Abstract
α-Amylases are important enzymes for starch degradation in plants. However, it has been a long-running debate as to whether α-amylases are localized in plastids where starch is stored. To study the subcellular localization of α-amylases in plant cells, a rice (Oryza sativa) α-amylase, αAmy3, with or without its own signal peptide (SP) was expressed in transgenic tobacco (Nicotiana tabacum) and analyzed. Loss-of-function analyses revealed that SP was required for targeting of αAmy3 to chloroplasts and/or amyloplasts and cell walls and/or extracellular compartments of leaves and suspension cells. SP was also required for in vitro transcribed and/or translated αAmy3 to be cotranslationally imported and processed in canine microsomes. αAmy3, present in chloroplasts of transgenic tobacco leaves, was processed to a product with Mr similar to αAmy3 minus its SP. Amino acid sequence analysis revealed that the SP of chloroplast localized αAmy3 was cleaved at a site only one amino acid preceding the predicted cleavage site. Function of the αAmy3 SP was further studied by gain-of-function analyses. β-Glucuronidase (GUS) and green fluorescence protein fused with or without the αAmy3 SP was expressed in transgenic tobacco or rice. The αAmy3 SP directed translocation of GUS and green fluorescence protein to chloroplasts and/or amyloplasts and cell walls in tobacco leaves and rice suspension cells. The SP of another rice α-amylase, αAmy8, similarly directed the dual localizations of GUS in transgenic tobacco leaves. This study is the first evidence of SP-dependent dual translocations of proteins to plastids and extracellular compartments, which provides new insights into the role of SP in protein targeting and the pathways of SP-dependent protein translocation in plants.
Two types of starch exist in plants: transitory (assimilatory) starch, which is located in chloroplasts, and reserve starch, which is deposited in amyloplasts. Degradation of starch can be either hydrolytic, mainly catalyzed by α-amylase, β-amylase, and debranching enzymes, or phosphorolytic, catalyzed by starch phosphorylase. Degradation of reserve starch in cereal grains is mainly hydrolytic, whereas degradation of transitory starch in leaves can be hydrolytic and/or phosphorolytic (Beck and Ziegler, 1989). In germinating cereal grains, α-amylases are the most abundant starch-degrading enzymes. The enzymes are secreted by aleurone cells into the starchy endosperm where they degrade the starch grains (Jacobsen et al., 1995). Whether α-amylases also play an essential role in starch degradation in photosynthetic tissues and in tissues other than endosperm is not clear. In most plants, starch can be found in pollens, seeds, leaves, stems, roots, and other tissues. In spinach (Spinacia oleracea), α-amylases are the only enzymes that have been demonstrated to attack starch granules isolated from chloroplasts (Steup et al., 1983).
Determination of the subcellular distribution of α-amylases is essential for understanding the physiological function of these enzymes in starch degradation. Using techniques based on subcellular fractionation, substrate-specific activities, and end-product analysis, α-amylase activities have been determined as being localized in leaf chloroplasts of spinach (Okita et al., 1979), pea (Pisum sativum), and wheat (Triticum aestivum; Ziegler and Beck, 1986), and Arabidopsis (Lin et al., 1988). However, several studies with pea and barley (Hordeum vulgare) indicate that α-amylase is either absent or has a very low activity in chloroplasts (Levi and Preiss, 1978; Kakefuda et al., 1986). Regardless of the plant species, it appears that the majority of amylase activity in leaf tissues is extrachloroplastic (Kakefuda et al., 1986; Beers and Duke, 1988; Lin et al., 1988), distinct from the site of starch accumulation and degradation. α-Amylase in pea stems has been shown to localize in apoplasts (Beers and Duke, 1988); however, the precise subcellular locations of α-amylases in plant cells are still a controversial subject.
We previously analyzed the subcellular localization of α-amylases in cultured rice (Oryza sativa) suspension cells and revealed that α-amylases are localized in cell walls as well as in starch granules within amyloplasts (Chen et al., 1994). The dual localization of α-amylases disagrees with the general belief that the translocation of proteins to chloroplasts or amyloplasts and the extracellular compartments is carried out by different targeting signals and via different pathways (Verner and Schatz, 1988). Import into chloroplasts of a nuclear-encoded protein from the cytoplasm requires an N-terminal transit peptide as a targeting signal (Schmidt and Mishkind, 1986; Keegstra, 1989). However, the deduced N-terminal amino acid sequences of nine rice α-amylases all contain typical signal peptides (SPs) characteristic for translocation of proteins across the endoplasmic reticulum (ER) membrane (Chen et al., 1994). Previously, we also showed in an assay using transformed rice suspension cells, that the SP of a rice α-amylase isozyme, αAmy8, directed the translocation of β-glucuronidase (GUS) to ER, with subsequent secretion into the culture medium (Chan et al., 1994). Questions were thus raised regarding the translocation pathways that α-amylases employ to achieve dual location targeting in rice cells. Additional questions consisted of whether individual α-amylases are differentially targeted to plastids or the extracellular compartment and whether a single α-amylase is targeted to two distinct subcellular locations.
In this study, we chose αAmy3 as a model to study its cellular localization in plant cells. By both loss- and gain-of-function analyses in transgenic tobacco (Nicotiana tabacum) and rice, we demonstrate that αAmy3 is localized in both plastids and the extracellular compartment, and SP is necessary and sufficient for targeting of αAmy3 and cargo proteins to these dual locations.
RESULTS
Expression of Rice αAmy3 in Transgenic Tobacco
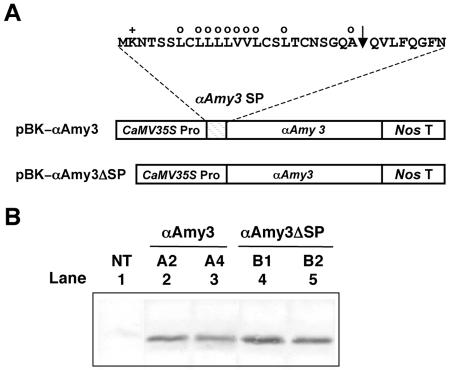
In order to study the targeting of an individual α-amylase and eliminate the complexity that occurs by the expression of multiple endogenous α-amylases in rice cells, we used transgenic tobacco lines that constitutively express a single rice α-amylase. Pilot experiments demonstrated that the rice α-amylase cDNA probe did not cross-hybridize to the native tobacco α-amylase genes and that the anti-rice α-amylase antibodies, which were raised against total rice α-amylases, did not cross-react with the native tobacco α-amylases. We constructed a chimeric gene encoding αAmy3 with or without its own SP under the control of the CaMV35S promoter (Fig. 1A). αAmy3 without SP was designated as αAmy3ΔSP. Plasmids containing the chimeric genes were introduced into the tobacco genome and several transgenic lines were obtained for each construct. Two lines of each construct expressing similar levels of αAmy3 in leaves were selected and demonstrated by protein western-blot analysis. As shown in Figure 1B, rice αAmy3 was detected in transgenic tobacco (lanes 2–5) but not in the nontransformed control (lane 1). This result demonstrates that the anti-rice α-amylase antibodies recognized the two foreign enzymes but not the endogenous tobacco α-amylases and could therefore be used to study the cellular localization of αAmy3 in transgenic tobacco. It is interesting to note that rice αAmy3 proteins detected in transgenic tobacco leaves expressing αAmy3 or αAmy3ΔSP have the same size. This suggests that the SP in αAmy3 has been cleaved in transgenic plants.
Figure 1.
Expression of αAmy3 in transgenic tobacco leaves. A, Diagram shows the amino acid sequence of αAmy3 SP and constructs containing the CaMV35S promoter and coding regions of αAmy3 without SP. Arrowhead indicates the putative SP cleavage sites. Amino acids with hydrophobic (○) or positively charged (+) chain are indicated. B, Two independent transgenic tobacco plants expressing αAmy3 (A2 and A4) or αAmy3ΔSP (B1 and B2) were analyzed. Total proteins were isolated from tobacco leaves and subjected to western-blot analysis using anti-rice α-amylase antibodies (Chen et al., 1994). Fifty micrograms of total proteins were loaded in each lane. NT, Nontransformed control.
SP-Dependent Localization of αAmy3 in Plastids and Cell Walls of Transgenic Tobacco
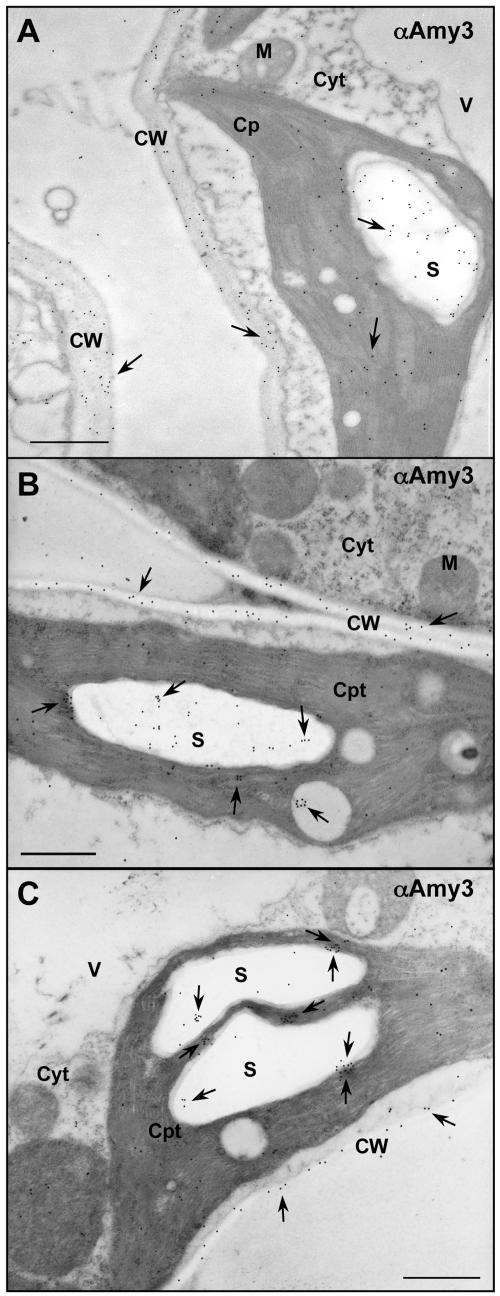
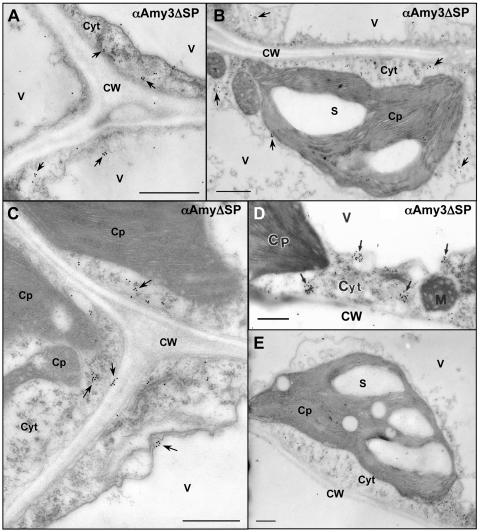
The subcellular localization of α-amylases in transgenic tobacco was examined using electron microscopic immunocytochemistry (EMI). In leaves expressing αAmy3, the antibody label was mainly found in cell walls and in stroma and over starch granules within the chloroplasts in mesophyll cells, and only background level of label was found in other cellular compartments (Fig. 2). In leaves expressing αAmy3ΔSP, the label was found in the cytoplasm of mesophyll cells (Fig. 3, A–D). No label was detected in any cellular compartment in nontransformed tobacco (Fig. 3E). Preimmune serum did not recognize the αAmy3 present in plastids and cell walls of transgenic leaves (data not shown).
Figure 2.
SP-dependent localization of αAmy3 in the chloroplast and cell wall of transgenic tobacco leaves. Immunocytochemical localization of αAmy3 in leaf samples using anti-rice α-amylase antibodies. A and B, Labeling of αAmy3 in leaf mesophyll cells of transgenic tobacco line A2 expressing αAmy3. C, Labeling of αAmy3 in leaf mesophyll cells of transgenic tobacco line A4 expressing αAmy3. αAmy3 detected in the stroma and over starch granules within a chloroplast and in cell walls. Arrowheads indicate positions of αAmy3. Scale bar represents 1 μm. Abbreviations: Cpt, chloroplast; Cyt, cytoplasm; CW, cell wall; M, mitochondria; S, starch granule; V, vacuole.
Figure 3.
SP-dependent localization of αAmy3 in the chloroplast and cell wall of transgenic tobacco leaves. Immunocytochemical localization of αAmy3 in leaf samples using anti-rice α-amylase antibodies. A to D, Labeling of αAmy3 in leaf mesophyll cells of transgenic tobacco line B2 expressing αAmy3ΔSP. αAmy3 mainly detected in the cytoplasm. E, The αAmy3 antibodies do not label starch grains of nontransformant. Arrowheads indicate positions of αAmy3. Scale bar represents 1 μm. Abbreviations: Cp, chloroplast; Cyt, cytoplasm; CW, cell wall; M, mitochondria; S, starch granule; V, vacuole.
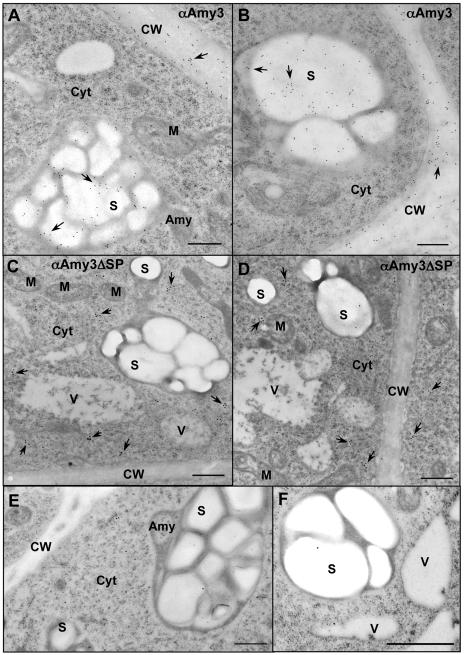
In tobacco suspension cells expressing αAmy3, the label was found over starch granules within amyloplasts and in cell walls (Fig. 4, A and B). In suspension cells expressing αAmy3ΔSP, the label was mainly found in the cytoplasm (Fig. 4, C and D). Preimmune serum did not recognize αAmy3 present in plastids and cell walls of transformed suspension cells (Fig. 4E). No label was detected in any cellular compartment in nontransformed tobacco (Fig. 4F).
Figure 4.
SP-dependent localization of αAmy3 in the amyloplasts and cell walls of transformed tobacco suspension cells. Immunocytochemical localization of αAmy3 in suspension cell samples using anti-rice α-amylase antibodies. A and B, Labeling of αAmy3 in suspension cells of transgenic tobacco line A2 expressing αAmy3. αAmy3 mainly detected over starch granules and in the cell wall. C and D, Labeling of αAmy3 in suspension cells of transgenic tobacco line B2 expressing αAmy3ΔSP. αAmy3 mainly detected in the cytoplasm. E, Preimmune serum does not label αAmy3 in starch grains and cell wall in transgenic tobacco line A2. F, The αAmy3 antibodies do not label starch grains of nontransformant. Arrowheads indicate positions of αAmy3. Scale bar represents 1 μm. Abbreviations: Amy, amyloplast; CW, cell wall; Cyt, cytoplasm; M, mitochondria; ML, middle lamella; S, starch granule; V, vacuole.
The density of the gold-labeled αAmy3 and αAmy3ΔSP in various cellular compartments of transgenic tobacco leaves was also analyzed. As shown in Table I, αAmy3 was mainly detected in chloroplasts and cell walls. By contrast, αAmy3ΔSP was mainly detected in cytosol. These findings demonstrate that in both leaves and cultured suspension cells of transgenic tobacco, αAmy3 is targeted to chloroplasts and/or amyloplasts and cell walls in an SP-dependent manner.
Table I.
Density of gold-labeled α-amylase in various cellular compartments of transgenic tobacco leaves
| Labeling Density
|
||||
|---|---|---|---|---|
| Cellular Compartmenta | Anti-α-Amylasea Serum | Preimmunea Serum | Correctedb | |
| gold particles/μm2 ± se | ||||
| αAmy3 | Chloroplast | 104.7 ± 8.0 | 6.8 ± 3.7 | 97.9 ± 4.3 |
| Cytosol | 7.7 ± 4.7 | 3.7 ± 2.1 | 4.0 ± 2.6 | |
| Cell wall | 212.3 ± 20.5 | 1.7 ± 0.3 | 210.6 ± 20.2 | |
| Nucleus | 3.3 ± 2.1 | 1.3 ± 0.4 | 2.0 ± 1.7 | |
| Vacuole | 8.2 ± 2.7 | 3.9 ± 1.3 | 4.3 ± 1.4 | |
| αAmy3ΔSP | Chloroplast | 8.2 ± 4.2 | 6.8 ± 3.7 | 1.4 ± 0.5 |
| Cytosol | 61.3 ± 12.7 | 3.7 ± 2.1 | 57.6 ± 10.6 | |
| Cell wall | 2.7 ± 0.8 | 1.7 ± 0.3 | 1.0 ± 0.5 | |
| Nucleus | 2.4 ± 1.0 | 1.3 ± 0.4 | 1.1 ± 0.6 | |
| Vacuole | 5.6 ± 2.3 | 3.9 ± 1.3 | 1.7 ± 1.0 | |
| NT | Chloroplast | 3.1 ± 1.2 | 1.6 ± 0.8 | 1.5 ± 0.4 |
| Cytosol | 2.6 ± 0.7 | 1.5 ± 0.4 | 1.1 ± 0.3 | |
| Cell Wall | 4.4 ± 1.5 | 2.5 ± 0.9 | 1.9 ± 0.6 | |
| Nucleus | 5.0 ± 2.0 | 3.4 ± 1.3 | 1.6 ± 0.7 | |
| Vacuole | 3.0 ± 0.8 | 1.7 ± 0.8 | 1.3 ± 0.0 | |
For each cellular compartment and for each antiserum, gold-labeling density was examined in a number of samples accounting for a total area exceeding 20 μm2 (NT, nontransformant).
Corrected = (labeling density for anti-α-amylase serum) − (labeling density for control preimmune serum).
In Vitro SP-Dependent Transport of αAmy3 into Microsomes
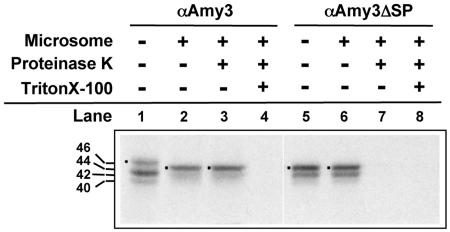
The predicted amino acid sequence of αAmy3 contains a typical SP at its N terminus (Fig. 1A). To determine whether the αAmy3 SP functions as a traditional ER targeting signal, αAmy3 with or without its own SP was in vitro transcribed and translated in the presence of canine microsomes and 35S-Met and then analyzed with SDS-PAGE and fluorography. The size of the predicted translation product of αAmy3 was 46 kD (Fig. 5, lane 1). Two smaller proteins of 42 and 40 kD, present in the translation products of αAmy3 and αAmy3ΔSP (Fig. 5, lanes 1 and 5), were likely produced through translation from two internal ATG codons which encode Met at amino acid residues 46 and 76 in αAmy3 downstream of the first ATG-encoded Met.
Figure 5.
SP-dependent import of αAmy3 into canine microsomes. RNAs encoding αAmy3 and αAmy3ΔSP were synthesized in vitro and then translated in rabbit reticulocyte lysate in the presence or absence of canine microsomes plus 35S-Met. The samples were treated with (+) or without (−) proteinase K and/or Triton X-100 and analyzed with SDS-PAGE and fluorography. Dots indicate positions of αAmy3. The molecular masses (kD) of the proteins are indicated to the left of the figure.
When αAmy3 was translated in the presence of microsomes, a 44-kD fragment was produced (Fig. 5, lane 2), which corresponded to a loss of the 25 SP residues and was identical in size to that of the translation product of αAmy3ΔSP (Fig. 5, lane 5). This 44-kD fragment derived from αAmy3 was not digested by proteinase K alone (Fig. 5, lane 3) but was digested by proteinase K in the presence of Triton X-100, which disrupts microsome membranes (Fig. 5, lane 4). The size of the 44-kD αAmy3ΔSP did not change when translated in the presence of microsomes (Fig. 5, lane 6) and was digested by proteinase K regardless of the presence or absence of Triton X-100 (Fig. 5, lanes 7 and 8). These results indicate that αAmy3 was imported into the microsomal vesicles, whereas αAmy3ΔSP was not.
αAmy3 Localized in Chloroplasts Is Processed
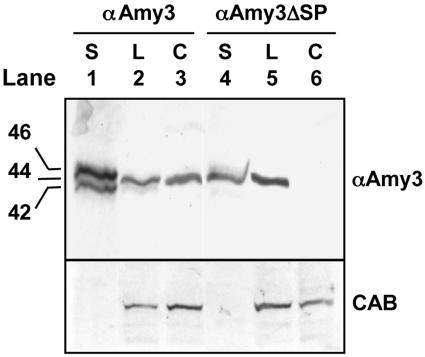
To determine whether αAmy3 is present within chloroplasts in a processed form, chloroplasts were isolated from tobacco leaves expressing αAmy3 or αAmy3ΔSP and total proteins were extracted and subjected to western-blot analyses using the α-amylase antibodies. As shown in the top section of Figure 6, the size of the in vitro translated αAmy3 was 46 kD (lane 1); however, the size of αAmy3 present in both leaves (lane 2) and chloroplasts (lane 3) was 44 kD, which was identical to that of αAmy3ΔSP derived from in vitro translation (lane 4) or present in leaves (lane 5), indicating that the αAmy3 present within chloroplasts has been processed. αAmy3ΔSP was not present in chloroplasts (Fig. 6, lane 6). Antibodies recognize a chloroplast marker protein, CAB, was reacted to the leaf and chloroplast extracts. CAB was detected in the leaf and chloroplast extracts of transgenic line expressing αAmy3 or αAmy3ΔSP (Fig. 6, bottom section), indicating comparable quality of the chloroplast extracts from two transgenic lines.
Figure 6.
αAmy3 localized in chloroplasts of transgenic tobacco leaves is processed. Leaves were collected from 1-month-old transgenic tobacco line A2 expressing αAmy3 and line B2 expressing αAmy3ΔSP. Chloroplasts were isolated from leaf extracts as described in “Materials and Methods.” Top section, RNA encoding αAmy3 or αAmy3ΔSP was in vitro transcribed from plasmid DNA and translated in the presence of 35S-Met as described in “Methods and Materials.” The 35S-labeled αAmy3 and αAmy3ΔSP were also resolved in the same SDS-PAGE along with leaf and chloroplast extracts, immunoblotted with the rice α-amylase antibodies, and viewed using fluorography. The x-ray film was overlaid onto the gel blot and photographed. Fifty micrograms of leaf extracts and 15 μg of chloroplast extracts were loaded in each lane. Lane 1, 35S-labeled αAmy3 (S); lanes 2 and 3, leaf (L) and chloroplast (C) extracts of line A2, respectively; lane 4, 35S-labeled αAmy3ΔSP; lanes 5 and 6, leaf and chloroplast extracts of line B2, respectively. Bottom section, Protein gel immunoblot analysis of the leaf and chloroplast extracts of lines A2 and B2 using CAB antibodies. Ten micrograms of leaf extract and 3 μg of chloroplast extracts were loaded in each lane.
To more precisely map the cleavage site of the αAmy3 SP, αAmy3 and αAmy3ΔSP were purified from isolated chloroplasts and leaves, respectively, of transgenic tobacco and subjected to amino acid sequence analysis. As shown in Table II, the N-terminal amino acid sequence of αAmy3 purified from chloroplasts is Ala (A), indicating SP was cleaved at a site one amino acid preceding the predicated SP cleavage site (Fig. 1A). The first amino acid of αAmy3ΔSP is Met (M), which was generated during construction of αAmy3ΔSP cDNA. This finding demonstrates that the SP of αAmy3 is cleaved during the process of translocation into chloroplasts.
Table II.
Analysis of the N-terminal amino acid sequences of αAmy3 and αAmy3ΔSP purified from chloroplasts and leaf, respectively, of transgenic tobacco
| Proteina | N-Terminal Sequenceb |
|---|---|
| αAmy3 (chloroplast) | AQVLFQGFN |
| αAmy3ΔSP (leaf) | MQVLFQGFN |
| Predicted N-terminal amino acid sequence of αAmy3: MKNTSSLCLLLLVVLCSLTCNSGQA ↓QVLFQGRNc | |
αAmy3 and αAmy3ΔSP were purified as described in “Materials and Methods.”
Analysis of N-terminal amino acid sequence was performed via automated Edman degradation on an ABI Procise 491 protein sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA).
Amino acids of SP are underlined; arrowhead indicates predicted cleavage site.
SP Is Sufficient for Directing Cargo Proteins to Plastids and Cell Walls
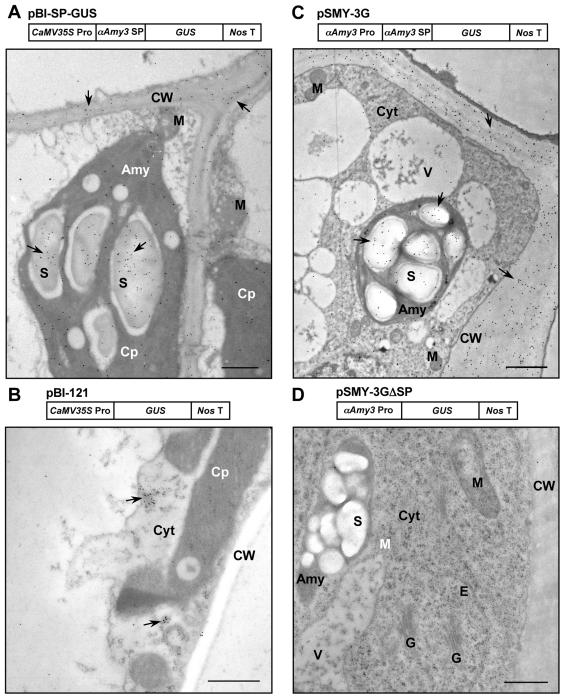
The function of αAmy3 SP was further analyzed in transgenic tobacco and rice for its capability in directing a cargo protein to plastids and the extracellular compartment. DNA encoding GUS with or without αAmy3 SP was fused downstream of either the CaMV35S promoter (Fig. 7, A and B) for expression in transgenic tobacco or the αAmy3 promoter (Fig. 7, C and D) for expression in transformed rice suspension cells. These chimeric genes were introduced into tobacco and rice genomes, and transgenic tobacco and transformed rice suspension cells were generated. EMI studies revealed that GUS expressed with SP was localized in stroma and over starch grains in chloroplasts and/or amyloplasts and in cell walls of transgenic tobacco leaves (Fig. 7A) and rice suspension cells (Fig. 7C). GUS expressed without SP was detected only in the cytoplasm of transgenic tobacco leaves (Fig. 7B) and transformed rice suspension cells (data not shown). No label was detected in any cellular compartment in nontransformed rice suspension cells (Fig. 7D). This study demonstrates that the αAmy3 SP is competent in targeting a cargo protein to both plastids and cell walls.
Figure 7.
The SP of αAmy3 is sufficient for directing GUS to plastids and cell walls. Immunocytochemical localization of GUS in transgenic tobacco leaf and transformed rice suspension cells using anti-GUS antibodies. A, Construct containing the CaMV35S promoter, αAmy3 SP, and GUS coding region was used for tobacco transformation. GUS was detected over starch grains within amyloplast and in the cell wall of leaf mesophyll cells of transgenic tobacco. B, Construct containing the CaMV35S promoter and GUS coding region was used for tobacco transformation. GUS was detected exclusively in the cytoplasm of leaf mesophyll cells of transgenic tobacco. C, Construct containing the αAmy3 promoter and SP and coding region of GUS was used for rice transformation. GUS was detected over starch grains within the amyloplast and in the cell wall. D, GUS was not labeled in any cellular compartment in nontransformed rice suspension cells. Arrowheads indicate positions of GUS. Scale bar represents 1 μm. Abbreviations: Amy, amyloplast; Cp, chloroplast; CW, cell wall; Cyt, cytoplasm; E, endoplasmic reticulum; G, Golgi apparatus; S, starch granule; V, vacuole.
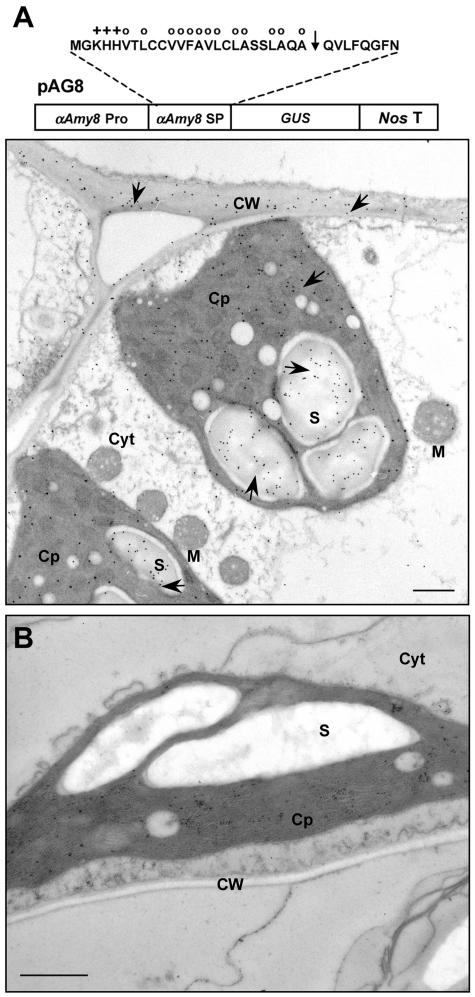
To determine whether other α-amylase SPs are also capable of directing a cargo protein into plastids, leaves of a transgenic tobacco line transformed with pAG8 (Chan et al., 1994), containing the rice αAmy8 promoter and its SP plus GUS coding region (Fig. 8A), was also examined. GUS expression with the αAmy8 SP was detected in chloroplasts and cell walls by anti-GUS antibodies (Fig. 8A) but not by the preimmune serum (Fig. 8B). This study indicates that at least one additional SP, from αAmy8, is also competent in targeting a cargo protein to both plastids and cell walls of plants.
Figure 8.
The SP of αAmy8 is sufficient for directing GUS to plastids and cell walls. Immunocytochemical localization of GUS in transgenic tobacco leaf using anti-GUS antibodies. A, Diagram shows the amino acid sequence of αAmy8 SP and the construct containing the αAmy8 promoter and SP and GUS coding region used for tobacco transformation. Arrowhead (↓) indicates the putative SP cleavage sites. Amino acids with hydrophobic (○) or positively charged (+) chain are indicated. GUS was detected over starch grains within the amyloplast and in the cell wall of leaf mesophyll cells in transgenic tobacco. B, Preimmune serum does not label GUS in leaf mesophyll cells of transgenic tobacco expressing GUS with SP. Arrowheads indicate positions of GUS. Scale bar represents 1 μm. Abbreviations: Cp, chloroplast; CW, cell wall; Cyt, cytoplasm; M, mitochondria; S, starch granule.
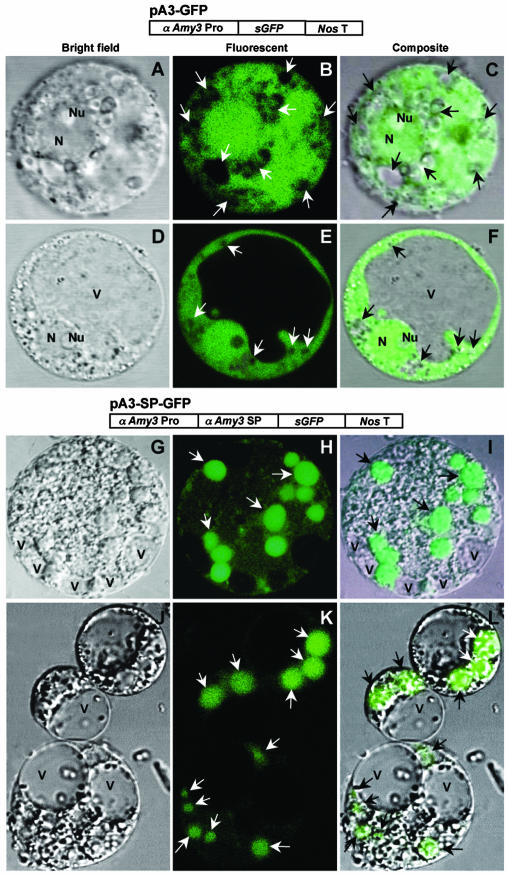
To confirm that the SP could indeed direct translocation of a cargo protein into amyloplasts, chimeric genes encoding green fluorescence protein (GFP) with or without the αAmy3 SP under the control of the rice αAmy3 promoter were constructed and introduced into the rice genome. Rice protoplasts were then isolated from the transformed calli, and the subcellular localization of GFP was analyzed using confocal microscopy. As the protoplasts derived from callus were a heterogenous population of cells, their morphology was not uniform. GFP without the SP was found throughout the cytoplasm as well as in the nucleus but not in vacuoles and organelles that appear to be amyloplasts containing starch grains (Fig. 9, A–F). As the nonfluorescent amyloplasts might overlap with the fluorescent cytoplasm, the image of amyloplasts appeared as irregular punctuated organelles. By contrast, GFP with the αAmy3 SP was targeted mainly to amyloplasts (Fig. 9, G–L). This study further demonstrates that SP is capable of directing a cargo protein to plastids.
Figure 9.
The SP of αAmy3 is sufficient for directing GFP to plastids. Rice calli were transformed with indicated constructs and selected with hygromycin. Protoplasts were isolated from the transformed calli expressing GFP. A to F, Construct containing the αAmy3 promoter and GFP coding region. GFP detected in cytoplasm and nucleus. G to L, Construct containing the αAmy3 promoter and SP and GFP coding region. GFP detected in amyloplasts. A, D, G, and J, Images of bright fields. B, E, H, and K, Images of fluorescent fields. C, F, I, and L, Composite images of fluorescence (green pseudo color) and transmission. Abbreviations: V, vacuole; N, nucleus; Nu, nucleolus. Arrowheads indicate positions of amyloplasts.
DISCUSSION
Dual Localizations of α-Amylases in Plant Cells
It has been previously reported that the rice α-amylases were found to localize in both cell walls and amyloplasts (Chen et al., 1994). As the rice α-amylases are encoded by a family of 10 genes (Huang et al., 1990), the individual members could be simultaneously or differentially targeted to two cellular compartments. In the present study, rice αAmy3 was shown to localize simultaneously in both plastids and cell walls and/or extracellular compartments of transgenic tobacco, suggesting that very likely αAmy3 also accumulates in the two compartments in rice cells. αAmy3 may not be the only α-amylase present in rice amyloplasts, as the αAmy8 SP has similar function as the αAmy3 SP. The capability of dual location targeting of αAmy3 and αAmy8 SPs, and possibly SPs of other α-amylases as well, may account for the accumulation of α-amylases in both amyloplasts and cell walls. The presence of α-amylases in plastids suggests they may play a role in plastidial starch degradation.
Two observations confirm the localization of αAmy3 in plastids. First, the anti-α-amylase antibodies only labeled the starch grains or plastids of cells expressing αAmy3 but not those of nontransformants or cells expressing αAmy3ΔSP. Second, by counting the numbers of gold particle-labeled αAmy3 present in the same unit area, we estimate that the relative amounts of αAmy3 present in chloroplasts, cell walls, and other cellular compartments are 31%, 66%, and 3%, respectively (Table I), indicating a large fraction of αAmy3 are localized in chloroplasts.
SP Is Necessary for Translocation of αAmy3 to Plastids and Extracellular Compartments
Two studies suggest that αAmy3 is transported extracellularly through the general secretory pathway and that the transportation is SP-dependent. First, SP is required for the in vitro import of αAmy3 into canine microsomal vesicles (Fig. 5). Second, SP is required for targeting of αAmy3 to cell walls and the extracellular compartment of transgenic tobacco leaves and suspension cells (Figs. 2 and 4, A and B).
The SP-dependent translocation of αAmy3 into plastids was unexpected, as in traditional model, a transit peptide is required for protein sorting to plastids (Schmidt and Mishkind, 1986; Keegstra, 1989). The deduced N-terminal amino acid sequences of the nine known rice α-amylases do not contain canonical chloroplast transit peptides. Instead, they all contain typical SP characteristics for translocation of proteins across the ER membrane (Chen et al., 1994). In this study, by loss-of-function analyses, SP was shown to be required for simultaneous translocation of αAmy3 to both plastids and cell walls of transgenic tobacco leaves and transformed tobacco suspension cells (Figs. 2–4). This study suggests the existence of a hitherto undiscovered pathway for targeting of proteins carrying SP to plastids in plant cells.
SP Is Sufficient for Directing Cargo Proteins to Plastids and Extracellular Compartments
Through gain-of-function analyses in transgenic tobacco and rice, we demonstrated that the αAmy3 SP is sufficient for directing GUS to plastids and extracellular compartments and GFP to plastids. SPs are known to carry proteins across the ER membrane prior to transport extracellularly. However, the expression of recombinant proteins fused with SPs, leading to dual targeting of the proteins to plastids and cell walls, is being increasingly observed. For example, assembled antibodies were found in chloroplasts, besides being detected in the ER, when mature light and heavy chains of a monoclonal antibody fused to a barley α-amylase SP were expressed in transgenic tobacco (During et al., 1990). Recently, the B-subunit of Escherichia coli heat-labile enterotoxin (LT-B) was found to be associated with starch grains in amyloplasts when this protein, carrying its native SP or fused with a maize (Zea mays) γ-zein SP, was overexpressed in transgenic maize endosperm (Chikwamba et al., 2003). Previously, αAmy8 SP was shown to direct extracellular translocation of GUS in transgenic tobacco, potato (Solanum tuberosum), and rice suspension cells (Chan et al., 1994). In this study, we showed that αAmy8 SP also directs the dual localizations of GUS in transgenic tobacco leaves. Recently, we also observed two bacterial derived enzymes, amylopullulanase and phytase, when fused to SPs derived from a rice glutelin and αAmy8, respectively, localized in both plastids and cell walls of transgenic rice endosperms and suspension cells (C.-M. Chiang, F.-S. Yeh, L.-F. Huang, T.-H. Tseng, C.-S. Wang, H.-S. Lur, H.-M. Lai, J.-F. Shaw, S.-M. Yu, unpublished data). All these studies demonstrate that a wide variety of SPs are competent for directing cargo proteins to both plastids and the extracellular compartment. It is unclear whether native proteins carrying these SPs are targeted to plastids and whether recombinant proteins carrying SPs are misrouted to plastids due to overexpression of these proteins. Nevertheless, the present study does suggest that, in addition to transit peptide, SP is capable of guiding protein translocation into plastids.
Pathway(s) for αAmy3 Translocation to Plastids and Extracellular Compartments
To our knowledge, our studies provide the first in vivo evidence for the transport of plant proteins into plastids via an SP-dependent pathway(s). From this study we now know (1) the αAmy3 SP is capable of transporting its cargo protein through the ER membrane (Fig. 5), (2) αAmy3 is processed prior to or upon entering chloroplasts (Fig. 6), (3) the αAmy3 SP is cleaved at a site only one amino acid preceding the predicted cleavage site (Table II), suggesting a possibility of processing carried out by a signal peptidase within the ER, and (4) the process of SP cleavage was rapid, as all detectable αAmy3 accumulating in transgenic tobacco leaves had a similar Mr as αAmy3ΔSP (Figs. 1B and 6).
The involvement of an SP and the ER in the targeting of proteins to photosynthetic plastids has been observed in algae. For example, Euglena chloroplasts are surrounded by three membranes, rather than two as found in higher plants and green algae, and this third chloroplast membrane is closely related to the ER membrane (Gibbs, 1981). In vivo protein labeling studies demonstrated that the Euglena light-harvesting chlorophyll a/b-binding protein of photosystem II (LHCPII) is transported as an integral membrane protein from the ER to the Golgi apparatus and then to the chloroplast (Sulli and Schwartzbach, 1995). All Euglena chloroplast protein precursors have been found to have functionally similar bipartite presequences composed of an N-terminal SP domain and a stromal targeting domain adjacent to the SP (Sulli et al., 1999). Examination of the entire peptide sequences of αAmy3 indicates that it does not contain a characteristic transit peptide domain adjacent to its SP, suggesting that algae and higher plants may employ different mechanisms for SP-dependent translocation of proteins to plastids.
How SPs direct protein translocation into plastids is unclear. One possible pathway could be that after entering the ER directed by SPs, most proteins are translocated extracelluarly as regular secretory proteins, but a significant fraction of these proteins are also targeted to the plastids. Alternatively, most proteins en route the ER and Golgi apparatus for extracellular transport, while a significant fraction of proteins is transported to plastids directly from the cytosol through an unknown SP-dependent pathway. In vitro chloroplast import assays were performed to test this hypothesis; however, no conclusion could be made for this possibility currently. A detailed mechanism for the import process requires further investigation. Whether the dual protein localizations to both plastids and extracellular compartments observed in this study is common for other secretory proteins, or is only unique to overexpressed proteins carrying SPs, also remains to be determined.
In summary, our study suggests the existence of a novel SP-dependent protein import pathway to plastids. Our study provides new insights into the role of SPs in protein targeting and the SP-dependent protein trafficking pathway in plants, as well as providing an ideal model system for future studies of these subjects.
MATERIALS AND METHODS
Plasmid Construction
For generation of αAmy3 cDNA with or without its SP sequence, a 1.6-kb full-length αAmy3 cDNA was isolated from a cDNA library made with RNA purified from cultured rice (Oryza sativa) suspension cells. The αAmy3 cDNA was subcloned into pBluescript SK+ (Stratagene, La Jolla, CA) to generate pBS-αAmy3. The DNA construct encoding αAmy3ΔSP was generated as follows. A 197-bp DNA fragment containing the 5′ region of αAmy3 cDNA, but without the coding region for the 25 SP residues (position 76 bp to 265 bp downstream of the translation initiation codon ATG), was generated by PCR. A Met was introduced as the first amino acid of αAmy3ΔSP. pBS-αAmy3 was truncated by removing the 5′end 272-bp region that contains the αAmy3 SP coding region. The 197-bp PCR product was then inserted into the truncated pBS-αAmy3, generating pBS-αAmy3ΔSP. The correct in-frame fusion of the nucleotide sequences was verified by DNA sequencing.
To prepare constructs for tobacco transformation, the cDNAs of αAmy3 with and without the SP sequence were excised from pBS-αAmy3 and pBS-αAmy3ΔSP and subcloned into pBK221, generating pBK-αAmy3 and pBK-αAmy3ΔSP, respectively. pBK221 (a gift from Dr. Teng-Yung Feng, Institute of Botany, Academia Sinica) is a binary vector and was generated by excising the GUS gene from pBI221 (CLONTECH, Palo Alto, CA). Therefore, expression of αAmy3 and αAmy3ΔSP is under the control of the CaMV35S promoter (Fig. 1A). pBI121 (CLONTECH) contains CaMV35S promoter-GUS chimeric gene (Fig. 7B). The αAmy3 SP was PCR synthesized and inserted between the CaMV35S promoter and GUS coding region in pBI121, generating pBI-SP-GUS (Fig. 7A).
To prepare constructs for rice transformation, plasmid p3G-132II containing the 1.7-kb rice αAmy3 promoter and SP sequence and GUS coding region (Lu et al., 1998) was linearized with PuvII and inserted into the same site in the binary vector pSMY1H (Ho et al., 2000), generating pSMY-3G (Fig. 7C). The αAmy3 SP was removed from p3G-132II, and the truncated plasmid was inserted into pSMY1H, generating pSMY-3GΔSP (Fig. 7D).
To generate the αAmy3 promoter-GFP chimeric gene, a 1.2-kb DNA fragment containing the promoter region of αAmy3 with or without the SP sequence of αAmy3 was PCR-amplified using plasmid p3G-132II (Lu et al., 1998) as the DNA template. The two DNA fragments were cloned into pBluescript SKII+ (Stratagene) to generate pBS-Amy3P (αAmy3 promoter only) and pBS-Amy3PSP (αAmy3 promoter plus SP sequence). The sGFP(S65T) cDNA encoding a modified GFP (Chiu et al., 1996) was inserted downstream of the αAmy3 promoter to make a transcriptional fusion or inserted downstream of the αAmy3 promoter and SP sequences to make a translational fusion, which were then inserted into the binary vector pSMY1H to generate pA3-GFP and pA3-SP-GFP (Fig. 9).
Transformation of Tobacco and Rice
Nicotiana tabacum L. cv Petit Havana SR1 and Oryza sativa cv Tainung 67 were used in this study. Plasmids were mobilized into Agrobacterim tumefaciens strain C58C1 carrying a rifamycin resistant gene using the freeze-thaw method (Holsters et al., 1978). Transgenic tobacco was obtained by transforming leaf discs with Agrobacterium according to the method of Horsch et al. (1988). Transformation of rice was performed as previously described (Ho et al., 2000). Suspension cell cultures of the transformed tobacco and rice were propagated as previously described (Yu et al., 1991).
Protein Western-Blot Analysis
Leaves were collected from 1-month-old transgenic tobacco, and total proteins were extracted from leaves as described (Yu et al., 1991). Protein western-blot analysis, using the rabbit anti-rice α-amylase polyclonal antibodies (Chen et al., 1994) diluted at 1:2,000, was performed as described (Yu et al., 1991).
EM Immunocytochemistry
Leaves of 3-month-old tobacco plants and cultured tobacco and rice suspension cells were fixed with 2% glutaraldyhyde and prepared for EMI as previously described (Chen et al., 1994). The primary antibody was the anti-rice α-amylase antibodies diluted 1:500, and control sections were incubated with preimmune serum similarly diluted. The GUS polyclonal antibodies were purchased from Molecular Probes (Eugene, OR). Colloidal gold (15 nm diameter)-conjugated goat anti-rabbit IgG (Biocell, Cardiff, UK) was used as the secondary antibody.
In Vitro Transcription and Translation
Plasmids pBS-αAmy3 and pBS-αAmy3ΔSP were linearized downstream of the α-amylase cDNA with KpnI. Transcription reactions were performed using T7 RNA polymerase in the presence of the cap analog m7G(5′) ppp(5′) G (Pharmacia, Piscataway, NJ) as described by Perry et al. (1991). The transcripts were subsequently translated in rabbit reticulocyte lysate (Promega, Madison, WI) in the presence of 35S-Met (Amersham, Buckinghamshire, UK) as suggested by the manufacturer.
In Vitro Protein Import into Microsomes and Post-Import Treatment
For import analysis of αAmy3 and αAmy3ΔSP, canine pancreatic microsomes (Promega) were included in the in vitro translation reactions described above, according to the method provided by the manufacturer. Proteinase K treatment was performed by incubating the microsomes after import with 1 mg/mL proteinase K on ice for 30 min. The reaction was terminated by adding 10 mm phenylmethylsulfonyl fluoride, and the reaction mixture was immediately boiled in SDS-PAGE sample loading buffer. α-Amylases were analyzed with SDS-PAGE using 12% gels. The gels were soaked in Amplify fluorography reagent (Amersham), dried, and exposed to x-ray film at −70°C.
Isolation of Chloroplasts and Purification of αAmy3
Chloroplasts were isolated as described (Perry et al., 1991) and treated with thermolysin (200 μg/mL) on ice for 30 min. The intact chloroplasts were reisolated through a 40% Percoll cushion. αAmy3 was purified from leaves or isolated chloroplasts using a method previously described (Chen et al., 1994).
Confocal Microscopy
Transformed rice calli were cultured in Murashige and Skoog medium containing 50 μg/mL hygromycin until the green fluorescence could be detected under a dissecting fluorescence microscope (Olympus, Tokyo). Transformed calli expressing GFP were selected and transferred to Suc-free medium for 2 d. Protoplasts were then isolated from the rice calli as previously described (Lu et al., 1998). The protoplasts expressing GFP were imaged with a Zeiss confocal microscope using a 488-nm laser line for excitation and a 515- to 560-nm long pass filter for emission.
Acknowledgments
We thank Dr. Neil Hoffman for the anti-pea CAB polyclonal antibodies, Dr. Tuan-Hua David Ho for critical review of the manuscript, and Drs. Lih-Jen Chen, Lin-Chih Yu, Shuh-Long Tu, and Sue-Ping Tsai for technical assistance.
This work was supported by Academia Sinica of the Republic of China.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042184.
References
- Beck E, Ziegler P (1989) Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40: 95–117 [Google Scholar]
- Beers EP, Duke SH (1988) Localization of α-amylase in the apoplast of pea (Pisum sativum L.) stems. Plant Physiol 87: 799–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M-T, Chao Y-C, Yu S-M (1994) Novel gene expression system for plant cells based on induction of alpha-amylase promoter by carbohydrate starvation. J Biol Chem 269: 17635–17641 [PubMed] [Google Scholar]
- Chen M-H, Liu L-F, Chen Y-R, Wu H-K, Yu S-M (1994) Expression of alpha-amylases, carbohydrate metabolism, and autophagy in cultured rice cells is coordinately regulated by sugar nutrient. Plant J 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Chikwamba RK, Scott MP, Mejia LB, Mason HS, Wang K (2003) Localization of a bacterial protein in starch granules of transgenic maize kernels. Proc Natl Acad Sci USA 100: 11127–11132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- During K, Hippe S, Kreuzaler F, Schell J (1990) Synthesis and self-assembly of a functional monoclonal antibody in transgenic Nicotiana tabacum. Plant Mol Biol 15: 281–293 [DOI] [PubMed] [Google Scholar]
- Gibbs SP (1981) The chloroplasts of some algal groups may have evolved from endosymbiotic eukaryotic algae. Ann N Y Acad Sci 361: 193–208 [DOI] [PubMed] [Google Scholar]
- Ho S-L, Tong W-F, Yu S-M (2000) Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol 122: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffmann N, Neidermeyer J, Rogers SG, Fraley RT (1988) Leaf disc transformation. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A5: 1–9
- Huang N, Sutliff TD, Litts JC, Rodriguez RL (1990) Classification and characterization of the rice alpha-amylase multigene family. Plant Mol Biol 14: 655–668 [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Gubler F, Chandler PM (1995) Gibberellin action in germinated cereal grains. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 246–271
- Kakefuda G, Duke SH, Hostak MS (1986) Chloroplast and extrachloroplastic starch-degrading enzymes in Pisum sativum L. Planta 168: 175–182 [DOI] [PubMed] [Google Scholar]
- Keegstra K (1989) Transport and routing of proteins into chloroplasts. Cell 56: 247–253 [DOI] [PubMed] [Google Scholar]
- Levi C, Preiss J (1978) Amylopectin degradation in pea chloroplast extracts. Plant Physiol 61: 218–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-P, Spilatro SR, Preiss J (1988) Subcellular localization and characterization of amylases in Arabidopsis leaf. Plant Physiol 86: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-A, Lim E-K, Yu S-M (1998) Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J Biol Chem 273: 10120–10131 [DOI] [PubMed] [Google Scholar]
- Okita TW, Greenberg E, Kuhn DN, Preiss J (1979) Subcellular localization of the starch degradative and biosynthetic enzymes of spinach leaves. Plant Physiol 64: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Li HM, Keegstra K (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol 34: 327–344 [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Mishkind ML (1986) The transport of proteins into chloroplasts. Annu Rev Biochem 55: 879–912 [DOI] [PubMed] [Google Scholar]
- Steup M, Robenek H, Melkonian M (1983) In-vitro degradation of starch granules isolated from spinach chloroplasts. Planta 158: 428–436 [DOI] [PubMed] [Google Scholar]
- Sulli C, Fang Z, Muchhal U, Schwartzbach SD (1999) Topology of Euglena chloroplast protein precursors within endoplasmic reticulum to Golgi to chloroplast transport vesicles. J Biol Chem 274: 457–463 [DOI] [PubMed] [Google Scholar]
- Sulli C, Schwartzbach SD (1995) The polyprotein precursor to the Euglena light-harvesting chlorophyll a/b-binding protein is transported to the Golgi apparatus prior to chloroplast import and polyprotein processing. J Biol Chem 270: 13084–13090 [DOI] [PubMed] [Google Scholar]
- Verner K, Schatz G (1988) Protein translocation across membranes. Science 241: 1307–1313 [DOI] [PubMed] [Google Scholar]
- Yu S-M, Kuo Y-H, Sheu G, Sheu Y-J, Liu L-F (1991) Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J Biol Chem 266: 21131–21137 [PubMed] [Google Scholar]
- Ziegler P, Beck E (1986) Exoamylase activity in vacuoles isolated from pea and wheat leaf protoplasts. Plant Physiol 82: 1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]