Abstract
A cDNA encoding a calcium (Ca2+)/calmodulin (CaM)-dependent protein kinase (CaMK) from tobacco (Nicotiana tabacum), NtCaMK1, was isolated by protein-protein interaction-based screening of a cDNA expression library using 35S-labeled CaM as a probe. The genomic sequence is about 24.6 kb, with 21 exons, and the full-length cDNA is 4.8 kb, with an open reading frame for NtCaMK1 consisting of 1,415 amino acid residues. NtCaMK1 has all 11 subdomains of a kinase catalytic domain, lacks EF hands for Ca2+-binding, and is structurally similar to other CaMKs in mammal systems. Biochemical analyses have identified NtCaMK1 as a Ca2+/CaMK since NtCaMK1 phosphorylated itself and histone IIIs as substrate only in the presence of Ca2+/CaM with a Km of 44.5 μm and a Vmax of 416.2 nm min−1 mg−1. Kinetic analysis showed that the kinase not previously autophosphorylated had a Km for the synthetic peptide syntide-2 of 22.1 μm and a Vmax of 644.1 nm min−1 mg−1 when assayed in the presence of Ca2+/CaM. Once the autophosphorylation of NtCaMK1 was initiated, the phosphorylated form displayed Ca2+/CaM-independent behavior, as many other CaMKs do. Analysis of the CaM-binding domain (CaMBD) in NtCaMK1 with truncated and site-directed mutated forms defined a stretch of 20 amino acid residues at positions 913 to 932 as the CaMBD with high CaM affinity (Kd = 5 nm). This CaMBD was classified as a 1-8-14 motif. The activation of NtCaMK1 was differentially regulated by three tobacco CaM isoforms (NtCaM1, NtCaM3, and NtCaM13). While NtCaM1 and NtCaM13 activated NtCaMK1 effectively, NtCaM3 did not activate the kinase.
Calcium as a universal second messenger regulates a range of cellular and physiological processes in plants and animals (Poovaiah and Reddy, 1993; Gilroy and Trewavas, 1994; Sanders et al., 2002). It is believed that changes in cytosolic Ca2+ are sensed by a group of Ca2+-binding proteins, including calmodulin (CaM)-like domain protein kinases (CDPKs) and CaM (Chin and Means, 2000; Harmon et al., 2000; Luan et al., 2002). CDPKs, well-characterized kinases, have been shown to be modulated by binding of Ca2+ to a carboxy-terminal CaM-like sequence with EF hands and mediate many of the effects of Ca2+ in plant cells (Poovaiah and Reddy, 1987; Roberts and Harmon, 1992; Harper et al., 1994; Harmon et al., 2000; Cheng et al., 2002).
CaM is a ubiquitous intracellular Ca2+ receptor involved in transducing a variety of extracellular signals (Bush, 1995; Crivici and Ikura, 1995; Zielinski, 1998; Chin and Means, 2000). Whereas mammalian cells, such as those of humans, contain only one CaM protein encoded by at least three differentially regulated CaM genes (Fischer et al., 1988), plant cells have multiple CaM genes that encode numerous CaM isoforms (Snedden and Fromm, 1998; Zielinski, 1998; Yamakawa et al., 2001). The existence of multiple divergent CaM isoforms in plants poses the question of whether or not they allow differential regulation of targets and can confer different Ca2+ sensitivity to CaM-binding enzymes or proteins. One group of CaM-binding proteins is composed of Ca2+/CaM-dependent protein kinases (CaMKs). In animals, a large number of Ca2+/CaMKs have been identified, including CaMKI, II, and IV (Fajisawa, 1990; Klee, 1991; Mochizuki et al., 1993). Among them, multifunctional CaMKII, a well-characterized kinase that can phosphorylate a broad range of proteins upon binding to Ca2+/CaM, is thought to play an important role in a variety of cellular events in mammals (Colbran and Soderling, 1990).
Although typical Ca2+/CaMKs with activities similar to CaMK in animals have not been identified in plants (Luan et al., 2002), several protein kinases that share sequence homology with CaMK have been identified in plants. While the CaM-binding abilities of these kinases have been demonstrated, the effects of CaM on their activities has not been established (Watillon et al., 1992, 1995; Lu et al., 1996; Yang et al., 2000; Wang et al., 2001; Zhang and Lu, 2003). A chimeric Ca2+/CaMK (CCaMK) with a visinin-like Ca2+-binding domain has been characterized from lily and tobacco (Nicotiana tabacum). Whereas CaM is required for substrate phosphorylation by this CCaMK, CaM suppresses its autophosphorylation (Takezawa et al., 1996; Ramachandiran et al., 1997; Liu et al., 1998; Poovaiah et al., 1999), indicating that CCaMK has different enzymatic and structural characteristics from classic animal CaMKs. Recently, a CaM-binding protein kinase (CBK), OsCBK, was isolated from rice (Oryza sativa). Although OsCBK can bind to CaM with high affinity, both autophosphorylation and substrate phosphorylation of this kinase are CaM independent (Zhang et al., 2002).
Here, we report on the isolation and identification of a cDNA from tobacco that encodes a CaMK (NtCaMK1). We show that substrate phosphorylation and autophosphorylation of NtCaMK1 is Ca2+/CaM dependent. Capillary electrophoresis of phosphoamino acids revealed that phosphorylation of NtCaMK1 occurs on Thr residues. Kinetic analysis showed that NtCaMK1 specifically bound CaM with high affinity (Kd = 5 nm) via a CaM-binding domain (CaMBD) having a 1-8-14 motif. Although the kinase was effectively activated by both NtCaM1 and NtCaM13, NtCaM3 failed to stimulate the enzymatic activation of NtCaMK1. These data indicate that NtCaMK1 is a novel Thr kinase modulated by Ca2+/CaM.
RESULTS
Cloning and Characterization of NtCaMK1 cDNA
A cDNA library constructed with the expression vector λZAPII and poly(A) RNA purified from tobacco cells (cv Wisconsin-38) was screened with 35S-labeled CaM as a ligand probe for the presence of CaM-binding proteins. A candidate recombinant phage was excised in vivo into recombinant pBluescript SK− plasmid pNtCaMK1 and the insert sequenced. Figure 1 shows the cDNA sequence of the insert and the deduced amino acid sequence designated as NtCaMK1. The nucleotide sequence of the NtCaMK1 cDNA has been lodged with GenBank (accession no. AF550608 for NtCaMK1). The NtCaMK1 cDNA contains an open reading frame (ORF) of 4,248 bp with a start codon at 67 to 69 bp and a stop codon at 4,312 to 4,314 bp. This clone was determined to be a full-length ORF based on the presence of an in-frame stop codon at 61 to 63 bp in the 5′ untranslated region (Fig. 1A). Northern-blot analyses revealed a single band of about 5.0 kb (Fig. 2), consistent with the predicted size of the cDNA. Southern-blot analysis with tobacco genomic DNA digested with EcoRV or KpnI showed only one band (Fig. 3), suggesting that NtCaMK1 is encoded by a single gene.
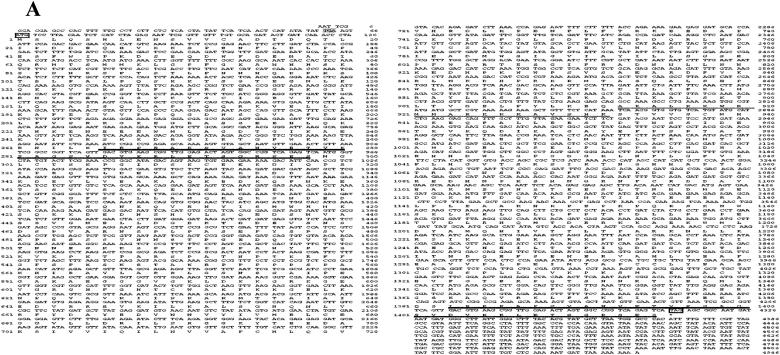
Figure 1.
Characterization of NtCaMK1 cDNA. A, Nucleotide and deduced amino acid sequences of NtCaMK1 (AF550608) from tobacco. Nucleotides and amino acid residues are numbered on the right. Start (ATG) and stop (TAG) codons are boxed. The stop codon (TGA) in frame in the 5′ untranslated region is indicated by gray squares at positions 61 to 63. The putative CaM-binding domain is underlined. The strong PEST sequences are in bold [452–466 (+8.75), 481–506 (+9.02), 1241–1272 (+8.63)]. The 33 amino acid repeats are double underlined. B, Alignment of NtCaMK1 with other kinases, including CaM-binding protein kinases and CDPKs. The kinases are: maize MCK1 (accession no. 1839597; Lu et al., 1996), rat brain CaMKII α-subunit (accession no. J02942; Lin et al., 1987), tobacco NtCBK2 (accession no. AF435452; Hua et al., 2003), maize MCKI (accession no. 1839597; Lu et al., 1996), Arabidopsis CRK5 (accession no. At3g50530), lily CCaMK (accession no. 860675; Patil et al., 1995), soybean CDPK (accession no. P28583; Harper et al., 1991), and Arabidopsis CDPK3 (accession no. 1220099; Urao et al., 1994). Identities between NtCaMK1 and other kinases are indicated by gray squares. C, Diagram showing genomic structure of NtCaMK1. The introns are shown by boxes and exons with lines. The numbers above the boxes indicate the size of the introns (in bp) and the numbers above the lines indicate the size of the exons (in bp). A larger version of this figure is available at http://www.plantphysiol.org/cgi/content/full/135/3/1280/DC1.
Figure 2.

Northern blot for NtCaMK1 in tobacco leaves. Total RNA (20 μg) from leaves was subjected to formaldehyde/agarose gel electrophoresis and blotted onto nitrocellulose. The blot was hybridized to α-32P-labeled DNA made from pNtCaMK1 and autoradiographed at −80°C.
Figure 3.
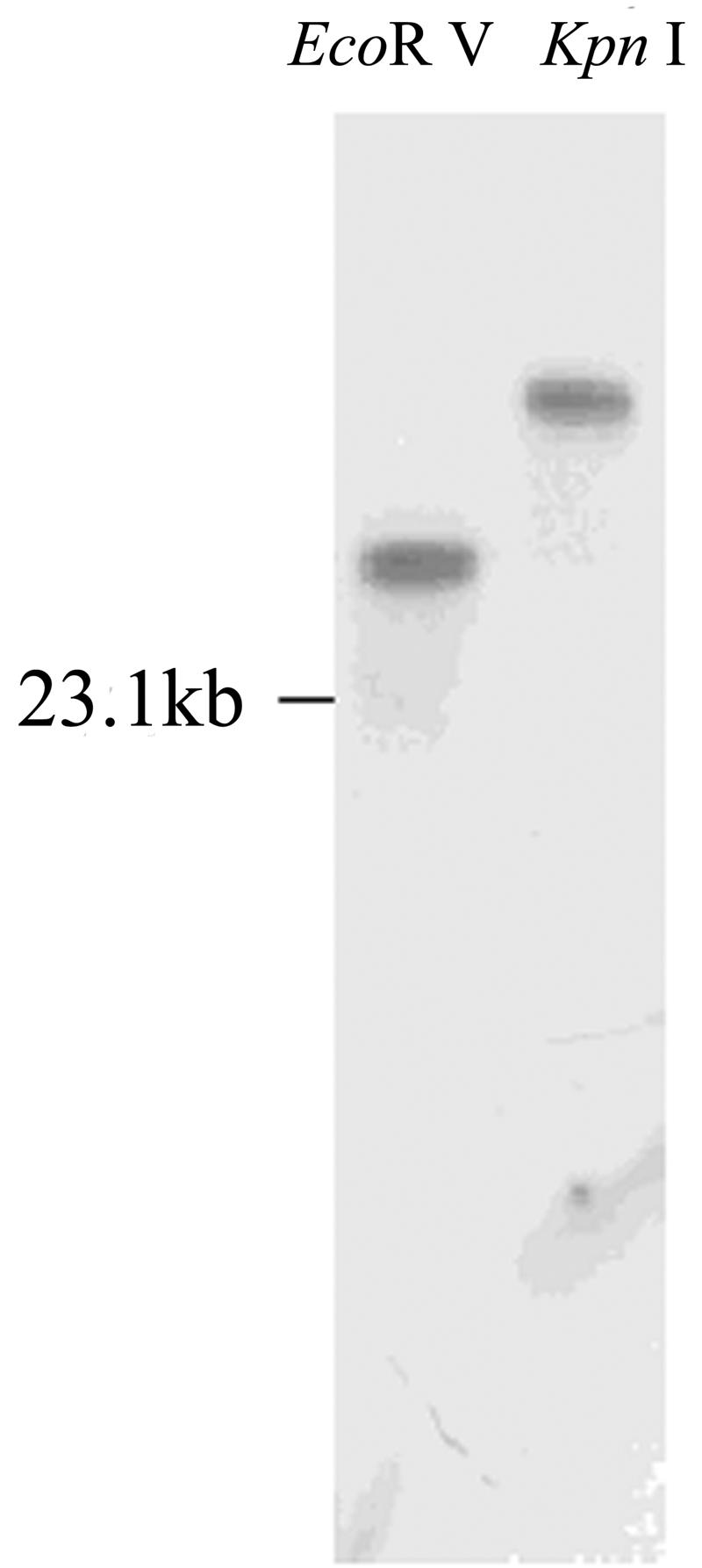
Genomic Southern-blot analysis. Ten micrograms of genomic DNA from tobacco were digested with EcoRV or KpnI, separated by agarose electrophoresis, and probed with the 32P-labeled NtCaMK1 cDNA.
The deduced amino acid sequence of NtCaMK1 consists of 1,415 residues with a calculated molecular mass of 156 kD, and contains all 11 subdomains characteristic of a protein kinase catalytic domain (Hanks et al., 1988). A database comparison to identify homologies between NtCaMK1 and other genes with known functions suggests that NtCaMK1 is a CaMK homolog (Fig. 1B). NtCaMK1 shares high sequence identity with many CBKs, including rat CaMKII α-subunit (30%; accession no. J02942; Lin et al., 1987), tobacco NtCBK2 (56%; accession no. AF435452; Hua et al., 2003), maize (Zea mays) MCKI (50%; accession no.1839597; Lu et al., 1996), lily CCaMK (31%; accession no. 860675; Patil et al., 1995); and CDPKs such as soybean (Glycine max) CDPK (44%; accession no. P28583; Harper et al., 1991), Arabidopsis AtCDPK3 (29%; accession no. 1220099; Urao et al., 1994), and Arabidopsis CRK5 (62%; accession no. At3g50530; Fig. 1B). Furthermore, the nonkinase part of NtCaMK1 shares 17% homology with a potato calmodulin-binding protein (accession no. AF378084; Reddy et al., 2002), a 5 kb-long sequence that encodes a protein of 1,309 amino acids. The NtCaMK1 sequence was also analyzed using programs found on the ExPASy Proteomics tools Web page (www.expasy.ch/tools). Using MotifScan, we found three strong PEST sequences with PEST scores (452–466 [+8.75], 481–506 [+9.02], 1241–1272 [+8.63]), respectively (Fig. 1A). Unlike CDPKs and CCaMKs, NtCaMK1 does not possess Ca2+-binding EF hands at its C terminus (Fig. 1B).
To rule out the possibility that NtCaMK1 resulted from some artifacts during the construction of the cDNA library, a PCR-based strategy was used to isolate the genomic sequence for NtCaMK1, and two PCR fragments, named P1 (13.3 kb) and P2 (13.5 kb), were obtained (data not shown). The sequence data (AY562224) indicated that these two PCR fragments have 2,266 bp of overlapping sequence, and the 24.6-kb genomic sequence has 21 exons interrupted by 20 introns in the full-length cDNA. The intron-exon borders were determined by comparison with the cDNA sequence, and they all conform to the canonical GT/AG 5′ and 3′ splice sites (Fig. 1C).
Definition of the CaMBD of NtCaMK1
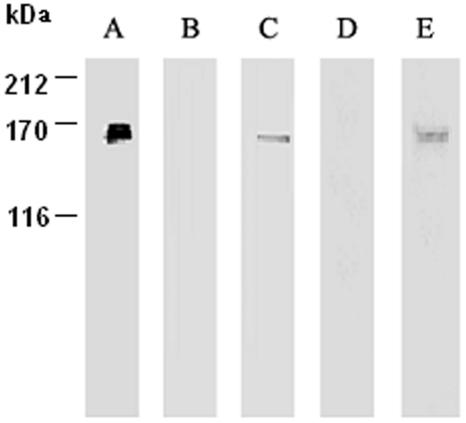
Since NtCaMK1 was isolated by screening a tobacco cDNA library with 35S-labeled CaM implying its CaM-binding ability, we characterized its ability to bind tobacco CaM (Fig. 4A). When purified NtCaMK1 was used for CaM binding with either 35S-labeled CaM or biotinylated CaM, CaM bound only in the presence of Ca2+ (Fig. 4, C and E). CaM binding was lost in the presence of EGTA (Fig. 4, B and D), indicating that NtCaMK1 is a CaM-binding protein that binds CaM in a Ca2+-dependent manner. This conclusion was also supported by the observation that, during purification, NtCaMK1 was eluted from CaM-Sepharose-4B only in the absence of Ca2+.
Figure 4.
CaM-binding assays of NtCaMK1. Purified NtCaMK1 was separated by SDS-PAGE and either silver stained (lane A) or transferred to membranes for CaM-binding assays with biotinylated CaM (lanes B and C) or 35S-labeled CaM (lanes D and E) in the presence of either 2 mm EGTA (lanes B and D) or 1 mm calcium (lanes C and E).
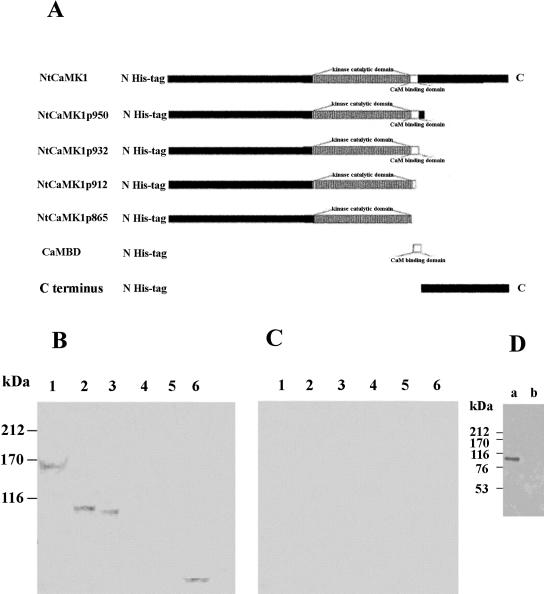
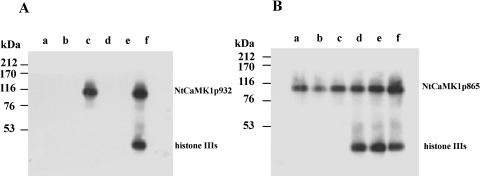
To further define the CaMBD of NtCaMK1, several expression plasmids were constructed to produce different truncated forms of NtCaMK1 that were used for CaM-binding assays (Fig. 5A). All recombinant forms of NtCaMK1 lacked CaM-binding ability in the presence of EGTA (Fig. 5C). However, in the presence of Ca2+, constructs NtCaMK1p932 and NtCaMK1p950 containing the putative CaMBD (amino acid residues 913–932) bound CaM (Fig. 5B, lanes 2 and 3), as did full-length NtCaMK1 (Fig. 5B, lane 1). Constructs NtCaMK1p865 and NtCaMK1p912 lacking the putative CaMBD lacked CaM-binding ability (Fig. 5B, lanes 4 and 5), indicating that the CaMBD is located among the 20 to 30 amino acid residues at the C terminus of NtCaMK1p932.
Figure 5.
Identification of the CaMBD of NtCaMK1. Several constructs for full and truncated NtCaMK1s were made and schematic representations of NtCaMK1 and the truncated forms are shown in A with numbers to indicate the truncated sites at the position of amino acid sequence of NtCaMK1. Proteins expressed from these constructs were assayed with 35S-labeled CaM in the presence of either 1 mm calcium (B) or 2 mm EGTA (C). Lane 1, NtCaMK1; lane2, NtCaMK1p950; lane 3, NtCaMK1p932; lane 4, NtCaMK1p912; lane 5, NtCaMK1p865, and lane 6, pCaMBD. D, The expressed proteins were assayed with 35S-labeled CaM in the presence of 1 mm calcium. Lane a, NtCaMK1p932; lane b, C terminus of NtCaMK1.
The prediction of secondary structure of the 20 amino acids from Ala-913 to Lys-932 of NtCaMK1 using the Anthwin 45 package (Microsoft, Redmond, WA) suggests a basic amphiphilic α-helix (data not shown). The helical wheel projection for NtCaMK1 CaMBD shows a typical CaMBD feature, with segregation of basic and hydrophobic residues on opposite sides of the helix (O'Neil and DeGrado, 1990; Putnam-Evans et al., 1990; Lu and Harrington, 1994). To experimentally identify this tentative CaMBD, construct pCaMBD encoding these 20 amino acid residues was made with vector pET32a and used for CaM-binding assay. The protein expressed by pCaMBD bound CaM in a Ca2+-dependent manner (Fig. 5B, lane 6, and C, lane 6), demonstrating that it is these 20 amino acid residues at positions 913 to 932 that are likely responsible for CaM binding. To test whether the truncated C terminus has a CaMBD, the C-terminal amino acid sequence from 933 to 1,415 was expressed and assayed for CaM binding. The results indicated that the C terminus of NtCaMk1 did not have CaM-binding ability (Fig. 5D).
Critical Residues of the CaMBD
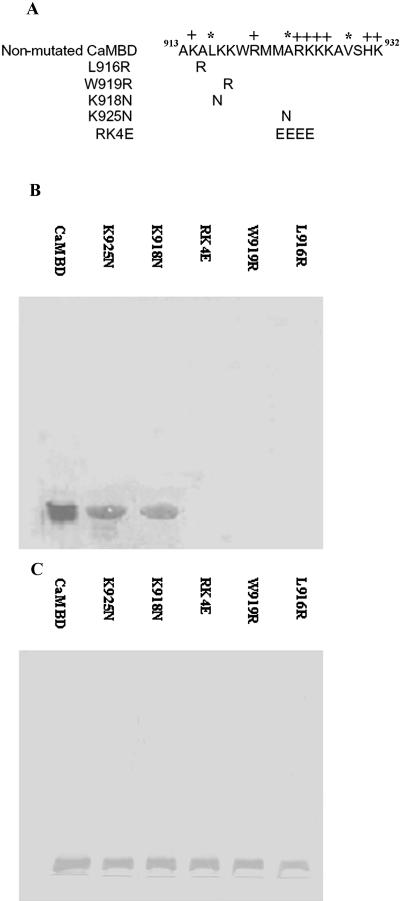
Comparative analyses of the CaM-binding regions of the many reported CaM-binding proteins has provided multiple sequence motifs required for CaM complex formation. Two motifs for Ca2+-dependent binding, named 1-8-14 and 1-5-10, were suggested based on the position of conserved hydrophobic residues (Rhoads and Friedberg, 1997). In the 1-8-14 motif of NtCaMK1, hydrophobic amino acids are present at positions 1 (Leu-916), 8 (Ala-923), and 14 (Val-929), and several basic residues (six Lys and two Arg) are interspersed between the hydrophobic residues. These characteristics of the CaMBD of NtCaMK1 match a consensus Ca2+-dependent CaM-binding motif designated 1-8-14 (Rhoads and Friedberg, 1997). A Trp residue (amino acid 919) known to play a critical role in the CaM binding of many CaM target proteins (Arazi et al., 1995; Diggle et al., 1999; Osawa et al., 1999) was also found within this region.
To confirm the role of these amino acid residues in CaM binding in NtCaMK1, amino acid substitutions at several residues that were expected to play a critical role in the interaction between CaM and NtCaMK1 were introduced. The hydrophobic 916Leu at the first position of the 1-8-14 motif was replaced by Arg (L916R), and 919Trp was replaced by Arg (W919R; Fig. 6A). Expression constructs for these mutations were introduced into Escherichia coli and the expressed proteins separated on SDS-PAGE and transferred to polyvinylidine difluoride (PVDF) membranes for CaM-binding assays with biotinylated CaM (Reddy et al., 1993; Zhang et al., 2002). Figure 6B shows that these single amino acid substitutions in NtCaMK1 abolished the CaM-binding ability (Fig. 6B, lanes L916R and W919R), demonstrating sequence-specific complex formation and the crucial roles of residues Leu-916 and Trp-919 for CaM binding in the NtCaMK1 CaMBD.
Figure 6.
CaM-binding assays of site-directed mutants in the CaMBD of NtCaMK1. Six constructs for nonmutant and site-directed mutated mutants in NtCaMK1 CaMBD were made with pET32a and the schematic representation of these mutants are shown in A with substituted amino residues shown in single-letter symbols, and (+) and (*) to indicate positive-charged amino acid residues and hydrophobic amino acid residues at positions 1, 8, and 14 of the 1-8-14 motif, respectively. The expressed proteins separated by SDS-PAGE were either transferred to PVDF membranes and assayed for CaM-binding assays with biotinylated CaM in the presence of either 1 mm Ca2+ (B) or stained with Coomassie Brilliant Blue (C).
We also modified the CaMBD of NtCaMK1 by introducing four negatively charged Glu residues instead of the positively charged Arg and Lys residues at positions Arg-924 and Lys927 (913AKALKKWRMMARKKKAVSHK932 to 913AKALKKWRMMAEEEEAV SHK932), resulting in the breakage of basic amphiphilic α-helix structure (Fig. 6A). CaM-binding assays with this modified CaMBD showed that activity was lost (Fig. 6B, lane RK4E). However, when positively charged amino acid residues were replaced by other positively charged amino acid residues, such as K918N or K925N (Fig. 6A), the substitutions did not affect complex formation between the CaMBD and CaM (Fig. 6B, lanes K918N and K925N), demonstrating the role of positive charges of amino acid residues at certain positions in the CaMBD of NtCaMK1.
Calculation of the Dissociation Constant of NtCaMK1/CaM
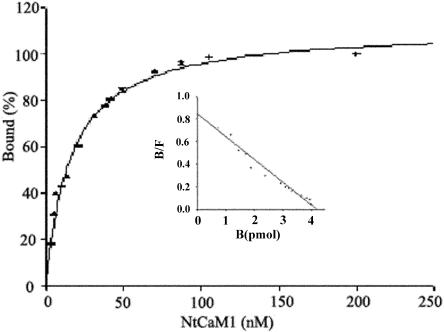
It has been reported that different CaM-binding proteins have different binding affinities for CaM, resulting in subtle regulation by different concentrations of either CaM and/or CaMBPs (Sikela and Hahn, 1987). To determine the binding affinity of CaM for NtCaMK1, 4 pm NtCaMK1 were assayed with different concentrations of 35S-labeled CaM in the presence of 1 mm CaCl2. Binding of CaM to NtCaMK1 was saturated at concentrations of about 105 nm CaM (Fig. 7), indicating the presence of a saturable, high-affinity binding site in NtCaMK1. The Kd of CaM for NtCaMK1 was estimated to be 5 nm, based on the saturation curve of 35S-CaM binding to NtCaMK1, and by fitting the data using SPSS EnzymeKinetics (SPSS, Chicago). The binding of CaM to NtCaMK1 was completely blocked in the presence of 2 mm EGTA.
Figure 7.
Assay for Kd of NtCaMK1 to CaM. The saturation curve of 35S-labeled CaM binding to purified NtCaMK1 was carried out as follows. Purified NtCaMK1 protein (4 pm) separated by SDS-PAGE was transferred to nitrocellulose and incubated with different amounts of 35S-labeled CaM. After washing in buffer without 35S-CaM, radioactivity on the filter was measured by liquid scintillation counting. Bound CaM at each CaM concentration was expressed as a percent of the maximal binding. The inset shows a Scatchard plot of data indicating that the binding ratio of CaM to NtCaMK1 is 1:1. Bound/free and bound CaM are expressed as B/F and B, respectively.
Autophosphorylation and Substrate Phosphorylation of NtCaMK1
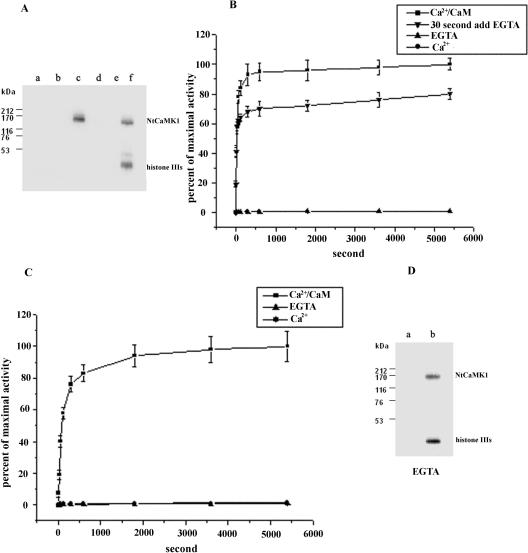
NtCaMK1 was purified and used to analyze autophosphorylation and substrate phosphorylation activity. Both autophosphorylation and histone IIIs phosphorylation activities were Ca2+/CaM-dependent (Fig. 8A, lanes c and f) and no phosphorylation activity was detected in the presence of either EGTA or calcium alone (Fig. 8A, lanes a, b, d, and e). This result was further supported by time course analyses for kinase activity of NtCaMK1. Time course analyses showed that NtCaMK1 rapidly phosphorylated itself and histone IIIs as substrate, but only in the presence of Ca2+/CaM (Fig. 8, B and C). The time to maximal autophosphorylation activity was 5 min and for maximal activity of substrate phosphorylation 30 min. Autophosphorylation and substrate phosphorylation can be catalyzed by autophosphorylated NtCaMK1 in a Ca2+/CaM-independent manner as shown in Figure 8B. These data show that autophosphorylation of NtCaMK1 occurred when it was initiated by Ca2+/CaM and then blocked with excess EGTA. Autophosphorylation of NtCaMK1 occurs when an autophosphorylation reaction was initiated for as little as 30 s in the presence of Ca2+/CaM, followed by incubation in excess EGTA, although maximal autophosphorylation activity was lower in the presence of excess EGTA than in the presence of Ca2+/CaM alone. Autophosphorylated NtCaMK1 showed substrate phosphorylation activity in the presence of EGTA (Fig. 8D).
Figure 8.
Phosphorylation assays of NtCaMK1. A, NtCaMK1 autophosphorylation (lanes a–c) and substrate phosphorylation using histone IIIs as substrate (lanes d–f) were performed in the presence of EGTA (lanes a and d), or Ca2+ (lanes b and e) or Ca2+/CaM (lanes c and f). The reaction products were analyzed by SDS-PAGE using a discontinuous 8%/20% separating gel. B, Time course of NtCaMK1 autophosphorylation in the presence of Ca2+/CaM (▪) or EGTA (▴) or Ca2+ (•). EGTA was added to the reaction mixture containing Ca2+/CaM after 30 s of incubation for initiation of NtCaMK1 autophosphorylation to a final concentration of 2 mm (▾). The data are averages of three determinations done in duplicate. C, Time course of phosphorylation of histone IIIs by NtCaMK1 in the presence of Ca2+/CaM (▪) or EGTA (▴) or Ca2+ (•). The data are averages of three determinations done in duplicate. D, Substrate phosphorylation of NtCaMK1 was performed with un-autophosphorylated NtCaMK1 in the presence of EGTA (a) or complete autophosphorylated NtCaMK1 by incubating the enzyme in the reaction mixture containing Ca2+/CaM for 6 min and then adding 2 mm EGTA following histone IIIs as substrate (b). The reaction products were analyzed by SDS-PAGE with a discontinuous 8%/20% separating gel.
Kinetic parameters of the kinase activity of NtCaMK1 were determined from double-reciprocal analyses of data for phosphorylation of various concentrations of histone IIIs in the presence of 100 μm ATP. The Km for histone IIIs was calculated to be 44.5 μm and the Vmax 416.2 nm min−1 mg−1 when Ca2+/CaM was added. A synthetic peptide substrate for CaMKII, syntide-2 (Hashimoto and Soderling, 1987), was also used as substrate in phosphorylation assays with NtCaMK1. Kinetic analysis showed that unphosphorylated NtCaMK1 had a Km for syntide-2 of 22.1 μm and a Vmax of 644.1 nm min−1 mg−1 when assayed in the presence of Ca2+/CaM.
Autophosphorylation and Substrate Phosphorylation of Truncated NtCaMK1
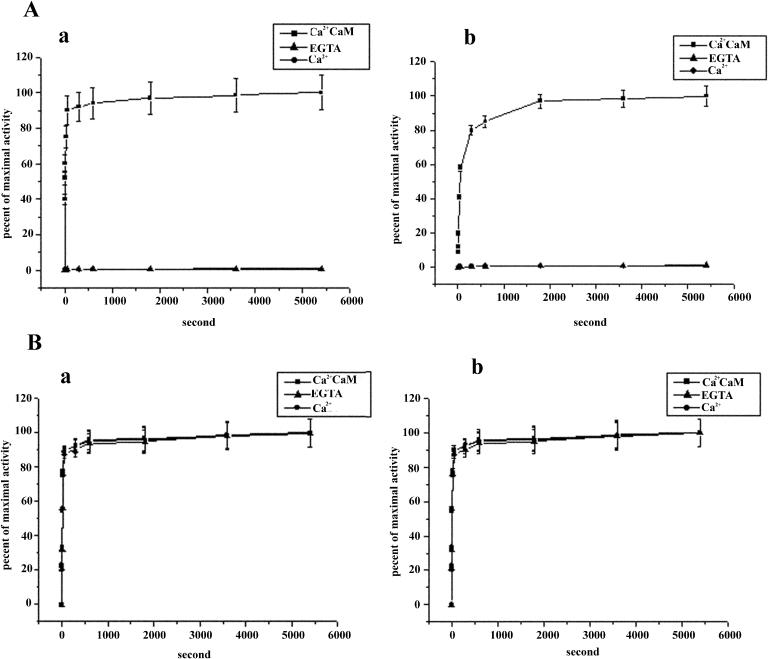
To explore the effects of the regulatory region, including the CaMBD, on the enzymatic properties of NtCaMK1, two truncated forms of NtCaMK1 were utilized for enzymatic assays. NtCaMK1p932, which includes the CaMBD, carried out autophosphorylation and substrate phosphorylation in the presence of Ca2+/CaM and lost kinase activity in the presence of either calcium or EGTA (Fig. 9A), showing the same Ca2+/CaM dependence as full-length NtCaMK1. Time course analyses showed that purified NtCaMK1p932 rapidly phosphorylated histone IIIs in the presence of Ca2+/CaM, with 5 min to maximal activity of autophosphorylation and 30 min to maximal activity of substrate phosphorylation (Fig. 10A), consistent with enzymatic characteristics of full NtCaMK1. NtCaMK1p865, lacking the CaMBD, carried out autophosphorylation and substrate phosphorylation in the presence of both Ca2+/CaM and EGTA (Fig. 9B). Time course analyses showed that purified NtCaMK1p865 rapidly phosphorylated histone IIIs in the presence of either EGTA or Ca2+ in the absence of CaM (Fig. 10B). Differences in the enzymatic characteristics of NtCaMK1p865 and NtCaMK1p932 suggest that amino acids 865 to 932 may inhibit NtCaMK1 activity in the absence of Ca2+/CaM, and that this inhibition can be abolished by binding CaM to the CaMBD of NtCaMK1.
Figure 9.
Phosphorylation assays of the truncated NtCaMK1. NtCaMK1p932 (A) and NtCaMK1p865 (B) autophosphorylation (lanes a–c) and substrate phosphorylation using histone IIIs as substrate (lanes d–f) were performed in the presence of either 2 mm EGTA (lanes a and d), or 1 mm Ca2+ (lanes b and e) or Ca2+/CaM (lanes c and f) and the products resolved by 10% SDS-PAGE.
Figure 10.
Time courses of autophosphorylation and substrate phosphorylation of the truncated forms of NtCaMK1. Time course of autophosphorylation (a) and phosphorylation of histone IIIs (b) by NtCaMK1p932 (A) or NtCaMK1p865 (B) in the presence of Ca2+/CaM (▪) or EGTA (▴) or Ca2+ (•). The data are averages of three determinations done in duplicate.
Activation of NtCaMK1 by NtCaM Isoforms
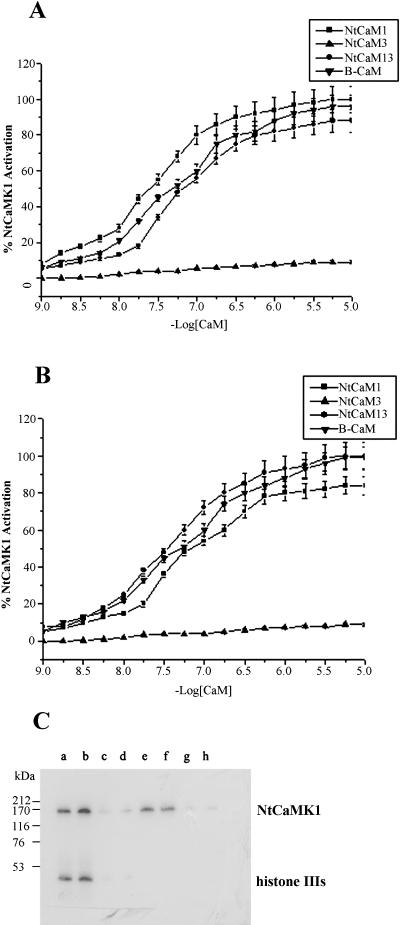
To investigate whether this NtCaMK1 is differentially regulated by various CaM isoforms, three tobacco CaM isoforms (NtCaM1, NtCaM3, and NtCaM13) and bovine brain CaM were used, and the Ka value of NtCaMK1 activity was assayed with the addition of increasing amounts of CaM in the presence of 1 mm Ca2+ (Fig. 11). Our experiments indicated that NtCaM1, NtCaM13, and bovine brain CaM half-maximally activated NtCaMK1 with a Ka of 26 nm, 60 nm, and 40 nm, respectively, using histone IIIs as substrate. However, NtCaM3 produced essentially no activation of NtCaMK1 (Fig. 11A). When syntide-2 was employed as substrate, NtCaMK1 can be activated by NtCaM1, NtCaM13, and bovine brain CaM, but with different Ka values (56 nm, 31 nm, and 45 nm, respectively). NtCaM3 still cannot stimulate the enzymatic activation of NtCaMK1 with syntide-2 (Fig. 11B). Thus, NtCaM13 was a better activator than NtCaM1 when syntide-2 was used as substrate, whereas NtCaM1 was better when histone IIIs was used. However, NtCaM3 failed to activate the kinase with both substrates. These data suggest differential regulation of the kinase by different CaM isoforms. This was further verified when the autophosphorylation and substrate phosphorylation products of NtCaMK1 were analyzed by SDS-PAGE. While both substrate phosphorylation and autophosphorylation activities were detected with NtCaM1 (Fig. 11C, lanes a and e) and NtCaM13 (Fig. 11C, lanes b and f) as activators, no phosphorylation activity was detected in the presence of 1 μm NtCaM3 (Fig. 11C, lanes c and g), even in the presence of extremely high concentrations (10 μm) of NtCaM3 (Fig. 11C, lanes d and h).
Figure 11.
Assay for CaM isoforms required for NtCaMK1 phosphorylation. Histone IIIs (A) or syntide-2 (B) as substrates were phosphorylated with NtCaMK1 (0.2 μg μL−1) in the presence of NtCaM1(▪), NtCaM3(▴), NtCaM13(•), or B-CaM (▾) in our standard phosphorylation conditions, except increasing amounts of NtCaM (10−9–10−5 m) at 30°C for 5 min. The data are averages of three determinations done in duplicate. C, NtCaMK1 autophosphorylation (lanes e–h) and substrate phosphorylation using histone IIIs as substrate (lanes a–d) were performed in the presence of Ca2+/NtCaM1 (lanes a and e), Ca2+/NtCaM13 (lanes b and f), Ca2+/NtCaM3 (1 μm; lanes c and g), or Ca2+/NtCaM3 (10 μm; lanes d and h) and resolved by SDS-PAGE with discontinuous 8%/20% separating gel.
Phosphoamino Acid Analysis
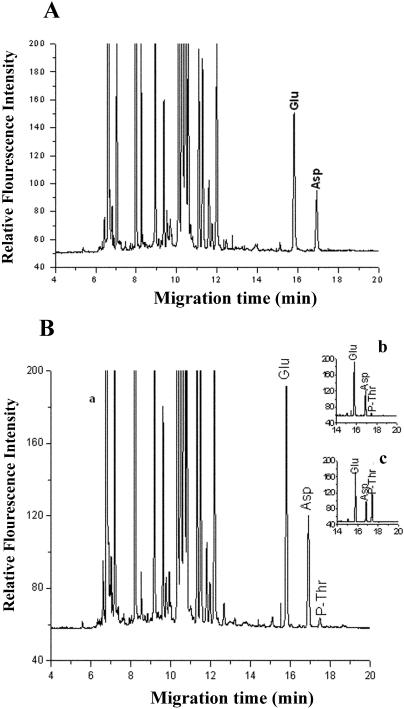
Capillary electrophoresis was used to identify the amino acids that become phosphorylated during autophosphorylation of NtCaMK1. When the hydrolysis products of purified NtCaMK1 that were not autophosphorylated in vitro were separated by capillary electrophoresis, no phosphoamino acids were detected, demonstrating that the purified NtCaMK1 used in our kinase activity assays was not phosphorylated in vivo (Fig. 12A). Capillary electrophoretic analyses of autophosphorylated NtCaMK1 showed that Thr residues were phosphorylated (Fig. 12B), indicating that NtCaMK1 was a Thr protein kinase, as suggested by our database analysis.
Figure 12.
Phosphoamino acid analyzed by capillary electrophoresis. A, Electropherogram of the hydrolyzed unphosphorylated NtCaMK1 with FITC labeling. B, Electropherograms of the hydrolyzed autophosphorylated NtCaMK1 with FITC labeling. a, Whole electropherogram; b, electropherogram intercepted from a and c; c, electropherogram with addition of standard P-Thr to phosphorylated NtCaMK1 sample to indicate phosphorylated Thr residue.
DISCUSSION
Plant cells generate Ca2+ signals with different amplitudes, frequencies, and durations in response to a variety of internal and external stimuli (Knight et al., 1991; Price et al., 1994; Bush, 1995; McAinsh and Hetherington, 1998). As a major calcium sensor and target modulator, CaM plays a vital role in transducing Ca2+ signals by regulating the activity of numerous target proteins, including CaMKs (Zielinski, 1998). CaMKs are key components in many calcium/CaM-mediated pathways in animals and yeast (Saccharomyces cerevisiae). However, typical Ca2+/CaMK similar to CaMKs have not been identified at the molecular and biochemical levels in plants. In contrast, plants contain a unique class of protein kinases, the CDPKs, that bind Ca2+ but not CaM (Poovaiah and Reddy, 1987; Roberts and Harmon, 1992; Harper et al., 1994). In this study, we have identified a novel plant protein kinase, NtCaMK1, that shares the features of animal and yeast CaMKs. The full length of cDNA for NtCaMK1 is 4.8 kb and contains an ORF consisting of 1,415 amino acid residues. This cDNA sequence is unusually long, but it does not result from a cloning artifact because our genomic sequence data demonstrate that this sequence is from one gene. At least one other plant CaM-binding protein has an unusually long cDNA. The cDNA of potato CaM-binding protein is about 5.0 kb long and shares 17% homology with NtCaMK1 (Reddy et al., 2002).
Several lines of evidence support our conclusion that NtCaMK1 is a protein kinase of the CaMK class and not a CDPK. Biochemical analyses indicate that autophosphorylation and phosphorylation of histone IIIs by NtCaMK1 occurs only in the presence of Ca2+/CaM, showing a regulatory role of CaM on NtCaMK1 activity. In contrast, plant CDPKs bind their own intramolecular CaM-like domain, but do not bind free CaM unless the C-terminal CaM-like domain is removed (Yoo and Harmon, 1996). Thus, CDPKs are calcium-dependent protein kinases that are not modulated by CaM (Yoo and Harmon, 1996). Further evidence supporting our conclusion that NtCaMK1 is a CaMK comes from the observation that NtCaMK1 lacks Ca2+-binding EF hands and has Ca2+/CaM-dependent autophosphorylation and substrate phosphorylation activity. With histone IIIs as substrate, NtCaMK1 exhibits half-maximal activity (Ka) of 40 nm for CaM, similar to that reported for CaMKII (20–100 nm; Schulman, 1988) and CaMKIV (26–150 nm; Kameshita and Fujisawa, 1993; Enslen et al., 1994). The similarity between the Ka of NtCaMK1 and animal CaMKs is probably a result of its dissociation constant for CaM (Kd of 5 nm), a value similar to that of animal CaMKs of around 1 to 10 nm (Sikela and Hahn, 1987). In addition, autophosphorylated NtCaMK1 showed substrate phosphorylation activity in the presence of EGTA (Fig. 8D), indicating that the autophosphorylated kinase has Ca2+-independent activity as CaMKII does (Lai et al., 1986).
CaM-binding proteins have characteristic CaMBDs that allow protein-protein interactions, and comparative analyses have revealed multiple sequence motifs required for CaM complex formation. Most of the CaMBDs identified in CaM-binding proteins are stretches of 16 to 35 amino acid residues that form α-helical wheels with the segregation of basic and polar residues on one side of the helix and hydrophobic amino acids on the other (Ikura et al., 1992; Meador et al., 1992; Crivici and Ikura, 1995). Rhoads and Friedberg (1997) have reported 1-8-14 and 1-5-10 motifs for Ca2+-dependent CaM binding in which the numbers indicate the positions of the conserved hydrophobic residues. Based on the sequence of the CaMBD of NtCaMK1, we have characterized the CaM-binding motif as a member of the 1-8-14 class. The CaMBD of NtCaMK1 is located in amino acid residues between Ala-913 and Lys-932 based on CaM-binding assays, with full-length and truncated forms of our kinase. We further classified the CaMBD as a 1-8-14 motif because of the importance of Leu-916 and Trp-919 for CaM binding, as demonstrated by our site-directed mutagenesis experiments. It was noted that truncated NtCaMK1p865 was constitutively active in the presence or absence of Ca2+/CaM, but activation of truncated NtCaMK1p932 required the presence of Ca2+/CaM (Fig. 9), suggesting that amino acids at positions 865 to 932 have an inhibitory effect on kinase activity in the absence of Ca2+/CaM. Kinase activity can be restored in NtCaMK1p932 by CaM binding to the CaMBD, an observation similar to that made for other CaMKs that have an autoinhibitory junction located between the N-terminal kinase catalytic domains and the C-terminal association domain (Kemp and Pearson, 1991; Hanson and Schulman, 1992). The precise amino acid sequence responsible for this inhibition remains to be determined experimentally.
Unlike mammalian systems, plant cells have many different isoforms of CaM. The differential expression and subcellular locations of plant CaMs in response to environmental stimuli and during plant development provide the mechanisms for altering Ca2+ signal transduction and the intrinsic means of selectively activating or inhibiting specific CaM target enzymes in plants (Lee et al., 1995; Heo et al., 1999; Yamakawa et al., 2001). Thus, understanding the way in which different CaM isoforms modulate their targets is essential to the understanding of signal pathways that are mediated by CaMs and their targets in plant cells. It has been reported that different CaM isoforms modulate their targets differentially (Lee et al., 2000; Yamakawa et al., 2001). Because NtCaMK1 is a Ca2+/CaMK lacking EF hands, it was used to investigate the possibility that different isoforms of CaM regulate its activity differentially. Three different CaM isoforms, NtCaM1, NtCaM3, and NtCaM13, as representatives of three groups of tobacco CaM isoforms, respectively, were used in our experiments. NtCaM1 belongs to group I, with high similarity to PCM1. NtCaM3 belongs to group II, including ordinary plant CaMs, such as Arabidopsis ACaM-2/3/5. NtCaM13 belongs to group III as SCaM-4 (Yamakawa et al., 2001). We show that NtCaMK1 is activated by some isoforms of CaM, including NtCaM13, but not by others, such as NtCaM3, suggesting that its CaMBD is differentially recognized by different types of plant CaMs. These results indicated that NtCaMK1, having a 1-8-14 CaMBD, interacts with NtCaM13 but not with NtCaM3 that binds to CaMBD as 1-5-10 motif. This could be predicted based on the report that, while NtCaM13 binds to 1-8-14 CaMBD, NtCaM3 interacts with 1-5-10 motif (Yamakawa et al., 2001; Choi et al., 2002). However, NtCaM1, a new and unique plant CaM isoform, also modulates NtCaMK1 activity. The differential regulation of NtCaMK1 by different CaM isoforms suggests the complexity in the activation of this kinase in plant cells.
Several other aspects of the sequence of NtCaMK1 are interesting. A 33 amino acid sequence (IRPRAKSVANVETVKSAGSVDVKRLETSGRSES) of NtCaMK1 at positions 266 to 298 of the N terminus is perfectly repeated at the C terminus at positions 1,383 to 1,415. This repeated sequence of unknown function is unique and so far not found in any other protein. We also find a number of PEST signals in NtCaMK1 that are present in rapidly degraded enzymes, transcription factors, and components of receptor signaling pathways (Rechsteiner, 1988). The presence of multiple PEST domains suggests that NtCaMK1 is rapidly turned over or plays a role in a signal pathway, although this speculation needs to be tested experimentally. The fact that NtCaMK1 binds CaM in a Ca2+-dependent manner is indicative of its involvement in a Ca2+/CaM signaling pathway. It was also noted that sequences 3,933 to 4,168 of NtCaMK1 share up to 90% homology at the nucleotide level with the reverse strand of a cyclic nucleotide-binding channel from tobacco (U65390).
In summary, this article describes the isolation and characterization of a novel CaMK from tobacco. Its CaMBD has high affinity for CaM and the CaMBD was mapped to a short stretch of amino acid residues between 913Ala and 932Lys, and was identified as a 1-8-14 motif. The protein kinase activity of NtCaMK1 is observed only in the presence of Ca2+/CaM and can be differentially regulated by different isoforms of CaM.
MATERIALS AND METHODS
Materials
Tobacco cells (Nicotiana tabacum L. cv Wisconsin-38) were grown in the dark at 23°C as suspension cultures in Gamborg's B-5 medium. Cells from mid-log-phase cultures (7 d old) were used in all experiments (Harrington and Alm, 1988). Tobacco plants were grown under natural conditions in a greenhouse at Wuhan University.
Construction and Screening of Tobacco cDNA Library
The construction and screening with 35S-labeled CaM of a cDNA library made with λZAP II expression vector (Stratagene, La Jolla, CA) and poly(A+) RNA isolated from tobacco cells were as described previously (Lu and Harrington, 1994). The candidate recombinant phage was in vivo excised into plasmid pNtCaMK1, and both strands of the NtCaMK1 cDNA insert were sequenced using an automatic DNA sequencer (ABI 377; Applied Biosystems, Foster City, CA). BLAST searches were performed at the National Center for Biotechnology Information Web site.
Isolation of the NtCaMK1 Genomic Clone
Based on the sequence of NtCaMK1 cDNA, four gene-specific primers with 5′-ATGTCCTTACAATCTCATCTAGAG-3′ and 5′-CTTTAAGTCCAAGAGTACGGC-3′ for 5′ end, and with 5′-GCGGACAAGATTCTGATGACC-3′ and 5′-CTATGACTCTGACCGGCCACTAG-3′ for 3′ end of NtCaMK1 were designed and used for PCR with tobacco genomic DNA as template. PCR was carried out with 150 ng each of primers, along with 200 μm of dNTPs, 5 units of TaKaRa LA tag, and 1 μg of tobacco genomic DNA as template in a 50-μL reaction volume. The PCR amplification conditions were 94°C for 1 min, 55°C for 1 min, and 72°C for 15 min, for 30 cycles. The amplified PCR products were separated in agarose gel, cloned into pGEM-T Easy Vector, and sequenced.
Preparation of 35S-Labeled and Biotinylated CaM
Tobacco calmodulin (NtCaM1, NtCaM3, and NtCaM13) cDNAs cloned into the pET15 expression vector (a gift from Dr. Y. Ohashi, National Institute of Agrobiological Sciences, Ibaraki, Japan; Yamakawa et al., 2001) were used to prepare 35S-labeled CaM as described (Fromm and Chua, 1992; Lu and Harrington, 1994). CaMs were biotinylated as described (Billingsley et al., 1985). Briefly, CaMs were dialyzed overnight at 4°C against 0.1 m phosphate buffer (pH 7.4). The dialyzate was supplemented with 1 mm CaCl2 and incubated with d-biotinoyl-ɛ-amidocaproic acid-N-hydroxysuccinimide ester (Sigma, St. Louis) dissolved in N,N-dimethylformamide at a final concentration of 1 mm. The incubation was performed for 2 h at 4°C with constant stirring. The biotinylated CaMs were further dialyzed in 0.1 m phosphate buffer (pH 7.4) for 48 h at 4°C and stored in 20% glycerol at −20°C. The amount of labeled NtCaM1, NtCaM3, and NtCaM13 proteins were measured as described (Bradford, 1976) using bovine serum albumin (BSA) as a standard. The concentration of NtCaM isoforms required for half-maximal activity (Ka) of NtCaMK1 was estimated as described previously (Lee et al., 2000).
RNA Isolation and RNA Analysis
Total RNA from tobacco was isolated with TRIZOL as described by the manufacturer (GIBCO, Carlsbad, CA) and northern blotting was carried out as described previously (Lu et al., 1996; Zhang et al., 2002). Briefly, 20 μg total RNA was separated on 1.5% formaldehyde agarose gels and blotted onto a nylon membrane (Nytran; Schleicher & Schull, Keene, NH). The membrane was incubated with a 32P-labeled probe made from pNtCaMK1 at 65°C overnight and washed under high stringency conditions.
Genomic Southern-Blot Analysis
Tobacco genomic DNA was extracted using the cetyl-trimethyl-ammonium bromide (CTAB) method (Wagner et al., 1987). Ten micrograms of DNA were digested with EcoRV or KpnI, fractionated on an 0.8% agarose gel, and then transferred onto a nylon membrane (Nytran). The membrane was probed with a 32P-labeled probe made from pNtCaMK1. Hybridization and washing conditions were as described above for northern-blot analysis.
Construction of C-Terminal Deletion Mutants of NtCaMK1 and Site-Directed Mutagenesis
For mapping the CaMBD of NtCaMK1, several C-terminal deletion constructs were prepared with a pET32a vector. The cDNAs for the full ORF and four truncated forms (NtCaMK1p865, NtCaMK1p912, NtCaMK1p932, and NtCaMK1p950) of NtCaMK1 (Fig. 5A) were amplified using Platinum Pfx DNA polymerase (GIBCO) with a 5′ primer (5′-ATGTCCTTACAATCTCATCTAGAGA-3′) and the following five 3′ primers: 3′ primer (5′-CTATGACTCTGACCGGCCACTAG-3′) for the full ORF of NtCaMK1; 3′ primer (5′-TGACGTCCTTAACCATGGATGACT-3′) for NtCaMK1p865 encoding the N-terminal 865 amino acid residues; 3′ primer (5′-CTGCTCCTTCAGATAAAACAGTTC-3′) for NtCaMK1p912 encoding the N-terminal 912 amino acid residues; 3′ primer (5′-TTTATGAGAAACAGCTTTCTTTTT-3′) for NtCaMK1p932 encoding the N-terminal 932 amino acid residues; and 3′ primer (5′-TTCTTCTAACAAAGCAAACTGCTC-3′) for NtCaMK1p950 encoding the N-terminal 950 amino acid residues. The C-terminal amino acid sequence from 933 to 1,415 was amplified using a 5′ primer (5′-TCAACGGTGGTGTCATGTATG-3′) and 3′ primer (5′-CTATGACTCTGACCGGCCACTAG-3′). All of these C-terminal deletion mutants were prepared with a pET32a vector at the EcoRV site. To precisely determine the putative CaMBD (amino acid residues at positions 913–932) of NtCaMK1, two complementary primers with BamHI and HindIII sites, respectively, were synthesized as follows: 5′-GATCCGCCAAAGCCCTGAAAAAGTGGCGTATGATGGCTCGTAAAAAGAAAGCTGTTTCTCATAAAA-3′ and 5′-ACGTTTATGAGAAACAGCTTTCTTTTTACGAGCCATCATACGCCACTTTTTCAGGGCTTTGGCG-3′, annealed and cloned into a BamHI- and HindIII-digested pET32a vector (denoted by pCaMBD).
To identify the critical amino acid residue(s) within the CaMBD of NtCaMK1 for the interaction between CaM and NtCaMK1, several point mutations were introduced into the putative CaMBD region. Substitutions of single amino acids were performed using the QuikChange XL site-directed mutagenesis kit (Stratagene) with pCaMBD and four pair of forward and reverse primers as follows: forward primer 5′-GCCAAAGCCCGGAAAAAGTGGCGTATG-3′ and reverse primer 5′-CATACGCCACTTTTTCCGGGCTTTGGC-3′ for L916R in which Arg was substituted for Leu at position 916; forward primer 5′-GCCCTGAAAAAGCGGCGTATGATGG CTCG-3′ and reverse primer 5′-CGAGCCATCATACGCCGCTTTTTCAGGGC-3′ for W919R in which Arg was substituted for Trp at position 919; forward primer 5′-GCCCTGAAAAATTGGCGTATGATGGC-3′ and reverse primer 5′-GCCATCATACGCCAATTTTTCAGGGC-3′ for K918N in which Asn was substituted for Lys at position 918; forward primer 5′-GGCGTATGATGGCTCGTAATAAGAAAGCTG-3′ and reverse primer 5′-CAGCTTTCTTATTACGAGCCATCATATACGCC-3′ for K925N in which Asn was substituted for Lys at position 925; and forward primer 5′-GGCGTATGATGGCTGAGGAGGAGGAGGCTGTTTCTC-3′ and reverse primer 5′-GAGAAACAGCCTCCTCCTCCTCAGCCATCATACGCC-3′ for RK4E in which 924Arg-Lys-Lys-Lys-927 was substituted for 924Glu-Glu-Glu-Glu-927 at positions of 924 to 927. All constructs mentioned above were confirmed by DNA sequencing.
Construction of Recombinant Virus for the Expression of NtCaMK1
The cDNAs for the full ORF and two truncated forms of NtCaMK1 (NtCaMK1p865 and NtCaMK1p932) were amplified with Platinum Pfx DNA polymerase and ligated into the EcoRI site of pFastBacHTb that was blunted by Klenow enzyme. After sequence confirmation, recombinant plasmids were transformed into DH10BAC competent cells containing the bacmid with a mini-att Tn7 target site and helper plasmid. The mini Tn7 element on the pFastBacHTb donor plasmid can transpose to the mini-att Tn7 element on the bacmid in the presence of transposition proteins provided by the helper plasmid. Clones containing recombinant bacmid were identified based on the disruption of the lacZ α-gene. The sf-9 cells were maintained as monolayers at 27°C in 10% fetal bovine serum supplemented with Grace's medium and transfected with the recombinant bacmid with Cellfectin reagent according to the manufacturer's instructions (Invitrogen). Recombinant virus was harvested after 72 h and identified by PCR with the primers described above.
Purification of NtCaMK1 Protein
Insect sf-9 cells were infected with the recombinant virus for 72 h and harvested at room temperature. Cells were washed once with Grace's medium, resuspended in 5 mL of lysis buffer (50 mm Tris-HCl, pH 7.5, 10% glycerol, 1% Nonidet P-40, 0.2 mm phenylmethanesulfonyl fluoride), and sonicated for 30 s, followed by centrifugation at 12,000g for 10 min. The supernatant was applied to Ni-NTA resin column pre-equilibrated with buffer A (50 mm potassium phosphate, pH 6.0, 300 mm KCl, 10% glycerol). After extensive washing with buffer A, followed by buffer A containing 25 mm imidazole, NtCaMK1 was eluted with buffer A containing 200 mm imidazole. The eluted protein was dialyzed against 25 mm Tris-HCl, pH 7.5, containing 2 mm CaCl2 for 6 h and loaded onto a CaM Sepharose-4B (Amersham Pharmacia Biotech, Uppsala) column pre-equilibrated with buffer B (25 mm Tris-HCl, pH 7.5, 2 mm CaCl2). After washing with buffer B plus 200 mm NaCl, NtCaMK1 was eluted with buffer C (25 mm Tris-HCl, pH 7.5, 2 mm EGTA). The purified NtCaMK1 was used immediately for SDS-PAGE and enzymatic analyses. Protein concentration was determined by the method of Bradford (1976) using BSA as a standard. All procedures were performed at 4°C unless otherwise noted.
CaM-Binding Assay
NtCaMK1 and its truncated forms expressed in Escherichia coli were separated by SDS-PAGE and blotted onto PVDF membrane. For biotinylated CaM-binding assay, the membrane was blocked in 2% BSA/TBS (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 50 mm MgCl2, plus 1 mm CaCl2 or 2 mm EGTA) and washed three times with TBS for 15 min each. After incubation in TBS containing biotinylated CaM for 3 h at room temperature and then washed with TBS, the membrane was treated with avidin-HRP conjugate (Bio-Rad, Hercules, CA) and dissolved in TBS for 1 h. Protein bound to biotinylated CaM was visualized by color development with 4-chloro-1-naphthol and H2O2. For 35S-CaM-binding assay, the proteins were electrophoretically transferred onto nitrocellulose filters and incubated in binding buffer (10 mm Tris-HCl, pH 7.5 ,150 mm NaCl, and 1% [w/v] nonfat dry milk) containing 35S-CaM (0.5 × 106 cpm/μg) plus either 1 mm CaCl2 or 2 mm EGTA as described by Lu and Harrington (1994). The membrane was washed with 25 mm Tris-HCl, pH 7.5, and 1 mm CaCl2 or 2 mm EGTA and exposed to x-ray film.
CaM-Binding Affinity Assay for NtCaMK1
CaM-binding affinity assays were carried out as described (Takezawa et al., 1996). Briefly, purified NtCaMK1 protein (4 pm) separated by SDS-PAGE was electrophoretically transferred onto nitrocellulose filters and incubated with overnight 0, 3.5, 5.25, 7.0, 10.5, 14.0, 21.0, 31.5, 38.5, 42.0, 49.0, 70.0, 87.0, 105, and 200 nm 35S-labeled CaM, all containing 1 mm CaCl2. After washing with 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 1 mm CaCl2, the radioactivities of the bound CaM on each filter and the free CaM in the incubation buffer collected before washing were measured using a liquid scintillation counter (Beckman LS 6500; Fullerton, CA). Average background counts were subtracted from the counts in NtCaMK1 protein samples when calculating the specific binding. The dissociation constant (Kd) was calculated from Scatchard plots (Takezawa et al., 1996). The data points are expressed as means of the results from three independent duplicated assays.
Autophosphorylation Assays
NtCaMK1 autophosphorylation was carried out in 100-μL reaction mixture containing 25 mm Tris-HCl, pH 7.5, 0.5 mm dithiothreitol, 10 mm magnesium acetate, 100 μm ATP, 10 μCi [γ-32P]ATP (5,000 Ci/mm) plus 1 mm CaCl2 or 1 mm CaCl2/1 μm CaM or 2 mm EGTA at 30°C. For time course assays, 0.2 μg of NtCaMK1 was used in a 100-μL reaction mixture. Aliquots for the zero time point were taken immediately after the addition of NtCaMK1 to initiate the reaction, and the reactions were terminated by adding one-fifth volume 5× SDS-PAGE sample buffer. All aliquots were separated on 10% SDS-PAGE. After staining with 0.1% Coomassie Brilliant Blue, gels were vacuum-dried and exposed to x-ray film at −80°C. The amount of phosphates transferred to the enzyme was determined by counting the radioactivities of the excised NtCaMK1 bands in a liquid scintillation counter (Beckman LS 6500). The experiments were repeated three times in duplicate.
Substrate Phosphorylation Assay
Substrate phosphorylation by NtCaMK1 was performed in a 100-μL reaction mixture containing 25 mm Tris-HCl, pH 7.5, 0.5 mm dithiothreitol, 10 mm magnesium acetate, 100 μm ATP, 10 μCi [γ-32P]ATP (5,000 Ci/mm), 100 μm histone IIIs or syntide-2 (Pro-Leu-Ala-Arg-Thr-Leu-Ser-Val-Ala-Gly-Leu-Pro-GLy-Lys-Lys; Sigma), a synthetic peptide substrate for CaMKII (Hashimoto and Soderling, 1987), plus 1 mm CaCl2 or 1 mm CaCl2/1 μm CaM or 2 mm EGTA with 0.2 μg of NtCaMK1 at 30°C.
For time course assays with histone IIIs as substrate, aliquots for the zero time point were taken immediately after the addition of NtCaMK1 to initiate the reaction and the reaction was terminated by adding one-fifth volume 5× SDS-PAGE sample buffer. Aliquots were separated by SDS-PAGE with a discontinuous 8%/20% separating gel and, after staining with 0.1% Coomassie Brilliant Blue, the gels were vacuum-dried and exposed to x-ray film at −80°C. The amount of phosphates transferred to histone IIIs was determined by counting the radioactivities of the excised histone IIIs bands in a liquid scintillation counter. The experiments were repeated three times in duplicate.
For time course assays with syntide-2 as substrate, the experiments with two parallel reactions for different treatments were repeated three times. Aliquots for the zero time point were taken immediately after the addition of NtCaMK1 to initiate the reaction, and the reaction was terminated by adding one-fifth volume 5× SDS-PAGE sample buffer. Aliquots from one reaction were separated by SDS-PAGE with 10% separating gel. The amount of phosphates transferred to the kinase was determined by counting the radioactivities of the excised NtCaMK1 bands in a liquid scintillation counter. Aliquots from another reaction were applied to P81 phosphocellulose filters (2 × 2-cm squares; Whatman, Clifton, NJ) for total 32P incorporation of the kinase and syntide-2. Filters were washed four times for 10 min each in 75 mm phosphoric acid and rinsed in 100% ethanol and air-dried. 32P incorporation was determined by liquid scintillation counting. The amount of phosphates transferred to syntide-2 substrate was determined by subtracting 32P incorporation of NtCaMK1 from total 32P incorporation of NtCaMK1 plus syntide-2.
Assay of NtCaMK1 Activity
The reaction for NtCaMK1 substrate phosphorylation was performed as described above. Aliquots (10 μL) were removed and applied to P81 phosphocellulose filters (2 × 2-cm squares; Whatman). Filters were washed four times for 10 min each in 75 mm phosphoric acid, rinsed in 100% ethanol, and air-dried. 32P incorporation was determined by liquid scintillation counting (Beckman LS 6500). The experiments were repeated three times in duplicate.
Phosphoamino Acid Assays
Unphosphorylated and autophosphorylated NtCaMK1 were hydrolyzed in 6 m HCl for 12 h at 110°C, then dried and dissolved in 20 μL of 10 mm borate buffer (pH 10.0). The hydrolyzed product was mixed with 20 μL of 1 mm fluorescein isothiocyanate (FITC), dissolved in acetone containing 0.05% pyridine (Sigma), and incubated in the dark for 12 h at room temperature.
To prepare FITC-tagged standard amino acids and phosphoamino acids, 2 μL standard solution containing each amino acid and phosphoamino acid (0.5 mm each) were mixed with 46 μL of 1 mm FITC, 100 μL of 20 mm borate buffer (pH 10.0), and 52 μL of water. The mixture was incubated in the dark for 12 h at room temperature.
FITC-tagged amino acids were analyzed by capillary electrophoresis as described by Liu et al. (2001). Data were collected by a computer with Spot Advanced software, and further processed with Scion Image and Origin software packages.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers GenBank AF550608 for NtCaMK1; GenBank AY56224 for genomic NtCaMK1.
Supplementary Material
Acknowledgments
We thank Dr Y. Ohashi for tobacco calmodulin cDNAs (NtCaM1, NtCaM3, and NtCaM13).
This work was supported in part by the National Natural Science Foundation of China (grant no. 30230050/30170449) and Major State Basic Research Program of China (grant no. 2002CCA00100 to Y.-T.L.), and by a grant from the National Science Foundation to (R.L.J. and Y-T.L.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041970.
References
- Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H (1995) Molecular and biochemical analysis of calmodulin interactions with the calmodulin-binding domain of plant glutamate decarboxylase. Plant Physiol 108: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley ML, Pennypacker KR, Hoover CG, Brigati DJ, Kincaid RL (1985) A rapid and sensitive method for detection and quantification of calcineurin and calmodulin-binding proteins using biotinylated calmodulin. Proc Natl Acad Sci USA 82: 7585–7589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bush DS (1995) Calcium regulation in plant cells and its role in signaling. Annu Rev Plant Physiol Plant Mol Biol 46: 95–122 [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J (2002) Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D, Means AR (2000) Calmodulin: a prototypical calcium sensor. Trends Cell Biol 10: 322–328 [DOI] [PubMed] [Google Scholar]
- Choi JY, Lee SH, Park CY, Heo WD, Kim JC, Kim MC, Chung WS, Moon BC, Cheong YH, Kim CY, et al (2002) Identification of calmodulin isoform-specific binding peptides form a phage-displayed random 22-mer peptide library. J Biol Chem 277: 21630–21638 [DOI] [PubMed] [Google Scholar]
- Colbran RJ, Soderling TR (1990) Calcium/calmodulin-dependent protein kinase II. Curr Top Cell Regul 31: 181–221 [DOI] [PubMed] [Google Scholar]
- Crivici A, Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct 24: 85–116 [DOI] [PubMed] [Google Scholar]
- Diggle TA, Seehra CK, Hase S, Redpath NT (1999) Analysis of the domain structure of elongation factor-2 kinase by mutagenesis. FEBS Lett 457: 189–192 [DOI] [PubMed] [Google Scholar]
- Enslen H, Sun P, Brickey D, Soderling SH, Klamo E, Soderling TR (1994) Characterization of Ca2+/calmodulin-dependent protein kinase IV. Role in transcriptional regulation. J Biol Chem 269: 15520–15527 [PubMed] [Google Scholar]
- Fajisawa H (1990) Calmodulin-dependent protein kinase II. Bioessays 12: 27–29 [DOI] [PubMed] [Google Scholar]
- Fischer R, Koller M, Flura M, Mathews S, Strehler-page MA, Krebs J, Penniston JT, Carafoil E, Strehler EE (1988) Multiple divergent mRNAs code for a single human calmodulin. J Biol Chem 263: 17055–17062 [PubMed] [Google Scholar]
- Fromm H, Chua N-H (1992) Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol Biol Rep 10: 199–206 [Google Scholar]
- Gilroy S, Trewavas A (1994) A decade of plant signals. Trends Genet 16: 677–682 [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domain. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Schulman H (1992) Neuronal Ca2+/calmodulin-dependent protein kinases. Annu Rev Biochem 61: 559–601 [DOI] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF (2000) CDPKs—a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang J-F, Llooyd SJ (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277 [DOI] [PubMed] [Google Scholar]
- Harper JF, Sussman MR, Schaller GE, Putanam-Evans C, Charbonneau H, Harmon AC (1991) A calcium-dependent protein kinase with a regulatory domain that is similar to calmodulin. Science 252: 951–954 [DOI] [PubMed] [Google Scholar]
- Harrington MH, Alm DM (1988) Interaction of heat and salt shock in cultured tobacco cells. Plant Physiol 88: 618–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HashimotoY, Soderling TR (1987) Calcium/calmodulin-dependent protein kinase II and calcium/phospholipid-dependent protein kinase activities in rat tissues assayed with a synthetic peptide. Arch Biochem Biophys 252: 418–425 [DOI] [PubMed] [Google Scholar]
- Heo WD, Lee SH, Kim MC, Kim JC, Chung WS, Chun HJ, Lee KJ, Park HY, Park HY, Choi JY, et al (1999) Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc Natl Acad Sci USA 96: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua W, Liang SP, Lu Y-T (2003) A tobacco (Nicotiana tabacum) calmodulin-binding protein kinase, NtCBK2, is regulated differentially by calmodulin isoforms. Biochem J 376: 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A (1992) Solution structure of a calmodulin-target peptide complex by multi-dimensional NMR. Science 256: 632–638 [DOI] [PubMed] [Google Scholar]
- Kameshita I, Fujisawa H (1993) Autophosphorylation of calmodulin-dependent protein kinase IV from rat cerebral cortex. J Biochem (Tokyo) 113: 583–590 [DOI] [PubMed] [Google Scholar]
- Kemp BE, Pearson RB (1991) Intrasteric regulation of protein kinases and phosphatases. Biochim Biophys Acta 1094: 67–76 [DOI] [PubMed] [Google Scholar]
- Klee CB (1991) Concerted regulation of protein phosphorylation and dephosphorylation by calmodulin. Neurochem Res 16: 1059–1065 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch, cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Lai Y, Nairn AC, Greengard P (1986) Autophosphorylation reversibly regulates the Ca2+/calmodulin-dependence of Ca2+/calmodulin-dependent protein kinase II (phosphorylation/dephosphorylation/thermal stability). Proc Natl Acad Sci USA 83: 4253–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Johnson JD, Walsh MP, Lierop JEV, Sutherland C, Xu A, Snedden WA, Kosk-Kosicka D, Fromm H, Narayanan N, et al (2000) Differential regulation of Ca2+/calmodulin-dependent enzymes by plant calmodulin isoforms and free Ca2+ concentration. Biochem J 350: 299–306 [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim JC, Lee MS, Heo WD, Seo HY, Yoon HW, Hong JC, Lee SY, Bahk JD, Hwang I, et al (1995) Identification of a novel divergent calmodulin isoform from soybean which has differential ability of activate calmodulin dependent enzyme. J Biol Chem 270: 21806–21812 [DOI] [PubMed] [Google Scholar]
- Lin CR, Kapiloff MS, Purgerian S, Tatemoto K, Russo AF, Hanson P, Schulman H, Rosenfeld MG (1987) Molecular cloning of a brain-specific calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci USA 84: 5962–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B-F, Zhang L, Lu Y-T (2001) Characterization of a novel protein kinase in rice cells by capillary electrophoresis. J Chromatogr A 918: 401–409 [DOI] [PubMed] [Google Scholar]
- Liu Z, Xia M, Poovaiah BW (1998) Chimeric calcium/calmodulin-dependent protein kinase in tobacco: differential regulation by calmodulin isoforms. Plant Mol Biol 38: 889–897 [DOI] [PubMed] [Google Scholar]
- Lu Y-T, Harrington MH (1994) Isolation of calmodulin binding protein cDNA clones and identification of a calmodulin binding domain. Plant Physiol Biochem 32: 413–422 [Google Scholar]
- Lu Y-T, Hidaka H, Feldman LJ (1996) Characterization of a calcium/calmodulin-dependent protein kinase homolog from maize roots showing light-regulated gravitropism. Planta 199: 18–24 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14: S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh MR, Hetherington AM (1998) Encoding specificity in Ca2+ signalling systems. Trends Plant Sci 3: 32–36 [Google Scholar]
- Meador WE, Means AR, Quiocho FA (1992) Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257: 1251–1255 [DOI] [PubMed] [Google Scholar]
- Mochizuki H, Ito T, Hidaka H (1993) Purification and characterization of Ca2+/calmodulin dependent protein kinase V from rat cerebrum. J Biol Chem 268: 9143–9147 [PubMed] [Google Scholar]
- O'Neil KT, DeGrado WF (1990) How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem Sci 15: 59–64 [DOI] [PubMed] [Google Scholar]
- Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M (1999) A novel target recognition by calmodulin revealed by its solution structure in complex with a peptide derived from Ca2+/calmodulin dependent protein kinase kinase. Nat Struct Biol 6: 819–824 [DOI] [PubMed] [Google Scholar]
- Patil S, Takezawa D, Poovaiah BW (1995) Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc Natl Acad Sci USA 92: 4879–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah BM, Reddy ASN (1987) Calcium messenger system in plants. CRC Crit Rev Plant Sci 6: 47–103 [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Reddy ASN (1993) Calcium and signal transduction in plants. Crit Rev Plant Sci 12: 185–211 [DOI] [PubMed] [Google Scholar]
- Poovaiah BW, Xia M, Liu ZH, Wang W, Yang TB, Sathyanarayanan PV, Franceschi VR (1999) Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209: 161–171 [DOI] [PubMed] [Google Scholar]
- Price AH, Taylor A, Ripley SJ, Griffiths A, Trewavas AJ, Knight MR (1994) Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 6: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam-Evans CL, Harmon AC, Cormier MJ (1990) Purification and characterization of a novel calcium-dependent protein kinase from soybean. Biochemistry 29: 2488–2499 [DOI] [PubMed] [Google Scholar]
- Ramachandiran S, Takezawa D, Wang W, Poovaiah BW (1997) Functional domains of plant chimeric calcium/calmodulin-dependent protein kinase: regulation by autoinhibitory and visinin-like domains. J Biochem (Tokyo) 121: 984–990 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M (1988) Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul 27: 135–151 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Day IS, Narasimhulu SB, Safadi F, Reddy VS, Golovkin M, Harnly MJ (2002) Isolation and characterization of a novel calmodulin-binding protein from potato. J Biol Chem 277: 4206–4214 [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Takezawa D, Fromm H, Poovaiah BW (1993) Isolation and characterization of two cDNAs that encode for calmodulin-binding proteins from corn root tips. Plant Sci 94: 109–117 [Google Scholar]
- Rhoads AR, Friedberg F (1997) Sequence motifs for calmodulin recognition. FASEB J 11: 331–340 [DOI] [PubMed] [Google Scholar]
- Roberts DM, Harmon AC (1992) Targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol 43: 375–414 [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman H (1988) The multifunctional Ca2+/calmodulin-dependent protein kinase. Adv Second Messenger Phosphoprotein Res 22: 39–112 [PubMed] [Google Scholar]
- Sikela JM, Hahn WE (1987) Screening an expression library with a ligand probe: isolation and sequence of a cDNA corresponding to a brain calmodulin-binding protein. Proc Natl Acad Sci USA 84: 3038–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Fromm H (1998) Calmodulin, calmodulin-related proteins and plant responses to the environment. Trends Plant Sci 3: 299–304 [Google Scholar]
- Takezawa D, Ramachndiran S, Paranjape V, Poovaiah BW (1996) Dual regulation of a chimeric plant serine/threonine kinase by calcium and calcium/calmodulin. J Biol Chem 271: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Urao T, Katagiri T, Mizoguchi T, Yamaguchi-Shinozaki K, Hayashida N, Shinozaki K (1994) An Arabidopsis thaliana cDNA encoding Ca2+-dependent protein kinase. Plant Physiol 105: 1461–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DB, Furnier GR, Saghai-Maroof MA, Williams SM, Dancik BP, Allard RW (1987) Chloroplast DNA polymorphisms in lodgepole and jack pines and their hybrids. Proc Natl Acad Sci USA 84: 2097–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liang S, Lu Y-T (2001) Characterization, physical location and expression of the genes encoding calcium/calmodulin-dependent protein kinases in maize (Zea mays L.). Planta 213: 556–564 [DOI] [PubMed] [Google Scholar]
- Watillon B, Kettmenn R, Boxus P, Burny A (1992) Cloning and characterization of an apple (Malus domestica L. Borkh) cDNA encoding a calmodulin-binding protein domain similar to the calmodulin-binding region of type-II mammalian Ca2+/calmodulin-dependent protein kinase. Plant Sci 81: 227–235 [Google Scholar]
- Watillon B, Kettmenn R, Boxus P, Burny A (1995) Structure of a calmodulin-binding protein kinase gene from apple. Plant Physiol 108: 847–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Mitsuhara I, Ito N, Seo S, Kamada H, Ohashi Y (2001) Transcriptionally and post-transcriptionally regulated response of 13 calmodulin genes to tobacco mosaic virus-induced cell death and wounding in tobacco plant. Eur J Biochem 268: 3916–3929 [DOI] [PubMed] [Google Scholar]
- Yang WN, Liang SP, Lu Y-T (2000) Immumohistochemical localization of Ca2+/calmodulin-dependent kinase in tobacco. Chin Sci Bull 45: 1747–1751 [Google Scholar]
- Yoo BC, Harmon AC (1996) Intramolecular binding contributes to the activation of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 35: 12029–12037 [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu B-F, Liang SP, Jones RL, Lu Y-T (2002) Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem J 368: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu Y-T (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zielinski RE (1998) Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 697–725 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.