Abstract
Phototropin is the blue-light receptor that mediates phototropism, chloroplast movement, and stomatal opening in Arabidopsis. Blue and red light induce chloroplast movement in the moss Physcomitrella patens. To study the photoreceptors for chloroplast movement in P. patens, four phototropin genes (PHOTA1, PHOTA2, PHOTB1, and PHOTB2) were isolated by screening cDNA libraries. These genes were classified into two groups (PHOTA and PHOTB) on the basis of their deduced amino acid sequences. Then phototropin disruptants were generated by homologous recombination and used for analysis of chloroplast movement. Data revealed that blue light-induced chloroplast movement was mediated by phototropins in P. patens. Both photA and photB groups were able to mediate chloroplast avoidance, as has been reported for Arabidopsis phot2, although the photA group contributed more to the response. Red light-induced chloroplast movement was also significantly reduced in photA2photB1photB2 triple disruptants. Because the primary photoreceptor for red light-induced chloroplast movement in P. patens is phytochrome, phototropins may be downstream components of phytochromes in the signaling pathway. To our knowledge, this work is the first to show a function for the phototropin blue-light receptor in a response to wavelengths that it does not absorb.
Blue light regulates a wide variety of photoresponses in plants, including chloroplast movement, inhibition of hypocotyl elongation, circadian timing, regulation of gene expression, and stomatal opening (Briggs and Huala, 1999; Christie and Briggs, 2001; Gyula et al., 2003). Three types of flavin-containing photoreceptors (phototropin, cryptochrome, and ZTL/FKF/ADO/LKP) have been identified as the blue-light receptors that mediate these responses (Briggs et al., 2001; Schultz et al., 2001; Briggs and Christie, 2002; Cashmore, 2003; Imaizumi et al., 2003; Lin and Shalitin, 2003; Liscum et al., 2003).
The phototropin gene PHOT1, first isolated in Arabidopsis, was shown to control phototropism (Huala et al., 1997). Phototropin contains a Ser/Thr protein kinase domain at the C terminus and two light, oxygen, or voltage (LOV) domains (LOV1 and LOV2) at the N terminus (Huala et al., 1997). The LOV domains function as binding sites for the chromophore FMN (Christie et al., 1999; Kasahara et al., 2002b) and belong to the PAS domain superfamily (Taylor and Zhulin, 1999). LOV1 and LOV2 are approximately 100 amino acids in length and are separated by a variable intervening sequence. Two phototropins, phot1 and phot2, function in Arabidopsis to control chloroplast movement and stomatal opening in addition to phototropism (Jarillo et al., 2001; Kagawa et al., 2001; Kinoshita et al., 2001; Sakai et al., 2001; Briggs and Christie, 2002).
Chloroplasts move to different locations in plant cells depending upon the intensity of light exposure. Under low-fluence-rate light conditions, chloroplasts spread over the cell surface perpendicular to the light direction, in order to harvest sufficient light and to maximize photosynthetic activity (Zurzycki, 1955). Under high-fluence-rate light conditions, chloroplasts become situated along the cell sides parallel to the direction of incident light, in order to minimize potential photodamage to the photosynthetic machinery (Kasahara et al., 2002a). These positions result from the movement of chloroplasts toward low-light-irradiated area (chloroplast accumulation movement) and away from strong-light-irradiated area (chloroplast avoidance movement; Haupt and Scheuerlein, 1990; Wada et al., 2003). In Arabidopsis, phot1 mediates the accumulation movement irrespective of fluence rates of blue light, whereas phot2 mediates the accumulation movement at low fluence rates and the avoidance movement at high fluence rates of blue light (Sakai et al., 2001; Kagawa and Wada, 2002; Wada et al., 2003). In the fern Adiantum capillus-veneris, blue light-induced chloroplast avoidance movement is also mediated by phot2 but not by phot1 (Kagawa et al., 2004).
In Physcomitrella patens, chloroplast movement is controlled by blue light. Different from seed plants, however, red light also induces this movement in P. patens (Kadota et al., 2000; Sato et al., 2001). This red light-induced movement is canceled by far-red light, indicating that phytochromes are the photoreceptors (Kadota et al., 2000). The motile systems for the chloroplast movement are both actin and tubulin based (Sato et al., 2001). Two cryptochromes (Ppcry1a and Ppcry1b) have been isolated in P. patens and are reported to regulate the blue light induction of side branches and gametophores (Imaizumi et al., 2002). Phototropins have not yet been isolated in P. patens.
Four phototropins from P. patens are reported in this study. Using phototropin disruptants, our data illustrate that both blue and red light-induced chloroplast movements are mediated by these phototropins. Several possible signaling pathways for chloroplast movement are discussed.
RESULTS
Isolation and Characterization of P. patens Phototropin Genes
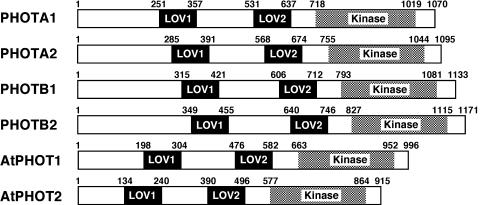
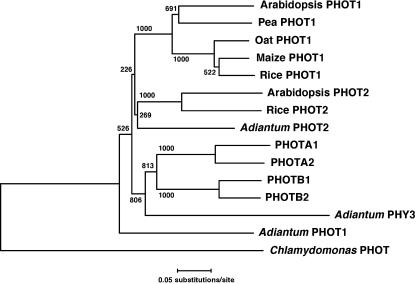
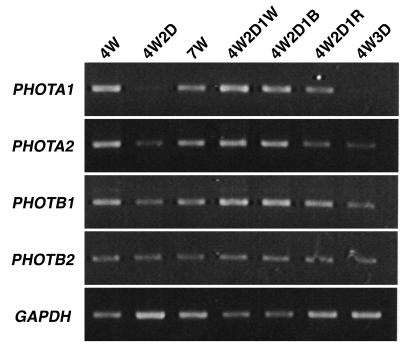
To analyze the function of the blue-light receptor phototropin in P. patens, four PHOT cDNA sequences were determined. They exhibited the typical domain organization of the phototropin family: two LOV domains (FMN-binding domains) at the N-terminal region and a Ser/Thr protein kinase domain at the C-terminal region (Fig. 1). The N-terminal extensions (from the initiation codon to the start of LOV1 domain) are longer than those of Arabidopsis (Fig. 1). A phylogenetic analysis showed that the four phototropins form a group that is independent from the PHOT1 and PHOT2 groups (Briggs et al., 2001). The P. patens group is further divided into two subgroups named PHOTA (1 and 2) and PHOTB (1 and 2) on the basis of their sequences (Fig. 2). The sequences were deposited in the GenBank/EBI/DDBJ database under accession numbers AB163420, AB163421, AB163422, and AB163423, respectively. A total of 8 × 105 plaques were screened from which multiple clones for the four PHOT genes were obtained, suggesting that the screening scale was large enough to isolate all of the PHOT genes that function in protonemal cells. And also, no other phototropin gene was found in an expressed sequence tag (EST) database, PHYSCObase (Nishiyama et al., 2003). Expressions of PHOT genes in protonemal cells were examined under different light conditions. PHOTA2, PHOTB1, and PHOTB2 were expressed under both light and dark conditions (Fig. 3). By contrast, PHOTA1 expression was induced by blue and red light but was drastically reduced under dark condition (Fig. 3).
Figure 1.
Domain organization of P. patens and Arabidopsis phototropins. Two LOV domains, LOV1 and LOV2, and Ser/Thr kinase domains are indicated. Amino acids of the start and the end of each domain are numbered.
Figure 2.
Phylogenetic tree of phototropins. The multiple sequence alignment and phylogenetic analysis were carried out using ClustalW program. The conserved domains in phototropins, i.e. LOV1, LOV2, and Ser/Thr protein kinase domains in Figure 1, were used for the phylogenetic analysis. Bootstrap values are indicated at the nodes.
Figure 3.
Expression of PHOT genes under different light conditions. Protonemata of P. patens were given different light treatments as described below: 4W, white light for 4 d; 4W2D, white light for 4 d and dark for 2 d; 7W, white light for 7 d; 4W2D1W, white light for 4 d, dark for 2 d, and white light for 1 d; 4W2D1B, white light for 4 d, dark for 2 d, and blue light for 1 d; 4W2D1R, white light for 4 d, dark for 2 d, and red light for 1 d; 4W3D, white light for 4 d and dark for 3 d. The light intensities were 20 W m−2 (approximately 80 μmol m−2 s−1) for white light, 4 W m−2 (approximately 16 μmol m−2 s−1) for blue light, and 5 W m−2 (approximately 28 μmol m−2 s−1) for red light. RT-PCR was performed using 5 μg total RNA and specific primers for each PHOT gene. Glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) was used as an internal control.
Generation of Disruptants of PHOT Genes
Four single disruptants (photA1, photA2, photB1, and photB2), two double disruptants (photA1photA2 and photB1photB2), and one triple disruptant (photA2photB1photB2) were generated by homologous recombination.
Single Disruptants
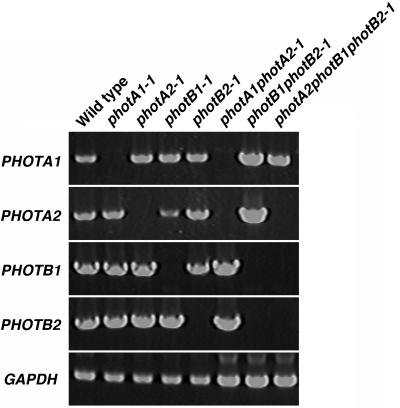
A fragment of each PHOT gene was amplified from the genomic DNA, cloned into a plasmid, and then the cloned PHOT genes were digested with appropriate restriction enzymes. The excised fragments were replaced by the nptII gene as a selection marker. After transformation of P. patens protoplasts, regenerated G418-resistant plants were screened by PCR using primers flanking the nptII cassette to find gene disruptants by homologous recombination events (Schaefer, 2001). From gene disruptants one DNA fragment having the expected size of the nptII cassette plus the flanking sequences of PHOT gene was amplified, but a wild-type copy of the PHOT gene was also amplified from mutants in which nptII cassette was additionally integrated randomly (data not shown). Southern-blot analysis of genomic DNA from the gene disruptants was performed with nptII gene as a probe to survey whether multiple targeting constructs might be integrated into the genome. Numbers of DNA fragments hybridized by the probe are shown in Table I. Not only single but also multiple integration occurred into the genome. Expression of PHOT genes in representative mutants by reverse transcription (RT)-PCR was analyzed to confirm that expected genes were targeted (Fig. 4).
Table I.
Phototropin mutants
| Type | Mutants | Antibiotics Resistancea | Number of Hybridized Fragmentsb |
|---|---|---|---|
| Single | photA1-1 | Km | 3 |
| photA2-1 | Km | 1 | |
| photA2-2 | Km | 6 | |
| photB1-1 | Km | 1 | |
| photB1-2 | Km | 1 | |
| photB2-1 | Km | 1 | |
| photB2-2 | Km | 3 | |
| Double | photA1photA2-1 | Km and Hyg | 2 |
| photA1photA2-2 | Km and Hyg | 2 | |
| photB1photB2-1 | Km and Hyg | 1 | |
| photB1photB2-2 | Km and Hyg | 1 | |
| Triple | photA2photB1photB2-1 | Km, Hyg, and Zeo | 1 |
| photA2photB1photB2-2 | Km, Hyg, and Zeo | 1 |
Km, kanamycin; Hyg, hygromycin; Zeo, Zeocin.
As hybridization probes, nptII, hpt, and Zeo genes were used for single, double, and triple mutants, respectively.
Figure 4.
Expression of PHOT genes in phototropin mutants. RNA samples were prepared from phototropin mutants grown under white light. RT-PCR was performed using 5 μg total RNA and specific primers for each PHOT gene. Glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) was used as an internal control.
Double Disruptants
Double disruptants in the same class of PHOT genes were produced. The photA2-1 and photB2-1 were transformed with the targeting constructs for PHOTA1 and PHOTB1 interrupted by a hygromycin cassette, respectively, resulting in the production of photA1photA2 and photB1photB2 double disruptants. After confirming the disruption of target genes by PCR, numbers of DNA fragments integrated into the genome were analyzed by Southern blot using a hygromycin resistance gene as a probe (Table I). Expression of PHOT genes in photA1photA2-1 and photB1photB2-1 was analyzed by RT-PCR (Fig. 4).
Triple Disruptants
Zeocin was used for antibiotic selection to generate triple disruptants of P. patens. Growth of protonemata was reduced or arrested with 50 μg mL−1 of Zeocin. However, P. patens protonemata containing a Zeocin resistance gene cassette grew normally on the medium with Zeocin, which could be used as a third selection marker in addition to kanamycin and hygromycin. The photB1photB2-1 mutant, which was chosen as a parental strain because PHOTB1 and PHOTB2 were disrupted without random integration (Table I), was transformed with the targeting construct for PHOTA2, in which the gene was interrupted with the Zeocin cassette. The integration of one copy of the Zeocin cassette into the genome of the resulting triple disruptants, photA2photB1photB2-1 and -2, were confirmed by Southern blot. The expression of PHOT genes in photA2photB1photB2-1 was analyzed by RT-PCR (Fig. 4).
Blue Light-Induced Chloroplast Movement in Basal Cells of Protonemata of Mutants
Chloroplast movement in protonemal cells was analyzed by microbeam irradiation with different fluence rates of blue light. The third or fourth cells from the apical tip cell of protonemal filaments were used for this analysis.
Wild Type, photA1-1, photB1-1, and photB2-1
As shown in Figure 5A, chloroplasts in a wild-type cell moved toward the area irradiated with weak blue light (seen in the panel at 80 min) and then moved away from the area irradiated with strong blue light (at 120 min). With weak light irradiation, chloroplasts, not only around the light irradiated area but also near the septums at the cell ends, accumulated toward blue light-irradiated area. The fluence-dependent responses of blue light-induced chloroplast movement is summarized in Table II. In wild-type cells, chloroplast accumulation movement was initiated at 0.002 W m−2, the transition point from accumulation movement to avoidance movement was at about 50 W m−2, and avoidance movement was observed at 100 W m−2. The photA1-1, photB1-1, and photB2-1 cells showed the same fluence rate response as that obtained in wild-type cells (Table II).
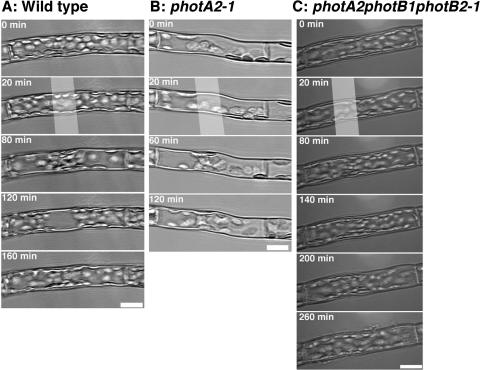
Figure 5.
Blue light-induced chloroplast movement in basal cells of wild-type, photA2-1, and photA2photB1photB2-1 protonemata. A, A wild-type cell was incubated in the dark (0–20 min), irradiated with 0.01 W m−2 (20–80 min) and 100 W m−2 blue light (80−120 min), and incubated in the dark (120–160 min). B, A photA2-1 cell was incubated in the dark (0–20 min) and irradiated with 0.01 W m−2 blue light (20–60 min) and 100 W m−2 blue light (60–120 min). C, A photA2photB1photB2-1 cell was incubated in the dark (0–20 min) and irradiated with 0.15 W m−2 (20–80 min), 1 W m−2 (80–140 min), 100 W m−2 (140–200 min), and 200 W m−2 blue light (200–260 min). The white bands indicate the position of the microbeam irradiation. Scale bars = 20 μm.
Table II.
Blue light-induced chloroplast movement in the basal cells of protonemata
| Strain | Fluence Rate of Blue Light (W m−2)
|
||||
|---|---|---|---|---|---|
| 0.001 | 0.002 | 50 | 100 | 200 | |
| Wild type | –a | Acb | Ac or weak Avc | Av | Av |
| photA1-1 | – | Ac | Ac or weak Av | Av | |
| photA2-1 | – | Ac | Ac | Ac | Ac |
| photB1-1 | – | Ac | Ac or weak Av | Av | |
| PhotB2-1 | – | Ac | Ac or weak Av | Av | |
| photA1photA2-1 | – | Ac | Ac | Ac | Ac |
| photB1photB2-1 | – | Ac | Ac | Ac | Av |
| photA2photB1photB2-1 | – | – | – | Weak Ac | Weak Ac |
–, No response.
Ac, Chloroplast accumulation movement.
Av, Chloroplast avoidance movement.
photA2-1 and photA1photA2-1
In photA2-1 cells, the lowest fluence rate at which accumulation movement was observed was 0.002 W m−2, which was similar to wild-type cells. However, in photA2-1 cells, chloroplasts stayed at the irradiated area even when light was switched to strong fluence rate (compare the panels at 120 min in Fig. 5, A and B). Unlike wild-type cells, avoidance movement was not induced at any fluence rate of blue light tested (Table II). Expression of PHOTA2 gene using cauliflower mosaic virus 35S promoter complemented the deficiency of avoidance movement in photA2-1 (data not shown). The photA1photA2-1 showed similar phenotype to the photA2-1 (Table II).
photB1photB2-1
In photB1photB2-1 the transition point from accumulation movement to avoidance movement was shifted to a value between 100 and 200 W m−2 (Table II). This value was several times higher than that of the wild type (50 W m−2).
photA2photB1photB2-1
The threshold required to induce accumulation movement in photA2photB1photB2-1 was 100 W m−2, which is 50,000-fold higher than that in the wild type (Table II). However, the accumulation movement was very weak and chloroplasts near the septums did not accumulate sufficiently to the light-irradiated area (200 and 260 min in Fig. 5C). Chloroplasts in basal cells hardly responded to blue light following the disruption of three PHOT genes. Chloroplast avoidance movement was not induced at any fluence rate of blue light tested.
Heterologous Expression of PHOTA2 and PHOTB2 in Acphot2 Mutant
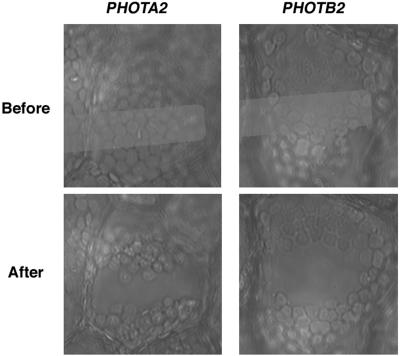
Kagawa et al. (2004) recently developed an assay system to access the function of phototropins as the photoreceptor for avoidance movement using A. capillus-veneris phot2 mutants, which are deficient in avoidance movement. To examine whether P. patens phototropins could function in the phot2 mutant, PHOTA2 and PHOTB2 were transiently expressed using this system. Prothallial cells of A. capillus-veneris phot2 mutants were cotransfected with cDNAs of PHOTA2 or PHOTB2 and green fluorescent protein (GFP) both driven by cauliflower mosaic virus 35S promoter. GFP was used as a marker to identify transformed cells. Chloroplasts moved away from areas irradiated with strong blue light (Fig. 6), showing that transient expression of PHOTA2 and PHOTB2 can complement the deficiency of avoidance movement of A. capillus-veneris phot2 mutants. PHOTA2 and PHOTB2 have the ability to induce avoidance movement in the heterologous system.
Figure 6.
Heterologous expression of PHOTA2 and PHOTB2 in A. capillus-veneris phot2 mutant cells. Prothallia of A. capillus-veneris phot2 mutants were transformed with cDNA of PHOTA2 or PHOTB2 using a particle bombardment delivery system (Kawai et al., 2003; Kagawa et al., 2004) as described in the text. Transformed cells were partially irradiated with a microbeam of 100 W m−2 blue light. The micrographs show the transformed cell before and after the irradiation. The white bands indicate the position of the microbeam irradiation.
Blue Light-Induced Chloroplast Avoidance Movement in the Tip Cells of Protonemata of Mutants
Although basal cells of photA2-1 and photA2photB1photB2-1 protonemata did not exhibit avoidance movement (Table II), the tip cells of these mutants did (Table III). However, avoidance movement did not occur in the tip cells of photA1photA2-1.
Table III.
Blue light-induced avoidance movement in the tip cells of protonemata
| Strain | Avoidance Movementa |
|---|---|
| Wild type | + |
| photA1-1 | + |
| photA2-1 | + |
| photA1photA2-1 | − |
| photA2photB1photB2-1 | + |
Chloroplast avoidance movement was analyzed in tip cells of protonemata. Cells were irradiated with blue-light microbeam at 200 W m−2.
Red Light-Induced Chloroplast Movement in Basal Cells of Protonemata of Mutants
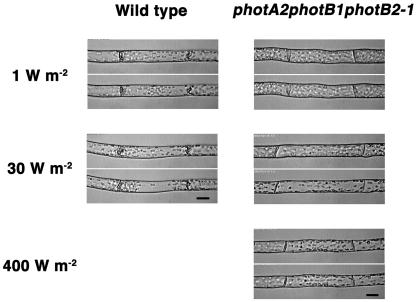
Red light as well as blue light induced chloroplast movement in protonemata of P. patens (Kadota et al., 2000; Sato et al., 2001). Because the induction by red light is canceled by far-red light irradiation, the photoreceptor for red light-induced chloroplast movement is most likely phytochrome (Kadota et al., 2000). In spite of this fact, we examined red light-induced chloroplast movement in phototropin mutants. In wild-type cells, red light at 1 and 30 W m−2 induced accumulation and avoidance movements, respectively (Fig. 7; Table IV). The photA2-1, photA1photA2-1, and photB1photB2-1 showed normal fluence rate responses (Table IV). However, in photA2photB1photB2-1, accumulation movement was not induced at 1 W m−2 but was induced at 30 W m−2 of red light, the intensity for inducing avoidance movement in wild-type cells (Fig. 7; Table IV). Because the accumulation of chloroplasts was very weak at 30 W m−2 (and even at 400 W m−2) of red light, chloroplasts located near the septums did not gather sufficiently around the light-irradiated area (Fig. 7). No avoidance movement occurred under red light in photA2photB1photB2-1.
Figure 7.
Red light-induced chloroplast movement in basal cells of wild-type and photA2photB1photB2-1 protonemata. Each section shows chloroplast distribution in the same cell before (top image) and after (bottom image) irradiation of red light. Fluence rates used (1, 30, and 400 W m−2) are shown. Scale bars = 20 μm.
Table IV.
Red light-induced chloroplast movement in the basal cell of protonema
| Strain | Fluence Rate of Red Light (W m−2)
|
||
|---|---|---|---|
| 1 | 30 | 400 | |
| Wild type | Aca | Avb | |
| photA2-1 | Ac | Av | |
| photA1photA2-1 | Ac | Av | |
| photB1photB2-1 | Ac | Av | |
| photA2photB1photB2-1 | –c | Weak Ac | Weak Ac |
Ac, Chloroplast accumulation movement.
Av, Chloroplast avoidance movement.
–, No response.
DISCUSSION
Four phototropin genes from the moss P. patens were isolated and divided into two groups (PHOTA and PHOTB) on the basis of the deduced amino acid sequences. This is similar to seed-plant phototropins, which are classified into two groups, PHOT1 and PHOT2 (Briggs et al., 2001). However, the P. patens phototropins form a separate clade from PHOT1 and PHOT2 groups in a phylogenetic tree (Fig. 2). Thus, the evolution of two groups of P. patens phototropins is likely to be independent of that of PHOT1 and PHOT2 groups of seed-plant phototropins.
It has been reported that phototropins control chloroplast movement in a seed plant, Arabidopsis (Jarillo et al., 2001; Kagawa et al., 2001; Sakai et al., 2001) and in a fern, A. capillus-veneris (Kagawa et al., 2004). The results in this article, using P. patens phot mutants, clearly show that chloroplast movements are also mediated by phototropins in this moss. This function of phototropin was conserved in plants.
When chloroplast movement in basal cells of protonemata was analyzed, photA1-1, photB1-1, and photB2-1 showed the same fluence rate response as the wild type, whereas photA2-1 lacked chloroplast avoidance movement at any fluence rate examined (Table II). This result illustrates that chloroplast avoidance movement is predominantly mediated by photA2 in basal cells. On the other hand, although single disruptants of PHOTB1 and PHOTB2 showed normal responses, photB1photB2 double mutants required a higher fluence rate than wild type to induce avoidance movement. photB1 and photB2 redundantly function and contribute, to some extent, to the photoperception for avoidance movement.
In Arabidopsis, phot2 mediates avoidance movement at high fluence rate of blue light, whereas phot1 mediates accumulation movement but not avoidance movement at any light fluence rate (Sakai et al., 2001). Because two classes (A and B classes) of P. patens phototropins can mediate avoidance movement, signaling mechanisms of phototropin itself or downstream pathway are likely more similar to those of Arabidopsis phot2. The ancestral origin of P. patens phototropins may be the phot2 type of Arabidopsis.
Although photA2-1 and photA2photB1photB2-1 were deficient in avoidance movement in basal cells, the response was induced in the tip cells of the same mutants with strong blue-light irradiation (Tables II and III). When examined in the tip cell of photA1photA2-1, avoidance movement did not occur. These results indicate that PHOTA1 contributes to the induction of avoidance movement much more in the tip cells than in the basal cells and might predominantly be expressed in the tip cells of protonemal filaments.
Chloroplast movement is regulated by blue light in most plants, and is regulated by red light (in addition to blue light) in the moss P. patens, the fern A. capillus-veneris, and the algae Mougeotia scalaris (Haupt and Scheuerlein, 1990; Kadota et al., 2000; Sato et al., 2001; Wada and Kagawa, 2001). The photoreceptor controlling the red light-induced chloroplast movement of A. capillus-veneris is phy3, a chimera protein consisting of a phytochrome chromophore-binding domain and phototropin (Nozue et al., 1998; Kawai et al., 2003). In P. patens, because the red-light induction of chloroplast movement is canceled by far-red light irradiation, the photoreceptor for this response is probably phytochrome (Kadota et al., 2000). Because no phy3-type chimera protein was isolated from P. patens in our screening (data not shown), conventional phytochromes, four of which are registered in the GenBank/EBI/DDBJ database and a previous report (Kolukisaoglu et al., 1993), are likely the primary photoreceptor for red light-induced chloroplast movement of P. patens.
Unexpectedly, the photA2photB1photB2-1 was deficient in both red light-induced accumulation and avoidance movements (Table IV). This result indicates that phototropins may be components of signal transduction pathways for the phytochrome-dependent chloroplast movement in P. patens. It should be noted that phytochrome genes were not disrupted by random integration of targeting constructs because no randomly integrated DNA fragments were detected in the photA2photB1photB2-1 (Table I). It is also possible that phototropins could control expression of phytochrome genes. However, the phytochrome-dependent chloroplast movement of P. patens takes place in red light-grown cells, but not in white light-grown cells (Kadota et al., 2000), and is enhanced by pretreatment of cells in the dark for 1 d (Sato et al., 2001), suggesting that the phytochrome that mediates red light-induced chloroplast movement predominantly expresses under dark and red-light conditions. Thus, expression of the phytochrome seems to be independent of phototropin. It has also been shown that phototropins play little role in transcriptional regulation in Arabidopsis (Ohgishi et al., 2004). For these reasons, an effect of the phototropin mutations on the expression of phytochrome in P. patens is unlikely. In A. capillus-veneris, a point mutation in the protein kinase domain of phy3 abolishes red light-induced chloroplast movement, indicating that the red-light signal received by the phytochrome domain is transmitted to downstream signaling components via the phototropin protein kinase domain of phy3 (Kawai et al., 2003). Thus, signaling mechanism, which is intramolecularly performed by phy3 in A. capillus-veneris, is performed directly or indirectly with phytochrome and phototropin molecules in P. patens. Regardless of light quality, phototropin could be an essential component for light-induced chloroplast movement. It will be interesting to test this in M. scalaris.
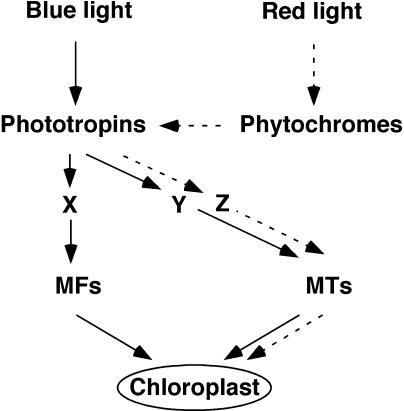
Sato et al. (2001) demonstrated that the blue light-induced chloroplast movement in P. patens utilizes both microfilaments and microtubules (MTs), whereas the red light-induced movement uses only MTs as traveling tracks. Figure 8 represents an updated model of the signaling mechanism of chloroplast movement in P. patens. There are two changes from a previous model (Sato et al., 2001): (1) the unidentified blue-light receptors are phototropins, and (2) the red-light signal received by phytochromes is transmitted to MTs system via phototropins (broken lines in Fig. 8). Although there are three pathways downstream of phototropins in this model, signaling components to control MTs system could be common in blue- and red-dependent pathways, that is Y = Z in Figure 8.
Figure 8.
A model for chloroplast movement. Blue and red light-induced signaling pathways are shown in plane and broken lines, respectively. X, Y, Z, Unknown signaling components; MFs, microfilaments; MTs, microtubules.
There are several reports showing an interaction between the phototropin and phytochrome signaling pathways. Chloroplast movement and phototropism are enhanced by red-light irradiation in Arabidopsis, and phytochromes are involved in both responses (Janoudi et al., 1997; Lasceve et al., 1999; Kagawa and Wada, 2000; DeBlasio et al., 2003). Phytochromes function as modulators because they are not essential for the induction of chloroplast movement and phototropic curvature. They modify the magnitude of the responses. On the other hand, the correlation between phytochromes and phototropins in the signaling pathway of red light-induced chloroplast movement in P. patens seems to be different. Because the disruption of phototropins abolishes the phytochrome-induced response, phototropins appear to be essential components, rather than modulators, for transmitting signals in the phytochrome pathway. This involvement of phototropins in phytochrome-signaling pathways is unique.
Chloroplast movement of P. patens is regulated by phototropins, like that of Arabidopsis. Availability of homologous recombination technique and easier observation of chloroplast movement because of simple cell organization are advantages for the use of P. patens as an experimental material. P. patens will be a good system to elucidate molecular mechanisms of chloroplast movement and phototropin signaling.
MATERIALS AND METHODS
Plant Materials
Protonemata of Physcomitrella patens subsp. patens were cultured aseptically at 26°C on BCDAT medium, which is BCD medium (1 mm MgSO4, 10 mm KNO3, 45 μm FeSO4, 1.8 mm KH2PO4, pH 6.5) supplemented with 1 mm CaCl2, 5 mm ammonium tartrate, and 0.8% (w/v) agar (Nishiyama et al., 2000).
Screening of P. patens Phototropin Genes
Partial phototropin fragments were amplified using cDNA prepared from protonemata of P. patens and degenerate primers specific to phototropin. Two sets of primer pairs used were F1 (5′-AARTTYATIGGIATGCARGTIGARGT-3′) and R1 (5′-CATYTCRTAIARIARIATICCIARIGCCCACCARTC-3′), or F2 (5′-GAYCCIMGIYTICCIGAYAAYCCIATIATITTYGC-3′) and R2 (5′-TCIGGIGCIATRTAYTCYTCIGTICCIACRAA-3′). A 1.5-kb fragment or a 1.1-kb fragment was amplified by PCR with F1 and R1, or F2 and R2, respectively. Sequencing analyses revealed that both amplified fragments had striking similarity to phototropin genes but were different from each other. We screened cDNA libraries produced from mRNA of P. patens protonemata using the PCR fragments as probes.
DNA Cloning, Sequencing, and Analysis
The positive clones were classified into four groups. The representative clones of each group were sequenced using the BigDye terminator sequencing kit using a DNA sequencer (model 377; Applied Biosystems, Foster City, CA). The 5′ regions of the cDNA sequences were obtained by 5′ RACE method using a kit (Invitrogen, Carlsbad, CA). BLAST search was performed using the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov) to analyze DNA sequences.
RT-PCR
Total RNA was prepared from protonemata with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RT of total RNA was carried out using oligo(dT) as a primer and SuperScript II RT (Invitrogen). PCR was performed using ExTaq polymerase (TaKaRa, Kyoto).
Construction of Phototropin Gene Disruption Vectors
The hygromycin cassette containing the E7133 promoter (Mitsuhara et al., 1996), the hygromycin B phosphotransferase (hpt) gene, and the nopaline synthase (nos) terminator was excised from pE7133-hpt vector, which was kindly gifted by R. Kofuji (Kanazawa University, Japan), and used as an antibiotics selection marker.
The BamHI (blunted)-NotI fragment of pE7133-hpt was cloned into the PstI (blunted)-NotI site of pMBL5 (Nishiyama et al., 2000). The resulting vector was named pE7133-nptII. The kanamycin cassette containing the E7133 promoter, the neomycin phosphotransferase II gene (nptII), and the 35S terminator of cauliflower mosaic virus (T35S) was excised from the pE7133-nptII vector and used as an antibiotics selection marker.
Zeocin resistance gene was amplified by PCR using the primers 5′-ATGGATCCATGGCCAAGTTGACCAGT-3′ (P1) and 5′-AGAGTCCCGCTCAGTCCTGCTCCTCGG-3′ (P2) and pCR-BluntII-TOPO (Invitrogen) as template. T35S was amplified using the primers 5′-GCAGGACTGAGCGGGACTCTGGGGTTC-3′ (P3) and 5′-ATCTCGAGGATCCCCGTCACCGGTG-3′ (P4) and pMBL5 as template. The PCR products of Zeocin resistance gene and T35S were mixed, and PCR was performed using the primers P1 and P4. The resulting PCR product was digested with BamHI and XhoI and cloned into BamHI-XhoI site of pE7133-hpt. The resulting plasmid pE7133-Zeo contains the E7133 promoter, Zeocin resistance gene, and T35S.
PHOTA1 targeting vector. The genomic DNA fragment of PHOTA1 was amplified using the primers 5′-ACGCAATGGTTGTTGAACTCTTC-3′ and 5′-GGATGATTCTTGATGTCGTTTGC-3′ and cloned into a vector, pGEM-T Easy (Promega, Madison, WI). The resulting vector was named pGEM-gPHOTA1. The kanamycin and the hygromycin cassettes were blunted and inserted into the blunted KpnI-XhoI site of the pGEM-gPHOTA1, and the resulting vectors were named pGEM-gPHOTA1-Km and pGEM-gPHOTA1-Hyg, respectively.
PHOTA2 targeting vector. The genomic DNA fragment of PHOTA2 was amplified using the primers 5′-GGACGAATTTGGGAGAGTGAGTT-3′ and 5′-TGTTTTCTGGCTTCAGGTCTCTG-3′ and cloned into a vector, pGEM-T Easy (Promega). The resulting vector was named pGEM-gPHOTA2. The kanamycin and the Zeocin cassettes were blunted and inserted into the SmaI-EcoRV site of the pGEM-gPHOTA2, and the resulting vectors were named pGEM-gPHOTA2-Km and pGEM-gPHOTA2-Zeo, respectively.
PHOTB1 targeting vector. The genomic DNA fragment of PHOTB1 was amplified using the primers 5′-CTACATTTGCAAGCAACGAGGAC-3′ and 5′-ACGAGACAAATGACTGCGAAAAA-3′ and cloned into a vector, pGEM-T Easy (Promega). The resulting vector was named pGEM-gPHOTB1. The kanamycin and the hygromycin cassettes were blunted and inserted into the blunted HpaI-BglII site of the pGEM-gPHOTB1, and the resulting vectors were named pGEM-gPHOTB1-Km and pGEM-gPHOTB1-Hyg, respectively.
PHOTB2 targeting vector. The genomic DNA fragment of PHOTB2 was amplified using the primers 5′-TGATGTTTGACTTTGGTGTGGTG-3′ and 5′-GGGTCCCTACAAAACCACACATT-3′ and cloned into a vector, pGEM-T Easy (Promega). The resulting vector was named pGEM-gPHOTB2. The kanamycin cassette was blunted and inserted into the blunted EcoRV-MunI site of the pGEM-gPHOTB2.
Transformation of Moss Protoplasts
Isolation of protoplasts and polyethylene glycol-mediated transformation were performed according to Nishiyama et al. (2000). To obtain stable transformants, in which single phototropin gene is disrupted, all phototropin targeting vectors containing the kanamycin resistant cassette were digested with NotI to separate its insert fragment from the vector sequences, and the digested plasmids were introduced into protoplasts. Transformed protoplasts were incubated for 4 to 5 d on BCDAT medium supplemented with 8% (w/v) mannitol without antibiotics and then transformed to BCDAT medium containing 50 μg mL−1 of G418 sulfate (Geneticin; Invitrogen) for 3 weeks.
To obtain double phototropin gene disrupted mutants, pGEM-gPHOTA1-Hyg and pGEM-gPHOTB1-Hyg were digested with NotI, and the digested plasmids were introduced into protoplasts prepared from photA2-1 or photB2-1 mutants, respectively. Transformed protoplasts were cultured as described above except for using 30 μg mL−1 of hygromycin B for antibiotic selection.
To obtain triple phototropin gene disrupted mutants, pGEM-gPHOTA2-Zeo was digested with NotI, and the digested plasmid was introduced into protoplasts prepared from photB1photB2-1 mutants. Transformed protoplasts were cultured as described above except for using 50 μg mL−1 of Zeocin (Invitrogen) for antibiotic selection.
Assay of Chloroplast Movement
Protonemal cells were inoculated between two layers of agar-gelatin film on a coverslip. The film was made from 0.5% (w/v) agar and 0.05% (w/v) gelatin. They were cultured under continuous dim red light for 1 to 2 weeks in the liquid BCDAT medium (Sato et al., 2001).
For partial irradiation of individual cells, a microbeam irradiation system (Olympus BX50; Tokyo) or one previously described (Kadota et al., 2000) were used. Monochromatic blue and red light was obtained through interference filters (Vacuum Optics of Japan, Tokyo), which have their transmission peaks at 452.4 and 663.2 nm and half-band widths of 23.3 and 32 nm, respectively. Neutral density filters of ND50, ND25, ND13, and ND3 (Hoya, Tokyo) were used when necessary. Fluence rate of the microbeam was measured using a siliconphotodiode (S1227-66BR; Hamamatsu Photonics K. K., Hamamatsu, Japan). One W m−2 of blue- and red-light microbeams used in this experiment are approximately 3.8 and 5.5 μmol m−2 s−1, respectively. Chloroplast movement induced by microbeam irradiation was monitored and recorded using an IR-sensitive video camera (C2400-07ER; Hamamatsu Photonics K. K.), a Macintosh computer (Power Macintosh 8600; Apple Japan, Tokyo), and the public domain NIH Image program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/) as described in Kagawa et al. (2004).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AB163420, AB163421, AB163422, and AB163423.
Acknowledgments
We thank Yuji Hiwatashi, Masae Umeda (National Institute for Basic Biology, Okazaki, Japan), and Takato Imaizumi (Scripps Research Institute, San Diego, CA) for helpful technical instructions about experimental systems of P. patens and Chieko Namba (NIBB, Okazaki, Japan) for careful help with P. patens culture. We also thank Edward B. Tucker (Baruch College, New York) for critical reading of the manuscript.
This work was partly supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Sports, Science and Technology of Japan (on Priority Areas, grant no. 13139203, and A, grant no. 13304061 to M.W.), by Solution Oriented Research for Science and Technology, Japan Science and Technology Corporation (grant to T. Kagawa), and by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (grant to Y.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042705.
References
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A, et al (2001) The phototropin family of photoreceptors. Plant Cell 13: 993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33–62 [DOI] [PubMed] [Google Scholar]
- Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- Christie JM, Briggs WR (2001) Blue light sensing in higher plants. J Biol Chem 276: 11457–11460 [DOI] [PubMed] [Google Scholar]
- Christie JM, Salomon M, Nozue K, Wada M, Briggs WR (1999) LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA 96: 8779–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio SL, Mullen JL, Luesse DR, Hangarter RP (2003) Phytochrome modulation of blue light-induced chloroplast movements in Arabidopsis. Plant Physiol 133: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P, Schafer E, Nagy F (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6: 446–452 [DOI] [PubMed] [Google Scholar]
- Haupt W, Scheuerlein R (1990) Chloroplast movement. Plant Cell Environ 13: 595–614 [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Janoudi AK, Gordon WR, Wagner D, Quail P, Poff KL (1997) Multiple phytochromes are involved in red-light-induced enhancement of first-positive phototropism in Arabidopsis thaliana. Plant Physiol 113: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410: 952–954 [DOI] [PubMed] [Google Scholar]
- Kadota A, Sato Y, Wada M (2000) Intracellular chloroplast photorelocation in the moss Physcomitrella patens is mediated by phytochrome as well as by a blue-light receptor. Planta 210: 932–937 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M (2004) Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol 45: 416–426 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Wada M (2000) Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol 41: 84–93 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Wada M (2002) Blue light-induced chloroplast relocation. Plant Cell Physiol 43: 367–371 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002. a) Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Swartz TE, Olney MA, Onodera A, Mochizuki N, Fukuzawa H, Asamizu E, Tabata S, Kanegae H, Takano M, et al (2002. b) Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol 129: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Kanegae T, Christensen S, Kiyosue T, Sato Y, Imaizumi T, Kadota A, Wada M (2003) Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421: 287–290 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu HU, Braun B, Martin WF, Schneider-Poetsch HA (1993) Mosses do express conventional, distantly B-type-related phytochromes: phytochrome of Physcomitrella patens (Hedw.). FEBS Lett 334: 95–100 [DOI] [PubMed] [Google Scholar]
- Lasceve G, Leymarie J, Olney MA, Liscum E, Christie JM, Vavasseur A, Briggs WR (1999) Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol 120: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Liscum E, Hodgson DW, Campbell TJ (2003) Blue light signaling through the cryptochromes and phototropins: so that's what the blues is all about. Plant Physiol 133: 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin IT, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100: 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh KC, Lagarias JC, Wada M (1998) A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc Natl Acad Sci USA 95: 15826–15830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci USA 101: 2223–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Wada M, Kadota A (2001) Choice of tracks, microtubules and/or actin filaments for chloroplast photo-movement is differentially controlled by phytochrome and a blue light receptor. J Cell Sci 114: 269–279 [DOI] [PubMed] [Google Scholar]
- Schaefer DG (2001) Gene targeting in Physcomitrella patens. Curr Opin Plant Biol 4: 143–150 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Kagawa T (2001) Light-controlled chloroplast movement. In D-H Häder, M Lebert, eds, Photomovement. Elsevier, Amsterdam, pp 897–924
- Wada M, Kagawa T, Sato Y (2003) Chloroplast movement. Annu Rev Plant Biol 54: 455–468 [DOI] [PubMed] [Google Scholar]
- Zurzycki J (1955) Chloroplasts arrangement as a factor in photosynthesis. Acta Soc Bot Pol 24: 27–63 [Google Scholar]