF532 binding.
Cloning cDNAs Encoding CMLs and Expression of the Recombinant Proteins
The genes encoding the CMLs examined in this study, with the exception of CML10, do not contain introns (McCormack and Braam, 2003) and thus were PCR-amplified from Arabidopsis genomic template DNA using high-fidelity Pfu DNA polymerase (MBI Fermentas) according to the manufacturer's instructions. CML10 was PCR-amplified from Arabidopsis seedling first strand cDNA as described under “RT-PCR.” GenBank accession numbers for the respective CML cDNAs are provided in the legend to Figure 3. PCR primers used to amplify the CML cDNAs were CML2 forward (5′-ACCATATGGATCGTGGAGAATTGAGTAGAG-3′), CML2 reverse (5′-AATGTTTAATTGGAGCTAAGAGCAGC-3′), CML3 forward (5′-ACCATATGGATCAAGCGGAGCTTGCC-3′) CML3 reverse (5′-TTACAAGTTTGATCCTAAGGCGG-3′), CML10 forward (5′-CATATGGCGAATAAGTTCACTAGACAAC-3′), CML10 reverse (5′-CATTTCAAGAAAACAACGCTTCGAAC-3′), CML18 forward (5′-CATATGAGCTGCGACGGAGGCAAAC-3′), CML18 reverse (5′-TCAACCCCAAGCATTATCAAACGC-3′), CML39 forward (5′-TCCATATGAAGAACACTCAACGTCAG-3′), CML39 reverse (5′-TTAGCGCATCATGAGGGCG-3′), CML42 forward (5′-ACCATATGGAGAGTAACAACAACGAG-3′), CML42 reverse (5′-GAATCAAGAAGAAGGGATGAC-3′), CML43 forward (5′-ACCATATGGAGATCAATAACGAGAAG-3′), CML43 reverse (5′-TTCAAGAAGGAACAACAACAG-3′), CML47 forward (5′-CGCATATGGAGGATTCGTCTCTTC-3′), and CML47 reverse (5′-GCTCACGAAAAGCTCTTCTCTATAAG-3′). All forward primers were designed to incorporate an NdeI site utilizing the first ATG codon. PCR products were directly ligated into pCR2.1 TOPO vector (Invitrogen) as per manufacturer's directions, then excised and directionally subcloned into the pET5a plasmid (Novagen) to facilitate recombinant protein expression. All cDNA inserts were confirmed by DNA sequencing (Cortec DNA Service Laboratories). Expression in E. coli strain BL21 (DE3) pLysS (Novagen) and purification of the recombinant CML proteins was essentially as described (Zielinski, 2002).
Immunoblot Analysis of Native CaM-Dependent NADK
CaM-dependent NADK was purified from Arabidopsis tissue (10 g) essentially as described above except for the following modifications. Briefly, buffer B was supplemented with 1 mm PMSF, 1 mm benzamidine, 50 μm leupeptin, and 10 μm E-64, and the DEAE nonbinding eluate was brought to 3 mm CaCl2 and passed three times over a CaM-agarose column (1 mL bed-volume). The column was washed sequentially with 5 mL of buffer F (50 mm Tris-Cl, pH 7.5, 10% [v/v] glycerol, 3 mm CaCl2, 1 mm PMSF, 1 mm benzamidine, 50 μm leupeptin, 10 μm E-64, and 1 mm DTT), then 5 mL of buffer G (buffer F containing 1 mm CaCl2 and 100 mm KCl), and finally 5 mL of buffer F. NADK activity was eluted with buffer F containing 2 mm EGTA. Fractions (0.5 mL) were collected, and the fraction containing peak NADK activity was lyophilized. Samples were boiled in SDS-loading buffer (Sambrook et al., 1989) and electrophoretic separation on SDS-PAGE gels and immunoblot analysis on nitrocellulose performed using standard procedures (Sambrook et al., 1989).
Production of Polyclonal Antibodies
Antibodies were raised in rabbits (Animal Care Services, Queen's University) against CaM-affinity purified NADK2 emulsified in Ribi adjuvant (Corixia, Seattle). Crude antiserum was cleaned against purified recombinant protein as described (Sambrook et al., 1989) and used for immunoblotting. Immunoreactive proteins were detected by visualization with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate (Promega, Madison, WI) using alkaline phosphatase-conjugated anti-rabbit secondary antiserum (Sigma).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF337912, BAB92868, BQ998926, AW620785, BE402274, and BE364471.
Supplementary Material
Acknowledgments
We thank Jaimie Moise (Queen's University, Kingston) for technical assistance in the preparation of the fluorescently labeled calmodulin.
This work was supported by the National Science and Engineering Council of Canada (NSERC). W.T. and J.W. were funded through a Premier's Research Excellence Award (recipient W.A.S.). B.V. was funded by an NSERC scholarship.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040428.
References
- Anderson JM, Charbonneau H, Jones HP, McCann RO, Cormier MJ (1980) Characterization of the plant nicotinamide adenine dinucleotide kinase activator protein and its identification as calmodulin. Biochemistry 19: 3113–3120 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer ELL (2000) The Pfam protein families database. Nucleic Acids Res 28: 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag DM, Rozycki MD, Edelstein SJ (1996) Protein Methods, Ed 2. Wiley-Liss, New York, pp 62–70
- Bouche N, Scharlat A, Snedden W, Bouchez D, Fromm H (2002) A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brooks SP (1992) A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques 13: 906–911 [PubMed] [Google Scholar]
- Brooks SP, Storey KB (1992) Bound and determined: a computer program for making buffers of defined ion concentrations. Anal Biochem 201: 119–126 [DOI] [PubMed] [Google Scholar]
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 [DOI] [PubMed] [Google Scholar]
- Churchill GC, Okada Y, Thomas JM, Genazzani AA, Patel S, Galione A (2002) NAADP mobilizes Ca2+ from reserve granules, lysosome-related organelles, in sea urchin eggs. Cell 111: 703–708 [DOI] [PubMed] [Google Scholar]
- Dieter P, Marme D (1984) A Ca2+, calmodulin-dependent NAD kinase from corn is located in the outer mitochondrial membrane. J Biol Chem 259: 184–189 [PubMed] [Google Scholar]
- Delumeau O, Renard M, Montrichard F (1998) NAD+ kinase activity, calmodulin levels during the growth of isolated cells from Lycopersicon pimpinellifolium and kinetic constants of the calmodulin-dependent NAD+ kinase. Plant Sci 138: 43–52 [Google Scholar]
- Delumeau O, Renard M, Montrichard F (2000) Characterization and possible redox regulation of the purified calmodulin-dependent NAD(+) kinase from Lycopersicon pimpinellifolium. Plant Cell Environ 23: 1267–1273 [Google Scholar]
- Epel D, Patton C, Wallace RW, Cheung WY (1981) Calmodulin activates NAD kinase of sea urchin eggs: an early event of fertilization. Cell 23: 543–549 [DOI] [PubMed] [Google Scholar]
- Fromm H, Chua N-H (1992) Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol Biol Report 10: 199–206 [Google Scholar]
- Gallais S, Pou De Crescenzo M-A, Laval-Martin DL (2000) Changes in soluble and membrane-bound isoforms of calcium-calmodulin-dependent and -independent NAD(+) kinase, during the culture of after-ripened and dormant seeds of Avena sativa. Aust J Plant Physiol 27: 649–658 [Google Scholar]
- Gallais S, Pou De Crescenzo M-A, Laval-Martin DL (2001) Characterization of soluble calcium calmodulin-dependent and -independent NAD(+) kinases from Avena sativa seeds. Aust J Plant Physiol 28: 363–371 [Google Scholar]
- Harding SA, Oh SH, Roberts DM (1997) Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J 16: 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Jarrett HW, Cormier MJ (1984) An enzymatic assay for calmodulins based on plant NAD kinase activity. Anal Biochem 141: 168–178 [DOI] [PubMed] [Google Scholar]
- Iwasa F, Mohri H (1983) Calmodulin-binding proteins in the cytosol extract of sea urchin eggs. J Biochem 94: 575–587 [DOI] [PubMed] [Google Scholar]
- Jarrett HW, Brown CJ, Black CC, Cormier MJ (1982) Evidence that calmodulin is in the chloroplast of peas and serves a regulatory role in photosynthesis. J Biol Chem 257: 13795–13804 [PubMed] [Google Scholar]
- Kawai S, Mori S, Mukai T, Hashimoto W, Murata K (2001. a) Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem 268: 4359–4365 [DOI] [PubMed] [Google Scholar]
- Kawai S, Mori S, Mukai T, Suzuki S, Hashimoto W, Yamada T, Murata K (2000) Inorganic polyphosphate/ATP-NAD kinase of Micrococcus favus and Mycobacterium tuberculosis H37Rv. Biochem Biophys Res Commun 276: 57–63 [DOI] [PubMed] [Google Scholar]
- Kawai S, Suzuki S, Mori S, Murata K (2001. b) Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol Lett 200: 181–184 [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim JC, Lee MS, Heo WD, Seo HY, Yoon HW, Hong JC, Lee SY, Bahk JD, Hwang I, et al (1995) Identification of a novel divergent calmodulin isoform from soybean which has differential ability to activate calmodulin-dependent enzymes. J Biol Chem 270: 21806–21812 [DOI] [PubMed] [Google Scholar]
- Lee SH, Seo HY, Kim JC, Heo WD, Chung WS, Lee KJ, Kim MC, Cheong YH, Choi JY, Lim CO, et al (1997) Differential activation of NAD kinase by plant calmodulin isoforms. The critical role of domain I. J Biol Chem 272: 9252–9259 [DOI] [PubMed] [Google Scholar]
- Lerner F, Niere M, Ludwig A, Ziegler M (2001) Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 288: 69–74 [DOI] [PubMed] [Google Scholar]
- Liao B, Gawienowski MC, Zielinski RE (1996) Differential stimulation of NAD kinase and binding of peptide substrates by wild-type and mutant plant calmodulin isoforms. Arch Biochem Biophys 327: 53–60 [DOI] [PubMed] [Google Scholar]
- Ling V, Zielinski RE (1993) Isolation of an Arabidopsis cDNA sequence encoding a 22 kDa calcium-binding protein (CaBP-22) related to calmodulin. Plant Mol Biol 22: 207–214 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Braam J (2003) Calmodulins and potential calcium sensors of Arabidopsis. New Phytol 159: 585–598 [DOI] [PubMed] [Google Scholar]
- McGuinness ET, Butler JR (1985) NAD kinase: a review. Int J Biochem 17: 1–11 [DOI] [PubMed] [Google Scholar]
- Muto S (1983) Kinetic nature of calmodulin-dependent NAD kinase from pea seedlings. Z Pflanzenphysiol 109: 385–393 [Google Scholar]
- Muto S, Miyachi S (1977) Properties of a protein activator of NAD kinase from plants. Plant Physiol 59: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto S, Miyachi S, Usuda H, Edwards GE, Bassham JA (1981) Light-induced conversion of nicotinamide adenine-dinucleotide to nicotinamide adenine-dinucleotide phosphate in higher-plant leaves. Plant Physiol 68: 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio L, Bewell MA, Siddiqua A, Dickinson GD, Galione A, Sanders D (2000) Calcium release from the endoplasmic reticulum of higher plants elicited by the NADP metabolite nicotinic acid adenine dinucleotide phosphate. Proc Natl Acad Sci USA 97: 8693–8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten CE, Culotta VC (2003) A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J 22: 2015–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Churchill GC, Galione A (2001) Coordination of Ca2+ signalling by NAADP. Trends Biochem Sci 26: 482–489 [DOI] [PubMed] [Google Scholar]
- Pou De Crescenzo MA, Gallais S, Leon A, Laval-Martin DL (2001) Tween-20 activates and solubilizes the mitochondrial membrane-bound, calmodulin dependent NAD+ kinase of Avena sativa L. J Membr Biol 182: 135–146 [DOI] [PubMed] [Google Scholar]
- Pugin A, Frachisse JM, Tavernier E, Bligny R, Gout E, Douce R, Guern J (1997) Early events induced by the elicitor cryptogein in tobacco cells: involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate pathway. Plant Cell 9: 2077–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy AS (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC (1992) Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol 43: 375–414 [Google Scholar]
- Roberts DM, Oh S-H, Besl L, Weaver CD, Stacey G (1990) Attenuation of calmodulin-dependent NAD kinase activation by post-translational methylation. Curr Top Plant Biochem Physiol 9: 67–84 [Google Scholar]
- Rutter GA (2003) Calcium signalling: NAADP comes out of the shadows. Biochem J 373: e3–e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer A, Robinson DG (1985) Calmodulin dependent NAD kinase is associated with both the outer and inner mitochondrial membranes in maize roots. Planta 166: 227–233 [DOI] [PubMed] [Google Scholar]
- Simon P, Bonzon M, Greppin H, Marme D (1984) Subchloroplastic localization of NAD kinase activity: evidence for a Ca2+, calmodulin-dependent activity at the envelope and for a Ca2+, calmodulin-independent activity in the stroma of pea chloroplasts. FEBS Lett 167: 332–338 [Google Scholar]
- Simon P, Dieter P, Bonzon M, Greppin H, Marme D (1982) Calmodulin-dependent and independent NAD kinase activities from cytoplasmic fractions of spinach (Spinacia oleracea L.). Plant Cell Rep 1: 119–122 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Blumwald E (2000) Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J 24: 317–326 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151: 1–35 [DOI] [PubMed] [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H (1996) Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem 271: 4148–4153 [DOI] [PubMed] [Google Scholar]
- Williams MB, Jones HP (1985) Calmodulin-dependent NAD kinase of human neutrophils. Arch Biochem Biophys 237: 80–87 [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2002) Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc Natl Acad Sci USA 99: 4097–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8: 505–512 [DOI] [PubMed] [Google Scholar]
- Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M (2000) Calmodulin target database. J Struct Funct Genomics 1: 8–14 [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu BF, Liang S, Jones RL, Lu YT (2002) Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem J 368: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–725 [DOI] [PubMed] [Google Scholar]
- Zielinski RE (2002) Preparation of recombinant plant calmodulin isoforms. In HJ Vogel, ed, Calcium-Binding Protein Protocols, Vol 1. Humana Press, Totawa, NJ, pp 143–149 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.