Abstract
The oligopeptide transporter (OPT) family contains nine members in Arabidopsis. While there is some evidence that AtOPTs mediate the uptake of tetra- and pentapeptides, OPT homologs in rice (Oryza sativa; OsGT1) and Indian mustard (Brassica juncea; BjGT1) have been described as transporters of glutathione derivatives. This study investigates the possibility that two members of the AtOPT family, AtOPT6 and AtOPT7, may also transport glutathione and its conjugates. Complementation of the hgt1met1 yeast double mutant by plant homologs of the yeast glutathione transporter HGT1 (AtOPT6, AtOPT7, OsGT1, BjGT1) did not restore the growth phenotype, unlike complementation by HGT1. By contrast, complementation by AtOPT6 restored growth of the hgt1 yeast mutant on a medium containing reduced (GSH) or oxidized glutathione as the sole sulfur source and induced uptake of [3H]GSH, whereas complementation by AtOPT7 did not. In these conditions, AtOPT6-dependent GSH uptake in yeast was mediated by a high affinity (Km = 400 μm) and a low affinity (Km = 5 mm) phase. It was strongly competed for by an excess oxidized glutathione and glutathione-N-ethylmaleimide conjugate. Growth assays of yeasts in the presence of cadmium (Cd) suggested that AtOPT6 may transport Cd and Cd/GSH conjugate. Reporter gene experiments showed that AtOPT6 is mainly expressed in dividing areas of the plant (cambium, areas of lateral root initiation). RNA blots on cell suspensions and real-time reverse transcription-PCR on Arabidopsis plants indicated that AtOPT6 expression is strongly induced by primisulfuron and, to a lesser extent, by abscisic acid but not by Cd. Altogether, the data show that the substrate specificity and the physiological functions of AtOPT members may be diverse. In addition to peptide transport, AtOPT6 is able to transport glutathione derivatives and metal complexes, and may be involved in stress resistance.
As other eucaryotic cells and procaryotes, plant cells have the ability to transport peptides across membranes. Peptide transport is important for storage and mobilization of reduced nitrogen (Higgins and Payne, 1977), and it is also involved in a wide range of cellular processes, including quorum sensing in bacteria, yeast mating, and the immune response in mammals (Stacey et al., 2002b). Furthermore, peptide-derived antibiotics (Dantzig et al., 1992) and anticancer agents (Hori et al., 1993) are also transported by peptide transporters.
The presence of multiple peptide transporters in the Arabidopsis genome and the known functions of peptide transport in bacteria, fungi, and animals suggest that peptide transporters may also play a key role in plant growth and development (Stacey et al., 2002a). Three gene families have been shown to transport various peptides in Arabidopsis, i.e. the peptide transporter (PTR) family, the oligopeptide transporter (OPT) family, and the multidrug resistance-associated protein (MRP) family.
The Arabidopsis genome contains 51 PTR family members (Stacey et al., 2002a). AtPTR2, the best known member of the family, mediates a cotransport of protons with dipeptides and tripeptides but not with larger peptides (Steiner et al., 1994, 1995; Song et al., 1996). AtPTR2 antisense plants exhibit delayed flowering and are arrested in seed maturation (Song et al., 1997). The Arabidopsis genome also contains nine members of the OPT family that are genetically and physiologically distinct from the PTR proteins (Koh et al., 2002). The AtOPT members encode proteins predicted to have 12 to 14 transmembrane domains and exhibit two conserved motifs (NPG and KIPPR) in protein regions predicted to be hydrophilic (Koh et al., 2002). Growth studies with a yeast Leu auxotroph mutant complemented by the AtOPT members indicated that seven members of the family (all but AtOPT2 and 3) transport tetra- and pentapeptides, presumably with proton cotransport (Koh et al., 2002). OPT homologs have also been characterized in other plants and in yeast. The Saccharomyces cerevisiae OPT1 protein was reported to transport the pentapeptides YGGFL and YGGFM (Leu- and Met-enkephalin; Hauser et al., 2000), reduced glutathione (GSH), oxidized glutathione (GSSG), and glutathione conjugates (Bourbouloux et al., 2000). Likewise, BjGT1, an OPT member from Indian mustard (Brassica juncea), and OsGT1, an OPT homolog from rice (Oryza sativa), were shown to transport GSSG and GS conjugates (Bogs et al., 2003; Zhang et al., 2004). In this respect, the OPT family resembles the last family of Arabidopsis peptide transporters, AtMRP1-4, which mediates the transport of GS conjugates (Lu et al., 1997, 1998; Sanchez-Fernandez et al., 1998). In terms of substrate specificity, the OPT family thus seems to combine properties of the PTR family, which is specific for di- and tripeptides, and of the MRP family, which is specific for GS derivatives.
However, although the ability of Arabidopsis OPT members to transport tetra- and pentapeptides has been well established, the possibility that they may also transport glutathione and its derivatives has not yet been investigated in detail. This study demonstrates that AtOPT6 is able to transport glutathione in strains grown under sulfur-deprived conditions, whereas AtOPT7 is not. Furthermore, it is shown that expression of AtOPT6, which was characterized by β-glucuronidase (GUS) reporter gene experiments, RNA blots, and real-time reverse transcription (RT)-PCR, is sensitive to the herbicide primisulfuron and to abscisic acid (ABA). These data shed new light on the possible physiological roles of the AtOPT family and suggest that AtOPT6 may be involved in stress resistance.
RESULTS
AtOPT6 and AtOPT7 full-length cDNAs were amplified from whole Arabidopsis seedlings by RT-PCR with specific primers and transferred to the yeast shuttle vector pDR 195 or pDR 196, respectively. The pDR/AtOPT6 and AtOPT7 constructs were used to transform the ABC 822 (MATa ura3-52 leu2-Δ1 lys2-801 his3- Δ200 trp1- Δ63 ade2-101 hgt1Δ::LEU2) and the ABC 817 yeast (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hgt1Δ::LEU2) yeast mutants deficient in glutathione transport. The requirement of the ABC 817 strain for a source of exogenous reduced sulfur is expected to be higher than that of ABC 822 due to disruption of a gene essential for Met synthesis.
We first assessed the effects of complementation of ABC 817 by either pDR/AtOPT6, pDR/AtOPT7, or TEF-HGT1 by studying the growth phenotype on synthetic dextrose (SD) solid media containing different organic sulfur sources (Met, Cys, GSH) in the presence of sulfate in the medium. While the hgt1met15 mutant complemented by the yeast glutathione transporter HGT1 can grow in the presence of GSH, the transformants carrying either the AtOPT6 or AtOPT7 cDNA required the presence of either Met or Cys in the medium, but their growth was not restored by the presence of GSH alone. Similar results were obtained with the hgt1met15 mutant complemented with BjGT1 or OsGT1 (data not shown). Furthermore, in the presence of Met, addition of high GSH concentrations (400 μm or 1 mm) did not affect the growth of the ABC 817 mutant complemented by the plant genes (compared to control without GSH), but it drastically inhibited the growth of the mutant carrying the TEF-HGT1 construct (data not shown). These data indicate that in this genetic background and under those conditions, the ability of the plant genes to mediate GSH transport in yeast was not sufficient, unlike that of the yeast transporter HGT1.
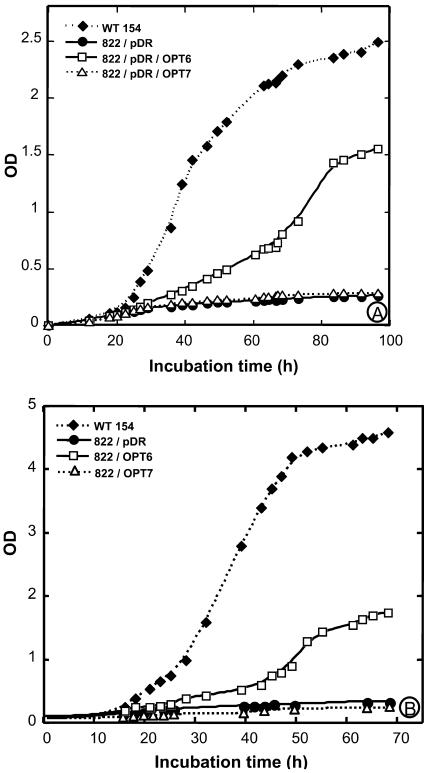
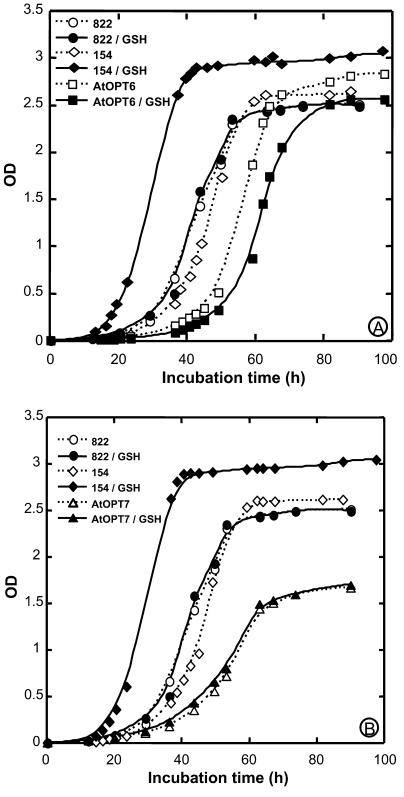
Further experiments were run with the ABC 822 strain, which, unlike the ABC 817 mutant, is affected in glutathione uptake, but not on Met synthesis. The ability of AtOPT6 to mediate uptake of glutathione was tested both by growth experiments and by direct measurements of glutathione uptake. The ABC 822 yeast strain disrupted on the OPT1 (HGT1) gene did not grow on a sulfur-free minimal medium containing either GSH or GSSG as the sole sulfur source (Fig. 1). However, complementation of this strain by the pDR/AtOPT6 construct allowed growth on the medium containing either GSH (Fig. 1A) or GSSG (Fig. 1B). Complementation of this strain by the empty vector or by the pDR/AtOPT7 construct did not allow growth in the presence of GSH (Fig. 1A). Similar results were obtained for growth assays in solid media (data not shown).
Figure 1.
Complementation of the ABC 822 (hgt1Δ) strain by AtOPT6 and AtOPT7. A, Growth assay in the presence of 500 μm GSH. The ABC 822 strain transformed with either the empty vector pDR or the pDR/AtOPT6 and AtOPT7 construct were grown in SD medium to OD600 = 0.6, washed three times in cold sterile water, and diluted to OD600 = 0.001 in SD-S medium containing 500 μm GSH. Their growth was compared with that of the wild-type strain ABC 154 under the same conditions. B, Growth assay in the presence of 500 μm GSSG.
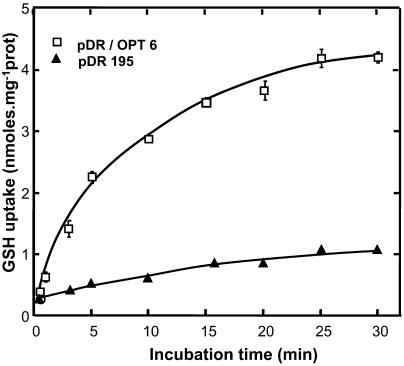
After complementation by the pDR/AtOPT6 construct, the ABC 822 strain was able to absorb labeled GSH, whereas it was not when transformed by the empty pDR vector. The uptake rate was constant during 5 min and thereafter decreased until 30 min (Fig. 2).
Figure 2.
Time course of [3H]GSH uptake by the ABC 822 (hgt1Δ) strain transformed with either the empty pDR vector or the pDR/AtOPT6 construct in a medium containing 100 μm [3H]GSH.
When the ABC 822 strain carrying the pDR/AtOPT6 construct was grown in synthetic complete medium, which contains many sulfur compounds, uptake of [3H]GSH was very low (data not shown). Thus, the glutathione uptake capacity was observed only when the yeasts were grown in a sulfur-deprived medium (SD-S) where the only source of sulfur is glutathione, confirming previous results (Bogs et al., 2003; Zhang et al., 2004).
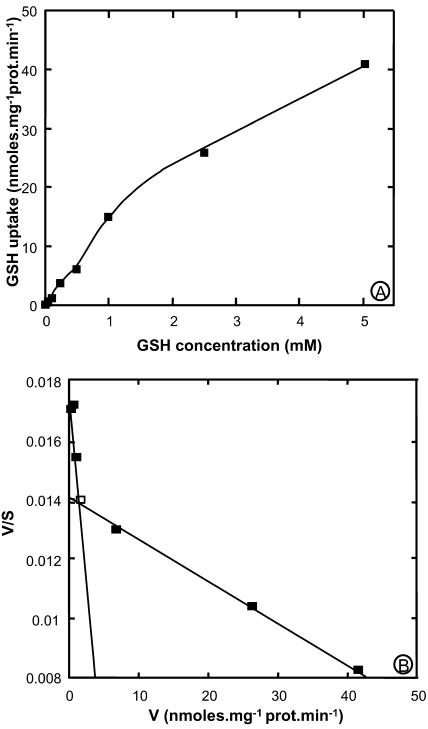
Concentration dependence studies showed that the initial rate of [3H]GSH uptake (measured within the first 3 min of incubation) was not fully saturable up to 5 mm (Fig. 3A). However, this rate decreased above 1 mm, and Eadie-Hostee plot analysis indicates that uptake kinetics may be interpreted as the superimposition of a low affinity (Km = 5 mm) and a high affinity (Km = 405 μm) component (Fig. 3B).
Figure 3.
Kinetic analysis of [3H]GSH uptake by yeast strain ABC 822 (hgt1) expressing AtOPT6. Initial rates of uptake of GSH were measured with yeasts carrying either the pDR/AtOPT6 construct or pDR alone. Background uptake measured with the strain carrying the empty plasmid was subtracted from uptake measured with the pDR/AtOPT6 strain, and the data were plotted according to Michaelis-Menten (A) and Eadie-Hofstee (B). The latter representation yields straight lines of slope −1/Km and ordinal intercept V/Km. V is given is nmol glutathione absorbed per min and per mg protein.
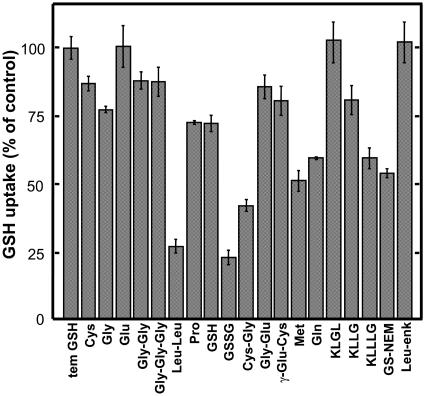
The substrate specificity of AtOPT6 was studied by competition experiments where the uptake of 50 μm [3H]GSH was challenged by a 10-fold excess of various potential competing substrates (Fig. 4). Various amino acids (Cys, Gly, Glu), dipeptides (Gly-Gly, Gly-Glu, γ-Glu-Cys), tripeptides (Gly-Gly-Gly), tetrapeptides (KLGL, KLLG), or the pentapeptide Leu-enkephalin (YGGFL) did not affect the uptake of [3H]GSH. By contrast, other amino acids (Pro, Met, Gln), dipeptides (Leu-Leu, Cys-Gly), and the pentapeptide KLLLG significantly decreased [3H]GSH uptake. Glutathione-N-ethylmaleimide conjugate and, even more, GSSG also decreased the uptake of labeled GSH. Addition of 500 μm unlabeled GSH significantly inhibited the uptake of 50 μm [3H]GSH, but the inhibition was not very strong, confirming that GSH did not completely saturate the transport system under these conditions. GSH uptake mediated by AtOPT6 in yeasts was strongly sensitive to protonophores and low temperature (data not shown).
Figure 4.
Effect of various compounds on GSH uptake in hgt1Δ/pDR/AtOPT6 yeast. The initial rates of uptake from a 50 μm GSH solution were determined in the presence of various compounds (500 μm) as indicated. Data were plotted as % of control (net initial rate of GSH uptake). The control initial rate of GSH uptake was 561 pmol mg prot−1 min−1. Results are the mean ± se of 8 to 12 samples from 2 or 3 independent experiments.
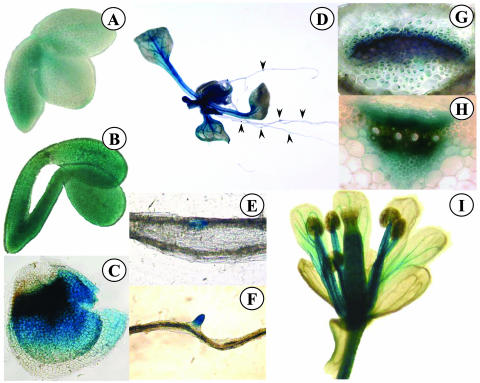
At least five independent transgenic lines expressing either an AtOPT6 promoter-GUS or AtOPT7 promoter-GUS fusion were analyzed histochemically to study the pattern of AtOPT6 and AtOPT7 expression during development. In young embryos, a weak AtOPT6-GUS-driven expression transiently appeared in the procambium (Fig. 5A). At a later stage, GUS expression was apparent over the whole surface of the embryos (Fig. 5B). AtOPT6 was also strongly expressed in the inner tegument, in the area adjacent to the micropyle (Fig. 5C). In adult plants, AtOPT6 expression was detected in the vascular bundles of the leaves, petioles, and stems (Fig. 5D). In the petiole (Fig. 5G) and in the stem (Fig. 5H), GUS activity was the strongest in the cambial zone of the vascular bundles. In the roots, AtOPT6-driven GUS expression was noticed in the regions of lateral root initiation (Fig. 5, E and F). In the flowers, the expression was detected at a relatively low level in the vascular network of the petals; by contrast, the stamen filaments and the gynoecium were strongly stained (Fig. 5I).
Figure 5.
Histochemical localization of GUS activity in Arabidopsis transformed with the AtOPT6 promoter region fused to the GUS reporter gene. A, Embryo; B, embryo; C, teguments left after embryo removal; D, whole plant; E and F, longitudinal views of roots; G, cross section of petiole; H, cross section of stem; I, flower.
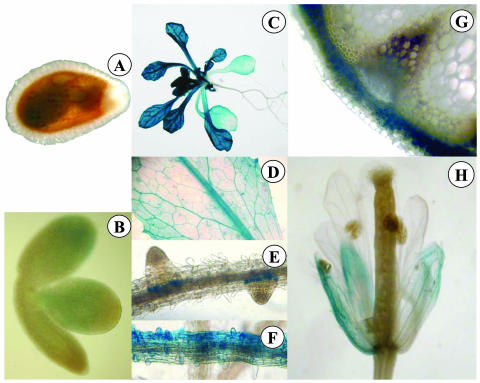
AtOPT7-driven GUS expression was not detected at the embryo stage (Fig. 6, A and B). In adult plants, this expression was more important in the aerial parts than in the root system (Fig. 6C) and somewhat stronger than AtOPT6 expression, although this was not quantified. In the leaves, the expression was detected in the major and the first order veins (Fig. 6D) and in the hydathodes. In the roots, AtOPT7 was expressed in circular zones surrounding lateral root primordia (Fig. 6E) and in some parts of the root epidermis (Fig. 6F). Unlike AtOPT6, AtOPT7 was expressed in the cortical tissues of the stem but not in the conducting bundles (Fig. 6G). In the flowers, AtOPT7 was expressed at a low level in the sepals, but neither in the petals nor in the reproductive tissues (Fig. 6H).
Figure 6.
Histochemical localization of GUS activity in Arabidopsis transformed with the AtOPT7 promoter region fused to the GUS reporter gene. A, Embryo tegument; B, embryo; C, whole plant; D, leaf area; E and F, longitudinal views of roots; G, cross section of stem; H, flower.
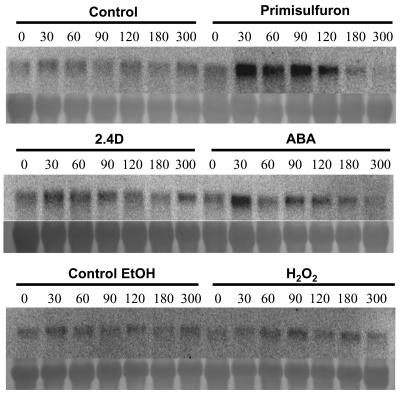
Attempts to detect AtOPT6 expression by northern blot in Arabidopsis plants grown in normal conditions were unsuccessful (data not shown). The inductibility of AtOPT6 expression by various compounds was tested in Arabidopsis suspension cells. Because glutathione has been reported to be involved in resistance against biotic and abiotic stress, and more particularly against oxidative stress (Noctor and Foyer, 1998), the effects of salicylic acid, H202, GSH, GSSG, and ABA were studied. The herbicide primisulfuron was also tested because the ABC transporters mediating the transport of GS conjugates across the tonoplast are strongly induced by herbicides (Tommasini et al., 1997). Cadmium (Cd) has been reported to affect BjOPT1 expression (Bogs et al., 2003) and was therefore also tested. Treatment by salicylic acid (10 μm), GSH (100 μm), GSSG (100 μm), H2O2 (5 mm), and Cd (50 μm) did not affect AtOPT6 expression within 5 h (data not shown). A weak increase of expression was observed after 2,4-dichlorophenoxyacetic acid (2,4-D) treatment (Fig. 7). AtOPT6 expression was significantly induced by ABA within 30 min. A strong induction was observed after 30 min of primisulfuron treatment and lasted for at least 90 min (Fig. 7).
Figure 7.
RNA blot analysis of AtOPT6 expression in Arabidopsis suspension cells treated with primisulfuron (80 nm), 2,4-D (10 μm), ABA (10 μm), ethanol (0.07%), and H2O2 (5 mm). The numbers refer to the duration of treatment expressed in min. Ethanol was used as a control for primisulfuron, which was added from an ethanolic stock solution. All treatments were repeated twice independently with similar results.
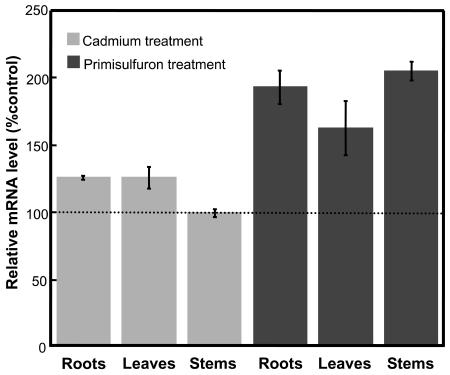
Real-time RT-PCR was conducted on various organs of hydroponically grown Arabidopsis plants treated either by cadmium or primisulfuron. Both compounds were added in the liquid medium 36 h before the plants were collected. Cadmium treatment did not affect AtOPT6 expression in roots, leaves, and stems (Fig. 8), thus confirming the data obtained with suspension cells. By contrast, primisulfuron almost doubled the expression of AtOPT6 in roots and stems but had no significant effect in leaves.
Figure 8.
RT-PCR analysis of AtOPT6 expression in Arabidopsis plants treated with cadmium (50 μm) or primisulfuron (80 nm). The data are means of three measurements ±se.
Glutathione is known to chelate heavy metals (Perrin and Watt, 1971), and we recently showed that Cd inhibited the expression of BjGT1 (Bogs et al., 2003). This led us to investigate possible interference between AtOPTs, glutathione, and heavy metals by studying yeast growth under different conditions (Fig. 9). In those experiments, all media contained cadmium, and the effects of glutathione addition, and of the presence of either pDR/AtOPT6 or pDR/AtOPT7 in the yeast, were studied. The addition of GSH in the medium accelerated the growth of the wild-type yeast, suggesting that glutathione conferred some protection against Cd for this strain (Fig. 9A). In the absence of GSH, the growth curve of the yeast mutant hgt1 was very similar to that of the wild type in the same conditions. However, in contrast to what was observed for the wild type, the addition of GSH did not affect the growth of the hgt1 yeast mutant. This was expected since this mutant is affected in glutathione uptake (Bourbouloux et al., 2000). Surprisingly, complementation of the yeast hgt1 mutant by AtOPT6 increased its sensitivity to Cd, since the growth curve was delayed compared to the mutant, and this sensitivity was even more increased when GSH was added to the medium (Fig. 9A). Although complementation by AtOPT7 increased the sensitivity of the mutant to Cd, as shown by a delay in the growth curve, GSH addition did not affect the growth of the AtOPT7 complemented strain (Fig. 9B), unlike what was observed after AtOPT6 complementation (Fig. 9A).
Figure 9.
Growth assays of various yeast strains in media containing 50 μm cadmium. A, Effect of transformation by AtOPT6 and/or GSH addition; 154 is the wild-type strain; 822 is a hgt1 mutant strain; AtOPT6 refers to the 822 strain carrying the pDR/AtOPT6 plasmid. GSH indicates that GSH was present at a final concentration of 50 μm in the medium. B, Effect of transformation by AtOPT7 and/or GSH addition. All experiments were repeated three times with similar results.
DISCUSSION
The importance of peptide transport in plants and the small information available on this process have been recently underlined (Stacey et al., 2002a). The OPT family in Arabidopsis has been described as an oligopeptide transporter family (Koh et al., 2002). Peptide growth assays demonstrated that AtOPT1-7 were able to mediate the uptake of Leu-containing peptides in yeasts and thus complemented Leu auxotrophy (Koh et al., 2002). Furthermore, AtOPT4 was shown to mediate the uptake of KLG-[3H]L, but at a much lower rate than that mediated by CaOPT1 from Candida albicans, so that KLGL appeared to be a poor substrate. The same paper mentioned that studies using [3H]glutathione failed to reveal any uptake when yeast cells were expressing any of the AtOPTs. By contrast, two plant homologs of yeast OPT1 have been reported to transport glutathione conjugates, GSSG, and to a lesser extent GSH. Glutathione transport was thus characterized for BjGT1 (Bogs et al., 2003) and OsGT1 (Zhang et al., 2004). This study, therefore, investigates the possibility that members of the AtOPT family may mediate glutathione uptake, and characterizes the localization of their expression and their inducibility.
AtOPT6 and AtOPT7 were cloned by PCR and introduced in the yeast mutants ABC 817 (hgt1met15) and ABC 822 (hgt1) by a 2-μ high copy number vector. Unlike the yeast glutathione transporter HGT1, none of the plant homologs tested (AtOPT6, AtOPT7, OsGT1, BjGT1) was able to restore growth of the hgt1met15 mutant on a medium containing GSH and sulfate. By contrast, complementation by AtOPT6, but not by AtOPT7, was able to restore growth of the hgt1 mutant and to mediate uptake of [3H]GSH in a sulfur-free minimal medium (Figs. 1 and 2). This indicates that the ability of the plant genes to complement glutathione auxotrophy is sufficient only in a genetic background where the need for reduced sulfur compounds is not too strong.
In contrast with AtOPT7, expression of AtOPT6 in the ABC 822 hgt1 yeast strain restored its ability to grow on a sulfur-free minimal medium containing either GSH or GSSG as the sole sulfur source (Fig. 1). In agreement with a previous report (Koh et al., 2002), this suggests that AtOPT7 is unable to transport GSH and GSSG at a substantial rate. This conclusion is further supported by the fact that the sensitivity of AtOPT7 yeast transformants to Cd is not enhanced by the presence of GSH (Fig. 9, and see discussion below).
The ability of the ABC 822/pDR/AtOPT6 yeast to absorb GSH was confirmed by direct uptake measurements under the same conditions (Fig. 2). The characteristics of [3H]GSH uptake mediated by AtOPT6 expression in the yeast were essentially the same as those described previously for BjGT1 and OsGT1. The initial rate of uptake mediated by AtOPT6 was about 0.3 nmol GSH mg−1 protein min−1, compared to about 1 nmol GSH mg−1 protein min−1 for BjGT1 (Bogs et al., 2003) and OsGT1 (Zhang et al., 2004). The initial rate of GSH uptake mediated by AtOPT6 is thus the lowest among the plant OPT transporters characterized so far and is about 6-fold less than the uptake rate mediated by the yeast transporter HGT1. As BjGT1 (Bogs et al., 2003) and OsGT1 (Zhang et al., 2004) but unlike HGT1 (Bourbouloux et al., 2000), AtOPT6 mediates GSH uptake in a way that does not obey simple kinetics. Eadie-Hofstee plots yielded a high affinity phase with a Km of about 400 μm, and a low affinity phase with a Km of about 5 mm. As discussed earlier (Zhang et al., 2004), it is difficult to know whether the low uptake rate and the biphasic kinetics of the plant transporters actually reflect their intrinsic properties or are due to improper or insufficient targeting of the plant transporters to the yeast plasma membrane. It is also noteworthy that GSH uptake by isolated wheat (Triticum aestivum) chloroplasts also obeyed biphasic kinetics with a high affinity phase (Km = 30−50 μm) and a low affinity phase with a Km in the mm range (Noctor et al., 2002).
The substrate specificity of AtOPT6 was studied indirectly by measurements of [3H]GSH uptake in the presence of an excess of potential competitors. An excess of unlabeled GSH inhibited uptake of [3H]GSH only moderately (about 30%) due to the fact that GSH transport may be mediated by the low affinity phase and thus is not completely saturated at 5 mm. In spite of this limitation, the main features observed with OsGT1 (Zhang et al., 2004) were also found for AtOPT6, i.e. a strong competition by GSSG, glutathione-N-ethylmaleimide conjugate, l-Met, l-Gln, and KLLLG. For both transporters, Leu-enkephalin did not compete with GSH uptake, whereas KLLG was a strong competitor. In addition, we show that KLGL (not studied previously) is not a good competitor of GSH uptake mediated by AtOPT6. The main differences derived from competition experiments are that l-Glu is a good competitor for OsGT1 but not for AtOPT6. By contrast, Leu-Leu is a strong inhibitor of GSH uptake mediated by AtOPT6, whereas it has little effect on OsGT1-mediated GSH uptake.
Both AtOPT6 and AtOPT7 expression in the ABC 822 (hgt1Δ) strain increased its sensitivity to Cd. This result is in agreement with a recent report showing that an OPT family member, AtOPT3, is regulated by metals and that its heterologous expression may rescue yeast mutants deficient in metal transports (Wintz et al., 2003). Furthermore, the AtOPT6-induced Cd sensitivity of the yeast is increased in the presence of GSH, whereas GSH does not affect AtOPT7-induced Cd sensitivity (Fig. 9). These results suggest that AtOPT6, unlike AtOPT7, is able to transport GSH-Cd complexes, in addition to GS conjugates. The effect of GSH on Cd sensitivity in yeasts expressing AtOPT6 may occur as follows: extracellular complexation of Cd by GSH, uptake of the complex by AtOPT6, release of Cd inside the cells. Whereas the amount of BjGT1 protein in B. juncea leaves is decreased by a 96-h exposure of the root system to Cd (Bogs et al., 2003), and whereas the amounts of AtOPT2 and AtOPT3 transcripts were strongly enhanced by iron deficiency (Wintz et al., 2003), we failed to detect any effect of Cd treatment of the level of AtOPT6 transcripts analyzed by quantitative RT-PCR. It is difficult to conclude to a general pattern of OPT regulation by metals because these data concern different members of the OPT family, and because there is not always a univocal correlation between the transcripts and protein amounts of OPTs, suggesting that this family may be subject both to transcriptional and translational regulations (Bogs et al., 2003; Zhang et al., 2004). Nevertheless, there are now several lines of evidence that at least some of the members may be able to transport metals and/or metal-peptide complexes. The expression of either AtOPT6 or AtOPT7 increases the sensitivity to Cd, even in the absence of GSH, compared to the mutant strain (Fig. 9). This suggests that both transporters may transport either Cd or, more likely, a complex between Cd and a component of the growth medium (His, for example).
Koh et al. (2002) studied the expression of various AtOPTs by RT-PCR. They found that both AtOPT6 and AtOPT7 were expressed in the flowers and, to a lower extent, in the roots. The data in this study extend these observations by a detailed analysis of GUS expression driven by either the AtOPT6 or AtOPT7 promoters. Thus, we show that in the flowers, the expression is mainly restricted to the filaments of the stamen and to the vascular bundles irrigating the young seeds. In the roots, AtOPT6 was mostly expressed in the regions of lateral root initiation. In this respect, it is interesting to note that the cad1 mutant blocked in glutathione synthesis is also impaired in the activation of cell division in root meristem (Vernoux et al., 2000). Furthermore, AtOPT6-driven GUS expression was also observed in the cambial zone of petiole and stem vascular bundles. The pattern of expression observed in the roots, stems, and petioles thus suggests that AtOPT6 is mainly expressed in areas where cell division is active. The pattern of AtOPT7 expression markedly differs from that of AtOPT6, in that it was poorly expressed in the conducting bundles (Fig. 6). This different pattern of expression, the different phenotypes conferred by AtOPT6 and AtOPT7 related to glutathione uptake, and Cd sensitivity in yeasts clearly indicate that these two transporters, in spite of their strong homology, exert different functions in the plant.
The metabolism of both primisulfuron and ABA is mediated via glucosylation. It is thus unexpected that both compounds induce the expression of a transporter so far characterized as a glutathione transporter. However, the same situation has been described for the induction of ABC transporters of the multidrug resistance proteins family by xenobiotics. Tommasini et al. (1997) studied the effects of primisulfuron and other biotics on the expression of four Arabidopsis ABC transporters homologous to MRPs. The abundance of one transcript called EST2 was strongly increased by primisulfuron and 1-chloro-2,4-dinitrobenzene, which are detoxicated as a Glc and a glutathione conjugate, respectively, but not by Cd (detoxicated by phytochelatins and glutathione complexes) and metolachlor (sequestered as glutathione conjugates). This indicated that plant may respond to toxic compounds with an induction of a large spectrum of proteins that may be related to various aspects of detoxification. The fact that AtOPT6 is strongly induced by primisulfuron and ABA suggests that it may play a role in detoxification and/or stress resistance, but the lack of effect of H2O2 indicates that oxidative stress is not directly involved.
In conclusion, these data and those described by Koh et al. (2002) suggest that AtOPT6 may transport peptides, glutathione conjugates, and glutathione complexes. However, as underlined by these authors, it is difficult to ascertain what are the most relevant substrates in vivo and thus the exact function of the AtOPT members. The comparison between AtOPT6 and AtOPT7 clearly indicates that the glutathione transport capacity is not shared by all the members of the family. Induction of AtOPT6 by primisulfuron and ABA suggests that this transporter may be involved in detoxification process.
MATERIALS AND METHODS
Cloning of AtOPT6 and AtOPT7 cDNAs
The PCR experiments were performed using cDNA prepared from 21-d-old plants and the Pfu turbo polymerase (Promega, Madison, WI). The AtOPT6 cDNA (GenBank accession no. AL035602.1) was cloned by PCR using primers AtOPT6-forward (5′CTCTTCTAAAACGATGGGAGAG3′) and AtOPT6-reverse (5′GTACAAAGCACTTGGTATATGC3′). The AtOPT7 (BAC clone T12H20, GenBank accession no. AF080119.1) coding sequence was cloned by PCR using primers AtOPT7-forward (5′GAAGACAACCATVGGAAGAATC3′) and AtOPT7-reverse (5′GGTACCAAATCCAACCACC3′). The PCR products were cloned into the pGEM-T Easy (Promega) vector and entirely sequenced.
Plasmid Constructs For Expression in Yeast
The 2.27-kb and 2.37-kb PCR fragments corresponding, respectively, to the open reading frame of AtOPT6 and AtOPT7 cDNAs were subcloned from the pGEM-T Easy vector into the EcoRI site of yeast expression vector pDR195 (AtOPT6) and into the NotI site of pDR196 (AtOPT7). The vectors pDR195 and pDR196 were kind gifts of Doris Rentsch (University of Bern, Switzerland).
Yeast Growth
The Saccharomyces cerevisiae strain ABC 822 bearing a deletion in HGT1 (MATa ura3-52 leu2-Δ1 lys2-801 his3-Δ200 trp1-Δ63 ade2-101 hgt1Δ::LEU2) and ABC 817 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hgt1Δ::LEU2) yeasts (Bourbouloux et al., 2000) are deficient in glutathione uptake and were used for complementation studies.
SD-S medium was prepared according to DIFCO's Bacto yeast nitrogen base without amino acids and ammonium sulfate recipe (DIFCO Laboratories, Detroit), with the modification that all sulfur containing reagents were substituted with equal amounts of the corresponding chloride salt (Zhang et al., 2004). The S. cerevisiae strain ABC 822 and the strain ABC 822 transformed with empty vectors (ABC 822/pDR) were grown in DIFCO's SD medium with ammonium sulfate, or in synthetic complete medium (Sherman, 1991), which contains ammonium sulfate and sulfur amino acids.
Transport Experiments
For growth assays in liquid and on solid medium, cells of ABC 822/pDR and ABC 822/pDR/AtOPT were grown overnight to an OD600 = 0.6 in minimal liquid SD medium containing ammonium sulfate and 2% Glc and the necessary amino acids. Uptake experiments were conducted as described by Zhang et al. (2004). Briefly, after a 5-min incubation of the cells at 28°C, [3H]GSH (1.9 TBq mmol−1; Amersham France, Les Ulis, France) was added to the buffered transport medium to a final concentration of 0.1 mm GSH (final specific activity 38 MBq mmol−1). At selected times, uptake was stopped by diluting the medium with a 20-fold volume of water (4°C), and cells were separated from the medium by filtration through a glass fiber filter (Sartorius AG, Goettingen, Germany). The cells were washed, the filter was dried, and the radioactivity was counted as described (Zhang et al., 2004).
Growth Test Experiments
The yeast were grown in yeast nitrogen base (DIFCO Laboratories) supplemented with ammonium sulfate, 2% Glc, and necessary auxotrophic supplements until an OD600 of 0.6. Twenty-five milliliters of fresh medium were inoculated to reach an initial OD600 of 9.7 10−3. The yeast were grown on a rotary shaker (200 rpm) at 28°C, and the OD600 was monitored for about 100 h by sampling 500-μL aliquots. In some experiments, CdCl2 (50 μm, final concentration), GSH, or GSSG (500 μm) were added to the medium as described in the “Results” section.
To check for glutathione auxotrophy, the yeasts were grown in SD-S liquid medium supplemented with either 500 μm GSH or GSSG.
Plant Growth
Seeds of Arabidopsis (Columbia ecotype) were grown hydroponically in nonsterile conditions. Cell suspensions of Arabidopsis (ecotype Columbia, strain T87) were cultured as described previously (Axelos et al., 1992).
Induction Experiments
The cultures were kept on a rotary shaker at 120 rpm and under 16-h photoperiod. Stress treatments were conducted 6 d after inoculation. Phytohormones (ABA) and growth regulators (2,4-D) were added at 10 μm (final concentration). The herbicide primisulfuron (80 nm), CdCl2 (50 μm), and hydrogen peroxide (5 mm) were directly administered into the culture medium. At the end of the treatment, plant cells were washed twice with deionized water and deep-frozen before RNA extraction.
In another set of experiments, Arabidopsis plants were grown hydroponically. Primisulfuron (80 nm) and CdCl2 (50 μm) were added to the liquid medium 36 h before the plants (3 weeks old) were sampled.
RNA Isolation and RNA Blot
Total RNA was isolated from yeasts as described by Logemann et al. (1987). Northern blots were performed following standard protocols (Sambrook et al., 1989), with 10 μg of total RNA in each lane. The full-length AtOPT6 cDNA was labeled by random primer labeling with 32P-dCTP and used as a probe.
Quantification of AtOPT Transcripts by Real-Time PCR
Real-time PCR-specific primers to AtOPT6 (forward, GAGTAGAACTCAATTCTTCTTAA; reverse, ACCAGAATTGATTTGGGATTGA) and to eIF1α (forward, TGTAACAAGATGCATGGCACT; reverse, ATGGGCACAAATGGGATTTT) were designed in the coding sequence of AtOPT6 and eIF1α. The specificity of the PCR product was checked by sequencing. For real-time PCR, total RNA was isolated from leaves, stems, and roots of 5-week-old hydroponically grown Arabidopsis plants. Either the herbicide primisulfuron or CdCl2 was added to the growth solution at final concentrations of 80 nm and 50 μm, respectively, 36 h before harvesting the plants. Five micrograms of the total RNA were reverse transcribed with a cDNA synthesis kit (MMLV reverse transcriptase; Gibco-BRL, Cleveland). Quantitative real-time PCR was carried out using an ABI Prism 7700 cycler (Applied Biosystems, Foster City, CA) and the SYBR Green PCR master mix (Applied Biosystems).
Histochemical GUS Assays
The promoter regions of AtOPT6 and AtOPT7 were amplified by PCR from genomic DNA. Genomic DNA was prepared from whole plants using the Wizard kit (Promega). The primers used were: ProAtOPT6-forward, ATCCGTGTGGTCATTATTTAGC, and ProAtOPT6-reverse, GAGTGAGTGTTCCGTTCTTTG; ProAtOPT7-forward, TGTTTTCTT TCCTACCACTGG, and AtOPT7-reverse, CTTCTTCTTCTTCTTGCTCTG. Amplification allowed to recover a 2.4-kb and a 2.0-kb fragment of the AtOPT6 and AtOPT7 promoter regions, respectively. These fragments were subcloned into the pSK vector and sequenced. They were subcloned into the XbaI/SmaI cloning site of the pCB308 vector containing the uidA reporter gene sequence (Xiang et al., 1999). The constructs were then introduced into Agrobacterium tumefaciens strain EHA105 by electroporation and selected for resistance to kanamycin (50 μg/mL). Arabidopsis ecotype Columbia was transformed by vacuum infiltration, and seeds were selected by spraying with glufosinate ammonium (BASTA, Frankfurt). Individual transformed plants were selfed to select for homozygous plants. At least five independent transformants were analyzed for each construct. One- to 3-week-old plants were stained by 5-bromo-4-chloro-3-indolyl-β-GlcUA by standard procedures. Samples were observed and photographed using a light microscope.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AL035602.1 and CAB38285.1 (AtOPT6), and AF080119 and AAC35527.1 (AtOPT7).
Acknowledgments
We thank Prof. Doris Rentsch (University of Bern, Switzerland) for gift of pDR plasmids, Prof. Alain Kitzis (University of Poitiers, France) for quantitative PCR facilities, and Dr. Nathalie Frangne (University of Poitiers, France) for help with GUS localization.
This work was supported by grants from the Indo-French Centre for the Promotion of Advanced Research and from the Association Franco-Chinoise pour la Recherche Scientifique et Technique.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039859.
References
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S (2003) Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant Cell Environ 26: 1703–1711 [Google Scholar]
- Bourbouloux A, Shahi P, Chakladar A, Delrot S, Bachhawat AK (2000) Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J Biol Chem 275: 13259–13265 [DOI] [PubMed] [Google Scholar]
- Dantzig AH, Tabas LB, Bergin L (1992) Cefaclor uptake by the proton-dependent dipeptide transport carrier of human intestinal Caco-2 cells and comparison to cephalexin uptake. Biochim Biophys Acta 1112: 167–173 [DOI] [PubMed] [Google Scholar]
- Guttierez-Alcala G, Gotor C, Meyer AJ, Fricker M, Vega JM, Romero LC (2000) Glutathione biosynthesis in Arabidopsis trichome cells. Proc Natl Acad Sci USA 97: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM (2000) Enkephalins are transported by a novel eukaryotic peptide uptake system. J Biol Chem 275: 3037–3041 [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW (1977) Characterization of active dipeptide transport by germinating barley embryos: effects of pH and metabolic inhibitors. Planta 136: 71–76 [DOI] [PubMed] [Google Scholar]
- Hori R, Tomita Y, Katsura T, Yasuhara M, Inui KI, Takano M (1993) Transport of bestatin in rat renal brush-border membrane vesicles. Biochem Pharmacol 45: 1763–1768 [DOI] [PubMed] [Google Scholar]
- Koh S, Wiles AM, Sharp JS, Naider FR, Becker JM, Stacey G (2002) An oligopeptide transporter gene family in Arabidopsis. Plant Physiol 128: 21–29 [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Drozdowicz YM, Hörtensteiner S, Martinoia E, Rea PA (1998) AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10: 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Rea PA (1997) AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP binding cassette transporter gene. Proc Natl Acad Sci USA 94: 8243–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH (2002) Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot 53: 1283–1304 [DOI] [PubMed] [Google Scholar]
- Perrin DD, Watt AE (1971) Complex formation of zinc and cadmium with glutathione. Biochim Biophys Acta 230: 96–104 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanchez-Fernandez R, Ardiles-Diaz W, Van Montagu M, Inze D, May MJ (1998) Cloning and expression anamlyses of AtMRP4, a novel MRP-like gene from Arabidopsis thaliana. Mol Gen Genet 258: 655–662 [DOI] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G (1997) Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol 114: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Steiner HY, Zang L, Naider F, Becker JM, Stacey G (1996) Cloning of a second Arabidopsis peptide transport gene. Plant Physiol 110: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, Koh S, Granger C, Becker JM (2002. a) Peptide transport in plants. Trends Plant Sci 7: 257–263 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker JM, Stacey G (2002. b) AtOPT3, a member of the oligopeptide transporter family is essential for embryo development in Arabidopsis. Plant Cell 14: 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner HY, Naider F, Becker JM (1995) The PTR family: a new group of peptide transporters. Mol Microbiol 16: 825–834 [DOI] [PubMed] [Google Scholar]
- Steiner HY, Song W, Zhang L, Naider F, Becker JM, Stacey G (1994) An Arabidopsis peptide transporter is a member of a new class of membrane transport proteins. Plant Cell 6: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Schmid J, Fromenteau M, Amrhein N, Martinoia E (1997) Differential expression of genes coding for ABC transporters after treatment of Arabidopsis thaliana with xenobiotics. FEBS Lett 411: 206–210 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inze D, et al (2000) The root meristemless1/cadmium sensitive 2 gene defines a glutathione dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Zhang MY, Bourbouloux A, Cagnac O, Shrikanth CV, Rentsch D, Bachhawat AK, Delrot S (2004) A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol 134: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]