Abstract
HsfA2 is a heat stress (hs)-induced Hsf in peruvian tomato (Lycopersicon peruvianum) and the cultivated form Lycopersicon esculentum. Due to the high activator potential and the continued accumulation during repeated cycles of heat stress and recovery, HsfA2 becomes a dominant Hsf in thermotolerant cells. The formation of heterooligomeric complexes with HsfA1 leads to nuclear retention and enhanced transcriptional activity of HsfA2. This effect seems to represent one part of potential molecular mechanisms involved in its activity control. As shown in this paper, the activity of HsfA2 is also controlled by a network of nucleocytoplasmic small Hsps influencing its solubility, intracellular localization and activator function. By yeast two-hybrid interaction and transient coexpression studies in tobacco (Nicotiana plumbaginifolia) mesophyll protoplasts, we found that tomato (Lycopersicon esculentum) Hsp17.4-CII acts as corepressor of HsfA2. Given appropriate conditions, both proteins together formed large cytosolic aggregates which could be solubilized in presence of class CI sHsps. However, independent of the formation of aggregates or of the nucleocytoplasmic distribution of HsfA2, its transcriptional activity was specifically repressed by interaction of Hsp17.4-CII with the C-terminal activator domain. Although not identical in all aspects, the situation with the highly expressed, heat stress-inducible Arabidopsis HsfA2 was found to be principally similar. In corresponding reporter assays its activity was repressed in presence of AtHsp17.7-CII but not of AtHsp17.6-CII or LpHsp17.4-CII.
Stress-induced gene expression leads to the rapid accumulation of heat stress proteins (Hsps). Many of them act as molecular chaperones with important functions not only for the protection of proteins against stress damage but also for their folding, intracellular distribution and degradation (Ellis, 2000; Dougan et al., 2002; Hartl and Hayer-Hartl, 2002; Haslbeck, 2002; Picard, 2002; Walter and Buchner, 2002; Young et al., 2003). Eukaryotic heat stress (hs)-inducible genes share conserved promoter elements with the palindromic consensus motif (AGAAnnTTCT; Pelham, 1982; Pelham and Bienz, 1982; Nover, 1987). They represent the recognition sites of the heat stress transcription factors (Hsf), which are the key regulators of the response conserved throughout the eukaryotic kingdom (Wu, 1995; Morimoto, 1998; Nover et al., 2001).
In plants the Hsf system is more complex than in any other organism investigated so far (for review, see Nover et al., 2001). Unfortunately, detailed investigations on the structural and functional specification of Hsfs are restricted to few examples of tomato (Lycopersicon esculentum) Hsfs A1, A2, A3, and B1 (Scharf et al., 1990; Treuter et al., 1993; Boscheinen et al., 1997; Bharti et al., 2000, 2004; Döring et al., 2000). Particularly striking are the properties of HsfA2, which is a strictly hs-inducible protein. Due to a strong C-terminal nuclear export signal (NES), it does not localize in the nucleus unless coexpressed with the constitutively present HsfA1, which was identified as master regulator of the hs response in tomato (Mishra et al., 2002). Evidently, HsfA1 is not only required for the hs-dependent expression of HsfA2, but also as coactivator and nuclear retention factor by formation of HsfA1/A2 heterooligomers (Scharf et al., 1998a; Heerklotz et al., 2001).
The network of protein interactions influencing the function and intracellular distribution of HsfA2 has a second aspect. In the course of a heat stress response, the ongoing accumulation of HsfA2 and other hs-inducible proteins results in a unique storage form of the transcription factor in cytoplasmic chaperone complexes composed of the 40-nm heat stress granules (HSG; Nover et al., 1989; Scharf et al., 1998a). No other Hsf so far identified in tomato cells (HsfA1, HsfA3, HsfB1) was found in the HSG complexes (Scharf et al., 1998a, Bharti et al., 2000), which are mainly formed of the cytosolic small Hsp (sHsp) classes CI and CII and Hsp70.
From the point of view of its abundance as well as its high activator potential, HsfA2 becomes the dominant Hsf in tomato after hs treatment (Mishra et al., 2002). Hence the activity control and intracellular localization of HsfA2 is an important aspect of plant performance in the daily cycles of heat stress and recovery experienced in a natural surrounding. In this study, we present results about a novel function of tomato Hsp17.4-CII as cytoplasmic retention factor and corepressor of HsfA2. In fact, the functional state and intracellular distribution of HsfA2 is influenced by a network of proteins involving HsfA1, Hsp17-CI, and Hsp17-CII.
RESULTS
Identification of Hsp17.4-CII as Specific Interaction Partner of Tomato HsfA2 in Yeast Two-Hybrid System
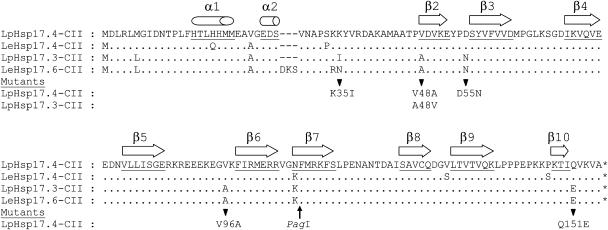
Yeast two-hybrid screening was used to identify tomato proteins interacting with heat stress transcription factor HsfA2 as bait. To this aim, the two previously identified activator motifs AHA1 and AHA2 in the C-terminal domain of HsfA2 had to be mutated (Döring et al., 2000). Because of residual activator function of this mutant bait pBDGal4xHsfA2.11.16B, we added 5 mm 3-aminotriazol (3-AT) to discriminate between strains showing residual growth on His-free media and those with strongly increased His biosynthesis due to HsfA2 interaction with other proteins. Among 104 positive clones, 49 preys encoded LpHsp17.4-CII (Figs. 1 and 2). Interestingly, the other type of tomato Hsp17-CII (LpHsp17.3-CII) was not detected in the two-hybrid screening. As shown in Figure 1, the sequences of the two isoforms of Hsp17-CII from L. peruvianum correspond to Hsp17.4-CII and Hsp17.6-CII of the culture tomato L. esculentum, and both forms are discriminated by only few amino acid residues marked in Figure 1.
Figure 1.
Sequence comparison of the two isoforms of Hsp17-CII proteins in tomato (Lp, Lycopersicon peruvianum; Le, L. esculentum). The positions of exchanged amino acid residues in mutant forms of LpHsp17.4-CII and LpHsp17.3-CII used in this manuscript are shown below the sequence alignment. Secondary structure predictions of α-helices in the N-terminal part and β-sheets in the C-terminal α-crystallin domain (β2–β9) are indicated on top of the corresponding amino acid residues (underlined). Database accession numbers of the four sequences are: AY608694 (LpHsp17.4-CII), AJ225049 (LpHsp17.3-CII), AF090115 (LeHsp17.4-CII), and U72396 (LeHsp17.6-CII).
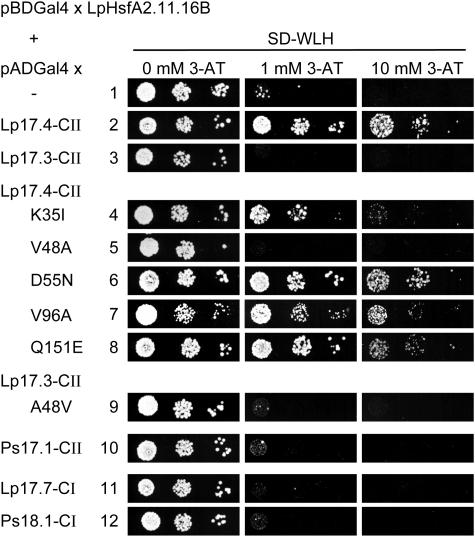
Figure 2.
Yeast two-hybrid interaction of HsfA2 in bait position with sHsps in prey position. For the bait construct, the C-terminal domain of LpHsfA2 was mutated in its two activator motifs (Döring et al., 2000). The residual growth of strains with the Gal4DBDxHsfA2.11.16B bait was abolished by adding 1 or 10 mm 3-amino-triazol (3-AT). In the prey position, we tested the indicated sHsps fused to the Gal4AD. Samples 4 to 9 represent tests with mutant forms of the two LpHsp17-CII isoforms (for sequence details see Fig. 1). In all cases, liquid cultures of the indicated yeast strains were grown overnight at 30°C in selective minimal medium without Trp and Leu (SD-WL). After adjustment to an OD600 of 0.1, 5 μL of these cultures as well as 2 subsequent dilutions by 1 to 10 were dropped on agar plates of SD-WL medium lacking His (SD-WLH). The His-auxotroph colony growth was monitored after incubation for 48 h at 30°C.
By directly testing different cytosolic sHsps as preys in combination with the C-terminal part of HsfA2 as bait, it was confirmed that the selectivity of interaction is very high (Fig. 2). Only Hsp17.4-CII interacts with HsfA2 (no. 2), but not Hsp17.3-CII (no. 3) nor any other sHsp of classes CI and CII from tomato or pea (Pisum sativum; nos. 10–12). To analyze the striking difference between the two types of Hsp17-CII, we created a number of mutant proteins of Hsp17.4-CII with single amino acid exchanges as indicated in Figure 1. The two-hybrid test clearly showed that only the mutant protein with exchange of V48>A in immediate vicinity of the α-crystalline domain lost the capability for interaction with HsfA2, whereas other amino acid exchanges discriminating the two different forms, i.e. K35>I, D56>N, V96>A, and Q151>E were unaffected in this respect (nos. 4 and 6–8). The V48>A exchange in Hsp17.4-CII abolished interaction, i.e. this loss-of-function mutation corresponds to the Hsp17.3-CII type of sHsps. We also tested the reverse exchange of A48>V in the Hsp17.3-CII background (no. 9). Although negative in the yeast two-hybrid test, this mutant form was shown to interact with HsfA2 in other test systems (see below).
Hsp17.4-CII Promotes Aggregation of HsfA2 in Tobacco Protoplasts
We used tobacco (Nicotiana plumbaginifolia) protoplasts to confirm and complement the results of the yeast two-hybrid system. In two important aspects the situation in these plant cells is more complex and fundamentally different from the yeast two-hybrid system: (1) expression of the full length proteins results in formation of their native oligomeric structures, i.e. any interaction observed under these conditions reflects the specific recognition between the HsfA2 as trimeric or multimeric complex and Hsp17-CII which is usually a dodecamer; and (2) The function of HsfA2 as activator of hs-inducible genes may contribute to the complexity of protein interactions because of the synthesis of the endogenous chaperones. Whenever necessary, this problem was minimized by using HsfA2 mutants exhibiting identical properties with respect to the intracellular localization and interaction with HsfA1 and Hsp17.4-CII, but with strongly reduced or lacking activator function. On the other hand, we tried to mimic the complex natural situation of HsfA2 as component of the chaperone and Hsf networks by coexpression of defined mixtures of the proteins in tobacco protoplasts.
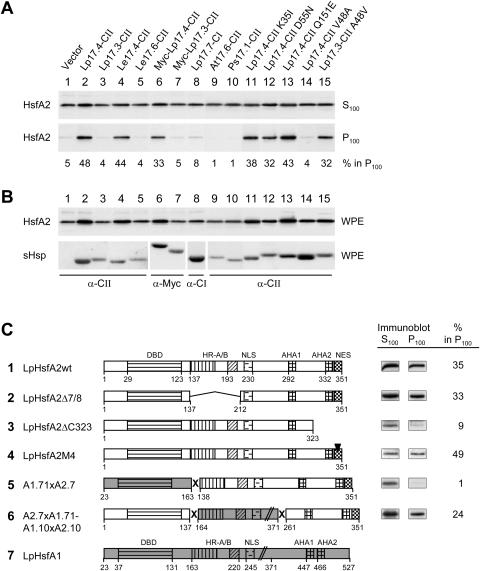
One intriguing aspect of the interaction of HsfA2 with Hsp17.4-CII is the formation of insoluble aggregates. To investigate this in more detail, we used differential centrifugation to characterize the high Mr, salt- and detergent-resistant aggregates of HsfA2. To avoid the complex situation of protein recruitment into HSG or HSG-like complexes, samples were incubated at room temperature. Whole cell extracts of tobacco protoplasts expressing the indicated proteins were prepared in buffer containing high-salt (500 mm NaCl) and detergent (0.5% Nonidet and 0.2% Sarcosyl). After centrifugation for 1 h at 100,000g (see “Materials and Methods”), the distribution of HsfA2 between the soluble form in the supernatant (S100) and the sedimentable form in the pellet (P100) fractions was determined by immunoblot analysis (Fig. 3). If expressed alone, only 5% of the total HsfA2 was detected in the pellet fraction (Fig. 3A, lane 1). However, a considerable portion (48%) of HsfA2 was sedimentable in the presence of Hsp17.4-CII but not of Hsp17.3-CII (lanes 2 and 3). The numbers at the bottom of the immunoblots of the P100-fraction give an estimate of the relative amount of HsfA2 in the pellet fractions (S100 + P100 = 100%). Similar results were obtained with the two proteins from the culture tomato, i.e. LeHsp17.4-CII as the interacting type and LeHsp17.6-CII as the noninteracting type (lanes 4 and 5; for sequences see Fig. 1). Interestingly, the oligomerization state of the two class CII sHsps was not crucial for the interaction with HsfA2. The N-terminally Myc-tagged forms of both proteins did not form the usual dodecamers of about 220 kD (data not shown), but the specific influence on the sedimentation behavior of HsfA2 was not altered (Fig. 3, lanes 6 and 7). None of the other sHsps tested, i.e. LpHsp17.7-CI (lane 8) nor Hsp17-CII proteins from Arabidopsis or pea (lanes 9 and 10), influenced the aggregation of HsfA2. For control, expression levels of the proteins were analyzed by immunoblots (Fig. 3B).
Figure 3.
Influence of different sHsps and functional domains of HsfA2 on the formation of soluble and sedimentable forms of HsfA2. HsfA2 was expressed in tobacco protoplasts together with the indicated sHsps. Whole protoplast extracts (WPE) in high salt and detergent buffer were centrifuged for 1 h at 100,000g, and the distribution of HsfA2 between supernatant (S100) and pellet fractions (P100) was estimated by immunoblot analysis. A, Numbers given at the bottom of the corresponding immunoblots represent the relative proportion of HsfA2 in the sedimentable form (S100 + P100 = 100%). B, Expression controls of HsfA2 and sHsps detectable in immunoblots of the WPE fractions. C, LpHsp17.4-CII was coexpressed with the indicated wild-type (sample 1) and mutant forms (samples 2–4) of HsfA2 as well as fusion proteins of HsfA2 with HsfA1 (5 and 6). Analysis of soluble (S100) and insoluble forms (P100) of the indicated Hsfs was performed and the signals of the immunoblot analyses as well as values for the amount of sedimentable HsfA2 calculated from the corresponding densitometer scans are given at the right. The block diagrams represent functional domains of HsfA1 (7, shaded) or HsfA2 (DBD, DNA-binding domain; HR-A/B oligomerization domain; NLS nuclear localization sequence; AHA1, 2, transcriptional activator motifs; NES, nuclear export signal).
We used the sedimentation assay also to test mutant forms of LpHsp17.4-CII and of the closely related LpHsp17.3-CII with exchanges of the amino acid residues discriminating the two forms (see sequences in Fig. 1). Similar to the results with the yeast two-hybrid test, only the Hsp17.4-CII mutant protein with V48>A (lane 14) but not the mutant forms with K35>I, D55>N, or Q151>E (Fig. 3, lanes 11–13) were disturbed in the interaction with HsfA2. Interestingly, in contrast to the results with the yeast two-hybrid system (Fig. 2, no. 9) the mutant of Hsp17.3-CII with A48>V showed a clearly detectable interaction with HsfA2 (Fig. 3, lane 15). These results confirmed the crucial role of the amino acid residue V48 for discriminating the interacting type (Hsp17.4-CII) from the noninteracting type (Hsp17.3-CII with A48).
The sedimentation assay offered also the opportunity for a simple check of different mutant forms of HsfA2 with respect to their interaction capacity (Fig. 3C). Although different to a certain extent, all forms of HsfA2 tested so far were affected in their sedimentation behavior in the presence of LpHsp17.4-CII (Fig. 3C, nos. 1, 2, and 4). The only exception was the C-terminally truncated form HsfA2ΔC323 (no. 3). Evidently, the interaction with HsfA2 and the resulting formation of high Mr forms require the C-terminal activation domain of HsfA2 but are not dependent on the oligomeric state. The HsfA2Δ7/8 mutant form lacking the oligomerization domain was not impaired in its interaction with Hsp17.4-CII (no. 2). The same was true for the mutant protein HsfA2M4 with defective nuclear export sequence. Despite the accumulation in the nucleus (Heerklotz et al., 2001), the sedimentation behavior in presence of Hsp17.4-CII was not affected (Fig. 3C, no. 4).
Particularly interesting are the results with two fusion proteins harboring the indicated parts of HsfA1 and HsfA2 respectively (Mishra et al., 2002). Although the C-terminal part of HsfA2 is necessary for the interaction with Hsp17.4-CII it is not sufficient for the formation of the sedimentable form (no. 5). Determinants in the N-terminal part with the DNA binding domain and its flanking sequences are required as well (compare results with fusion proteins 5 and 6 in Fig. 3C).
HsfA2 as Part of a Network of Interacting Proteins
Based on the results from the two-hybrid screening, HsfA2 interacts with HsfA1 (Scharf et al., 1998a) and with Hsp17.4-CII (this manuscript). However, in tomato cells the situation is much more complex. After hs induction all three proteins coexist, and their interactions are crucial for the intracellular distribution of HsfA2 between nucleus and cytoplasm as well as its function as heat stress transcription factor. Under heat stress conditions the cytoplasmic complexes of HsfA2 and Hsp17.4-CII are incorporated into HSG (Scharf et al., 1998a). The other dominant class of cytosolic sHsps, Hsp17-CI, also interacts with Hsp17-CII and keeps it in a soluble state under control temperature conditions (Siddique et al., 2003).
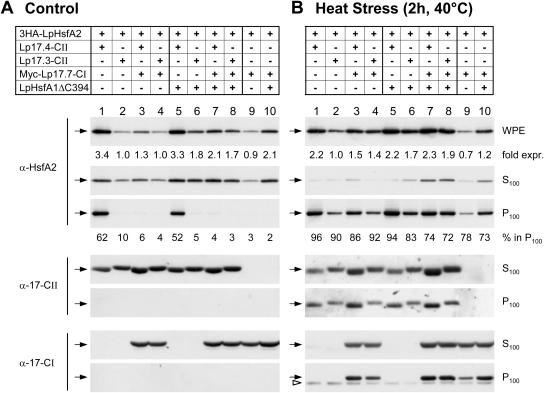
To elaborate more details of this network of protein interactions and mutual influences of the partners, we used tobacco protoplasts for coexpression of different combinations of Hsfs and members of the tomato sHsp family and tested the sedimentation behavior under control and hs conditions (Fig. 4). The results enlarge our perspectives of the influence of protein interactions on the stability and/or oligomerization state of the proteins involved. The major difference between the control (Fig. 4A) and hs samples (Fig. 4B) was the general tendency for structural binding of HsfA2 and Hsp17 classes CI and CII in the hs samples, irrespective of the mixture of proteins expressed. Selectivity was only observed in samples incubated under control temperature conditions, which are reflecting more likely the situation of tomato cells during recovery from a heat stress. We therefore concentrate our further discussion mainly on the results obtained with the control temperature samples (Fig. 4A).
Figure 4.
Coexpression of Hsp17 class CI prevents the formation of sedimentable HsfA2 complexes under control but not under heat stress conditions. Immunoblot analysis of HsfA2 expression and distribution between soluble (S100) and insoluble (P100) protein fractions after coexpression with the indicated forms of Hsp17-CI and/or Hsp17-CII in control (A) and heat stressed (B) protoplasts. To minimize possible interferences from the endogenous cellular heat stress response of the tobacco protoplasts, the inactive, 3HA-tagged version of HsfA2 was used. In some cases the plasmid mixture was complemented by addition of pRTLpHsfA1ΔC394 (samples 5–8 and 10 in part A, B). For heat stress treatment (B) protoplasts were incubated for 2 h at 40°C before harvesting. The analyzed proteins are indicated on the left margin by arrows and the corresponding antibodies. The open arrow head at the lower section in part B indicates the heat stress-induced expression of endogenous, tobacco Hsp17 class CI proteins. Variations in the steady state levels of HsfA2 expression were estimated on the basis of immunoblot signals in the WPE fractions and the values given at the bottom of the corresponding immunoblots are normalized to the expression levels in samples number 2 (=1.0) in A and B, respectively. For evaluation of the amounts of insoluble HsfA2 in the P100 fractions see legend to Figure 3.
Similar to previous experiments, the sedimentation of a considerable part of HsfA2 in high salt-resistant form was only observed in the presence of Hsp17.4-CII (lane 1 and 5) but not of Hsp17.3-CII (lane 2 and 6). Interestingly, nuclear retention of HsfA2 in the presence of HsfA1 (Scharf et al., 1998a; Heerklotz et al., 2001) does not change the amount of HsfA2 detected in the sedimentable form (Fig. 4A, lane 5). However, in samples including Hsp17.4-CII and Hsp17.7-CI (lanes 3 and 7), the formation of sedimentable HsfA2 was drastically reduced or abolished. This solubilizing effect of Hsp17.7-CI was also observed with the Myc-tagged as well as untagged form of Hsp17.7-CI or with other representatives of tomato class CI small Hsps, e.g. LeHsp17.8-CI and LeHsp17.6-CI (Fig. 5, A and B) as well as with corresponding proteins from Arabidopsis and pea (AtHsp18.1-CI, AtHsp17.6-CI, PsHsp18.1-CI, data not shown). Evidently, in contrast to the exclusivity of the interaction between HsfA2 and Hsp17.4-CII, the solubilizing effect of class CI sHsps is more general and, probably, essential for maintaining sHsps in a soluble state at normal growth temperatures.
Figure 5.
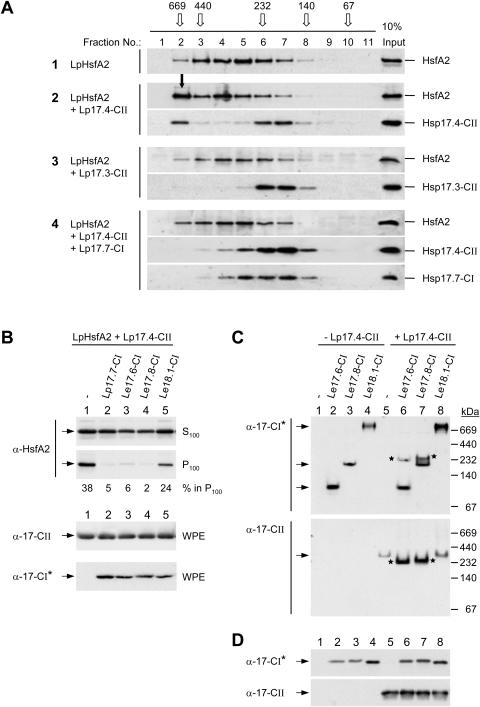
Influence of class CI sHsps on the formation of HsfA2 and sHsp class CII complexes. A, HsfA2 was expressed either alone (sample 1) or in the indicated combinations with sHsps (samples 2–4) in tobacco protoplasts and separated by size exclusion chromatography (SEC) of whole protoplast extracts. The elution profiles were analyzed by immunoblotting as described in “Materials and Methods”. For comparison, immunoblot signals corresponding to one-tenth of the SEC loading samples are shown in the right lane. The elution profile and size of molecular mass standards are given on top (open arrows). The full arrow (sample 2, fraction no. 2) points to the signal corresponding to the sedimentable HsfA2 formed in presence of Hsp17.4-CII. B, The indicated isoforms of tomato class CI sHsps (samples 2–5) were coexpressed with LpHsfA2 and LpHsp17.4-CII and the formation of sedimentable HsfA2 was analyzed as described in Figures 3 and 4. C, Separation of native oligomeric sHsp complexes formed by the indicated isoforms of tomato Hsp17-CI in absence (−) or presence (+) of LpHsp17.4-CII in polyacrylamide pore exclusion gels. Arrows point to specific positions of homooligomeric complexes of the individual Hsp17-CI isoformes (first section) or of Hsp17.4-CII (second section). Asterisks indicate the positions of comigrating Hsp17-CI and -CII complexes in the coexpression samples (6 and 7). Separation and sizes of molecular mass standards are indicated at the right margin. D, For expression control aliquots of the samples indicated in part C were separated by SDS-PAGE and processed for immunoblot analysis as described in “Materials and Methods”. Because of an unequal detection of the Hsp17-CI isoforms by the peptide-specific antibody, the class CI-specific polyclonal antiserum generated in guinea-pigs (α-Hsp17-CI*, see “Materials and Methods”) was used for the corresponding immunodetections in parts B, C, and D.
Two points are important to notice. First, in contrast to the situation under heat stress conditions where a considerable portion of both Hsp17-CI and Hsp17-CII together with HsfA2 are found in the high salt and detergent-resistant sedimentable fraction (Fig. 4B), this is not the case for samples maintained under control temperature conditions. Due to the specific buffer conditions used in the cosedimentation assays, the HsfA2 aggregates (Fig. 4A, lanes 1–5) contain only very small amounts of Hsp17.4-CII. This indicates that major structural differences might exist between HsfA2/Hsp17.4-CII complexes formed under control and hs conditions, respectively. Second, for these experiments we used the 3-HA-tagged form of HsfA2 and a C-terminally truncated form of HsfA1. Both have very low or no transcriptional activity. This special test situation avoids the synthesis of endogenous Hsp17-CI that evidently would influence the outcome of the experiment.
The results on the decisive role of class CI chaperones for the solubilization of aggregated HsfA2 and Hsp17 class CII complexes were confirmed by size exclusion chromatography of whole protein extracts from tobacco protoplasts expressing the indicated proteins or mixtures of proteins (Fig. 5). Expression of HsfA2 alone (sample 1) gave a broad elution peak with a maximum in fraction number 5 which corresponds to an apparent molecular size of about 350 kD for soluble oligomeric HsfA2 complexes. Coexpression with Hsp17.4-CII (sample 2) but not with Hsp17.3-CII (sample 3) shifted a considerable part of HsfA2 to fraction number 2, i.e. close to the exclusion volume. This fraction evidently represents the sedimentable aggregates of HsfA2 (see Fig. 5A) and contains also a significant portion of Hsp17.4-CII. As estimated on the basis of densitometer scans of the corresponding signals of protein imunoblots shown in Fig. 5A, the portions of HsfA2 and Hsp17.4-CII shifted to the high-molecular size fraction (Fig. 5A, sample 2, fraction no.2) correspond to approximately 27% and 22%, respectively, based on the total amounts of these proteins eluted from the size exclusion column. In contrast to the high salt and detergent conditions used for the sedimentation assay (Figs. 1–4), the lower stringency conditions used for the size exclusion chromatography were not sufficient to dissociate the salt/detergent-sensitive association of HsfA2 and Hsp17.4-CII. As expected from the solubilizing effects of class CI Hsps, these high-Mr aggregates of HsfA2 and Hsp17.4-CII were not observed upon coexpression with Hsp17.7-CI (sample 4).
When we tested three different isoforms of Hsp17-CI from the cultivated tomato L. esculentum for their capacity to solubilize aggregated HsfA2 complexes (Fig. 5B), we were surprised to find that only two of them, LeHsp17.6-CI and LeHsp17.8-CI, were comparable to LpHsp17.7-CI. In contrast to this, LeHsp18.1-CI was much less efficient in solubilizing the HsfA2 aggregates formed in presence of Hsp17.4-CII (Fig. 5B, lane 5).
The peculiarities of the three isoforms of tomato Hsp17-CI in this respect seem to be correlated with their capacity for interaction with Hsp17.4-CII, as detected by native gel electrophoresis (Fig. 5C). Hsp17.6-CI expressed alone formed small oligomers in the range of 100 kD, Hsp17.8-CI formed dodecamers of about 220 kD, whereas Hsp18.1-CI gave a complex of >700 kD. Upon coexpression with Hsp17.4-CII part of the former two class CI proteins were found in new complexes of about 235 kD containing all of the Hsp17.4-CII (bands marked with asterisks in lanes 6 and 7). However, coexpression of Hsp18.1-CI and Hsp17.4-CII gave no comparable shift of the Hsp17.4-CII signal.
It is worth repeating once more that all effects observed in tobacco protoplasts mimic at least parts of the natural situation in tomato cells in the recovery period. As reported earlier (Kirschner et al., 2000), interactions between class CI and class CII proteins were not detectable in the yeast two-hybrid system. They are evidently restricted to the oligomeric state of sHsps.
An interesting common aspect of the protein interactions documented in Figure 4 is the stabilization of HsfA2, both in the presence of HsfA1 and Hsp17.4-CII. The relative numbers given on top for the signal density of HsfA2 in whole protoplast extracts (WPE) indicate a 2- to 3-fold increased level of HsfA2 in all combinations with HsfA1 and/or Hsp17.4-CII (lanes 1, 5–8, and 10) but not in combinations with Hsp17.3-CII or Hsp17.7-CI (lanes 2, 4, and 9). The same was true for the samples used for the size exclusion chromatography (compare Fig. 5A, lanes for the input controls for samples 3 versus 2 and 4). Stabilization of HsfA2 in the insoluble cytoplasmic aggregates seems plausible, but this cannot explain the stabilization by interaction with HsfA1.
Intracellular Distribution and Aggregation State of HsfA2 as Visualized by Immunofluorescence
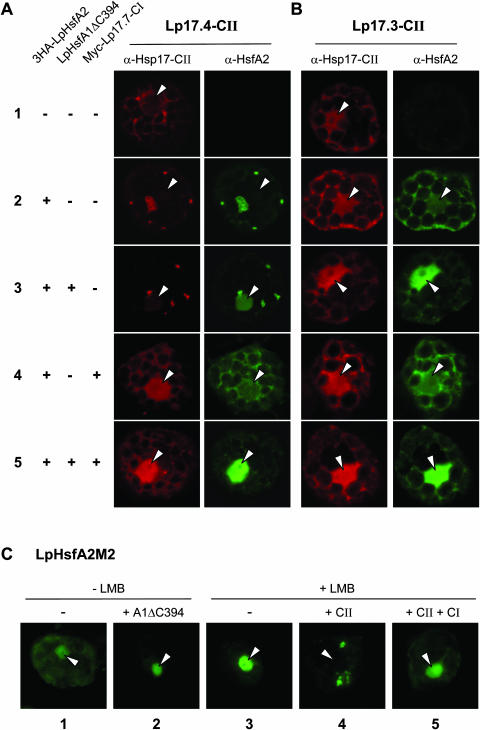
The different types of protein interactions characterized in Figures 3 to 5 may also have marked influences on the intracellular localization of HsfA2. To investigate this in detail, we used tobacco protoplasts expressing the indicated proteins under control temperature conditions (Fig. 6). In cells with Hsp17.4-CII alone (Fig. 6A, sample 1), the small Hsp was found mainly distributed throughout the cytoplasm, but upon coexpression with HsfA2 (sample 2) large cytoplasmic aggregates of both proteins were observed. In the triple combination of Hsp17.4-CII and HsfA2 with HsfA1 (sample 3), the situation was very similar. However, as a result of the interaction with HsfA1, part of the HsfA2 was now found in the nucleus. The observed solubilizing effect of Hsp17.7-CI was also visible at the cellular level (samples 4 and 5 of Fig. 6A). In both cases, the cytoplasmic aggregates of HsfA2 completely disappeared, and the nuclear portion of HsfA2 in the presence of HsfA1 clearly increased (compare samples 3 and 5). Interestingly, a similar increase in the nuclear localization was also visible for Hsp17.4-CII in the corresponding immunofluorescence images. Neither HsfA2 nor sHsp aggregates were observed in the presence of Hsp17.3-CII (Fig. 6B), and changes in the nucleocytoplasmic localization of HsfA2 in these samples were only influenced by coexpression of HsfA1.
Figure 6.
Immunofluorescence localization of HsfA2 and of the two forms of tomato Hsp17-CII in tobacco protoplasts. LpHsp17.4-CII (A) and LpHsp17.3-CII (B) were expressed alone (sample 1) or in presence of 3HA-tagged HsfA2 (samples 2–5). As indicated at the left margin, some samples contained HsfA1ΔC394 and/or Myc-tagged LpHsp17.7-CI. Hsp17-CII was detected in the red, HsfA2 in the green channel. Arrows mark positions of nuclei. For further explanations see text. C, The inactive DNA binding mutant of HsfA2 (HsfA2M2) was expressed either alone (sample 1 and 3) or coexpressed with HsfA1ΔC394 (sample 2), with LpHsp17.4-CII (sample 4), or with LpHsp17.4-CII and LpHsp17.7-CI (sample 5). One hour before harvesting, 20 ng mL−1 LMB were added to samples 3 to 5.
One obvious difference between the results in Figure 6A and those shown before in Figure 4A needs further explanations. Although the predominant portion of Hsp17.4-CII was clearly associated with the HsfA2 aggregates in situ (Fig. 6A, samples 2 and 3), this association is evidently not high salt/detergent-resistant and hence, it was not preserved under the high stringency conditions used for the sedimentation procedure (Fig. 4A, samples 1 and 5).
The functional significance of the different states of HsfA2 is particularly striking from the results shown in Figure 6C. Our earlier observations indicated that HsfA2 has nucleocytoplasmic localization but because of its strong NES the steady-state localization is more cytoplasmic (Heerklotz et al., 2001). Following this, we used leptomycin B (LMB) to block the nuclear export and to detect possible changes in the shuttling behavior of HsfA2 as a result of protein interactions with the sHsps (Fig. 6C). As expected, the more cytoplasmic localization (sample 1) was shifted to a strong nuclear localization of HsfA2 in presence of LMB (sample 3). Remarkably, this effect was blocked if HsfA2 was entrapped in cytoplasmic aggregates with Hsp17.4-CII (sample 4), and as seen before, this block was released in the presence of Hsp17.7-CI (sample 5).
Reporter Assays: Function of Hsp17.4-CII as Putative Corepressor of HsfA2
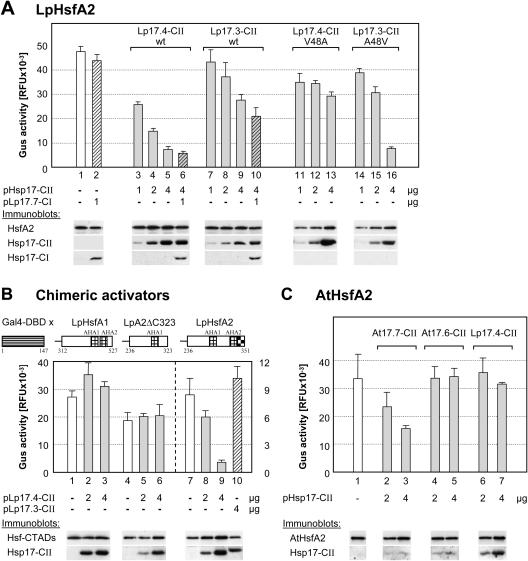
Is the specific interaction between HsfA2 and Hsp17.4-CII limited to the cytoplasmic part of the transcription factor and, if not, does binding of the small Hsp also influence the activator potential of HsfA2? To answer these questions, we used reporter assays in tobacco protoplasts and coexpressed HsfA2 in presence of the C-terminally deleted (inactive) form of HsfA1 to provide the basis for nuclear retention of the former (see Fig. 6C, sample 2). The high activator potential of HsfA2 (Fig. 7A, sample 1) was markedly reduced in the presence of increasing amounts of Hsp17.4-CII (samples 3–5). Interestingly, the repressor effect of Hsp17.4-CII on HsfA2 were not altered in the presence of Hsp17.7-CI (sample 6), i.e. they reflect the interaction of HsfA2 and Hsp17.4-CII in the nucleus, and they are largely independent of the aggregation state of cytoplasmic HsfA2 and Hsp17.4-CII (see immunofluorescence data in Fig. 6). As expected, the noninteracting type (Hsp17.3-CII) had only weak effects on the reporter activity (samples 7–10). Samples 11 to 16 (Fig. 7A) complement the picture by showing the results obtained with the two mutant sHsps (Hsp17.4-CII V48A and Hsp17.3-CII A48V) tested earlier for their influence on the interaction and aggregation of HsfA2 (Figs. 2 and 3). In support of the earlier results on the decisive role of the V48 residue for the interaction with HsfA2, the former mutant sHsp lost its capability to interfere with HsfA2 function (samples 11–13), whereas the latter gained this capability (samples 14–16). In all cases, the signals from immunoblot analyses at the bottom serve as expression controls for the interacting proteins.
Figure 7.
Influence of sHsps on the activator function of HsfA2. A, Hsf-dependent reporter assay with phsp17gus as reporter. To achieve optimum activity, 50,000 tobacco protoplasts were transformed with 2 μg of HsfA2 expression plasmid plus 0.2 μg of HsfA1ΔC394 expression plasmid (sample 1) and for samples 2 to 16 the indicated amounts of sHsp encoding plasmids were added to the transformation mix. Samples 11 to 13 contain the LpHsp17.4-CII V48A mutant, whereas samples 14 to 16 contain the LpHsp17.3-CII A48V mutant (for sequence details, see Fig. 1). Expression levels of the proteins were controlled by immunoblot analysis as indicated at the bottom. The GUS activity was determined in cell lysates harvested 11 h after transformation (see “Materials and Methods”). B, The pGal4DBDxgus reporter was used to monitor the activator potential of Gal4DBD fusion proteins with the indicated activation domains of tomato HsfA1, HsfA2ΔC323, and HsfA2. Note the differences in the activity scales for the first two activators (left) as compared to the Gal4DBDxHsfA2 activator (right). C, Hsf-dependent reporter assay of Arabidopsis AtHsfA2 in tobacco protoplasts. One microgram of AtHsfA2 expression plasmid was transformed alone (no. 1) or in mixtures with the indicated amounts of plasmids encoding the two isoforms of class CII sHsps from Arabidopsis (nos. 2–5) and the HsfA2-interacting Hsp17.4-CII of tomato (nos. 6, 7). The AtHsfA2 expression plasmid and the immunserum for detection of the protein were kindly provided by P. von Koskull-Döring (unpublished data).
To confirm the role of the C-terminal part with the transcriptional activator modules of HsfA2 for the specific interaction with Hsp17.4-CII, we used a Gal4p-dependent β-glucuronidase (GUS) reporter construct (Döring et al., 2000) and expression constructs encoding chimeric activator proteins consisting of the yeast Gal4p DNA binding domain and the C-terminal activator domains of tomato HsfA1 and HsfA2 (for details, see block diagrams at the top of Fig. 7B). The activity of the two chimeric activators with the activator domain of HsfA1 or the C-terminally truncated form of HsfA2 were not affected by the presence of Hsp17.4-CII (samples 1–6), whereas the activator with the complete C-terminal domain of HsfA2 (amino acids 263–351) was strongly repressed in its activity by increasing amounts of Hsp17.4-CII (samples 7–9) but not by Hsp17.3-CII (sample 10). The results extend our earlier conclusions. Interaction between HsfA2 and Hsp17.4-CII is not restricted to the aggregated state but is also observed with the soluble nuclear HsfA2. Hsp17.4-CII acts as corepressor of HsfA2 irrespective of the presence or absence of Hsp17.7-CI, and for this effect only the C-terminal activator domain of HsfA2 is required and sufficient.
To prove whether the specific repressor function of Hsp17.4-CII on the activity of HsfA2 is not only a peculiarity of the tomato system but rather presents a more general regulatory principle, we tested the activities of the orthologous proteins from Arabidopsis in the Hsf-dependent reporter assay. The results in Figure 7C confirm that the activity of AtHsfA2 was markedly repressed by coexpression with increasing amounts of AtHsp17.7-CII (Fig. 7C, lanes 1–3), but not in presence of AtHsp17.6-CII (lanes 4, 5). As observed for the tomato proteins, the functional interaction of AtHsfA2 with one of the two closely related members of class CII Hsps in Arabidopsis seems to be species-specific. Coexpression of AtHsfA2 with tomato Hsp17.4-CII had no effect on the level of GUS activity (lanes 6 and 7).
DISCUSSION
Although our knowledge is still rather fragmentary, there is experimental evidence that the maintenance of the inactive state of Hsfs requires additional proteins functioning as corepressors. On the one hand, a small Hsf-binding protein (HSBP1) in mammals and nematodes was reported to mask the HR-A/B region by coiled-coil interactions, and thus to stabilize the inactive, monomeric form of Hsfs (Satyal et al., 1998; Tai et al., 2002). Recently, a HSBP ortholog was identified in maize. The lack of HSBP in the mutant empty pericarp 2 (emp2) leads to an uncontrolled expression of hs genes and embryo lethality (Fu et al., 2002). On the other hand, Hsp70 and Hsp90 are evidently involved in the control of Hsf activity in yeast and vertebrates (Baler et al., 1996; Nair et al., 1996; Ali et al., 1998; Shi et al., 1998; Bharadwaj et al., 1999; Bonner et al., 2000; Guo et al., 2001), and the function of Hsp70 as coregulator of Hsf activity may be similar in plants as well (Lee and Schöffl, 1996; Kim and Schöffl, 2002). To our knowledge, a role of a sHsp for control of Hsf activity was never shown before.
The tomato Hsf system is characterized by an intriguing cooperation between two Hsfs, i.e. HsfA1 and HsfA2. As master regulator of the heat stress response, HsfA1 is responsible for the hs-induced synthesis of HsfA2 (Mishra et al., 2002). With ongoing heat stress, the latter accumulates to fairly high levels and becomes the dominant Hsf of tomato cells. Its strong activator potential and abundance markedly enhances the efficiency of hs gene expression, especially in combination with HsfA1. Earlier, we distinguished three major states of HsfA2 in tomato cell cultures (Scharf et al., 1998a; Heerklotz et al., 2001): (1) Nuclear HsfA2: Despite its strong nuclear export signal, HsfA2 accumulates in the nucleus in presence of HsfA1, probably in form of HsfA2/A1 heterooligomeric complexes. This corresponds to the behavior of HsfA2 if coexpressed together with HsfA1 in native tissues or in tobacco protoplasts (Scharf et al., 1998a). (2) HsfA2 in HSG complexes: With ongoing synthesis during hs, a considerable portion of HsfA2 can be detected in large cytoplasmic aggregates (heat stress granules) together with Hsp17-CI, Hsp17-CII, and probably denatured proteins (Scharf et al., 1998a; Mishra et al., 2002). (3) Cytoplasmic, soluble HsfA2: The hs-specific, high salt-resistant structural binding of HsfA2 in the HSG fraction is reversible. In the recovery period, most of the HsfA2 is found in soluble form in the cytoplasm.
Results in this manuscript demonstrate that the three states of HsfA2 not only depend on its interaction with HsfA1 mediated by the similarity of their oligomerization domains, but also with Hsp17.4-CII binding to the C-terminal domain of HsfA2. The latter interaction has two effects. On the one hand, Hsp17.4-CII acts as corepressor of HsfA2. On the other hand, it promotes formation of high Mr aggregates of HsfA2 and Hsp17.4-CII even under control temperature conditions (Figs. 3, 4, and 6). This aggregation represents an experimental artifact valuable to investigate details of the protein network responsible for the intracellular localization and function of HsfA2. In the reality of tomato cells in the recovery period following a short hs treatment, no aggregates of HsfA2 are observed because two proteins counteract the structural binding of HsfA2 and Hsp17.4-CII. First, interaction with HsfA1 not only prevents aggregation of HsfA2, but it also promotes nuclear accumulation and keeps HsfA2 in a transcriptionally competent state (Scharf et al., 1998a). Second, Hsp17-CI interacts with Hsp17.4-CII and inhibits the aggregation together with HsfA2 (Fig. 4A).
This solubilizing effect of Hsp17-CI under control temperature conditions is a class-specific but organism-independent character. It reflects the interaction between Hsp17.4-CII and Hsp17-CI in their oligomeric states as found upon coexpression in plant cells (Siddique et al., 2003). It can be visualized by shifts of the relative positions of oligomeric Hsp complexes in a native gel (Fig. 5C) and by disintegration of the HsfA2/Hsp17.4-CII aggregates (Figs. 4A and 6). The results in native plant cells are in contrast to the lacking interaction of class CI and CII proteins in the yeast two-hybrid system, because the dimer interfaces of both sHsps are class-specific and not compatible (Kirschner et al., 2000).
In contrast to the broad specificity of the class CI proteins, the interaction between HsfA2 and Hsp17.4-CII is highly selective. It was not observed with any other class CII protein tested so far. This is even true for the closely related tomato Hsp17.3-CII. A single amino acid exchange (V48>A) makes the crucial difference between the two proteins. Remarkably, this interaction can be detected in all test systems, e.g. in yeast two-hybrid tests (Fig. 2), in glutathione S-transferase (GST)-pull-down assays (data not shown) as well as in situ in protoplasts (Fig. 6) and in reporter assays (Fig. 7). Evidently, it is independent of the oligomeric state of the two proteins.
Analysis of the Arabidopsis orthologs, i.e. of HsfA2 and the two class CII sHsps, indicates that a comparable functional relationship might exist in this plant as well. Similar to tomato, the Arabidopsis HsfA2 is also strictly hs-inducible and, together with the small Hsps and other chaperones, it accumulates to high levels in cell cultures and leaves (Kotak et al., 2004). In the protoplast reporter assay, the activity of AtHsfA2 was clearly repressed upon coexpression with AtHsp17.7-CII, but not with the closely related AtHsp17.6-CII. However, unlike the situation with the tomato proteins, we could not detect this interaction in any of our other assays. One possible explanation for this failure could be that, in contrast with the reporter assays, the outcome of the other interaction tests is strongly influenced by the intrinsic properties of the tested partner proteins, i.e. only tomato HsfA2 has the intrinsic tendency for aggregation, which is lacking for Arabidopsis HsfA2. This is also supported by results from the analysis of modified forms of the tomato sHsps, e.g. the A48>V mutant form of LpHsp17.3-CII. It acts as corepressor of LpHsfA2 and promotes the formation of the sedimentable form, but it does not interact in the yeast two-hybrid assay (Figs. 7A and 2). Further, the results obtained with the chimeric constructs encoding the C-terminal activator domain of tomato HsfA2 fused to the DNA binding domain of HsfA1 showed no detectable formation of sedimentable forms in presence of LpHsp17.4-CII (Fig. 3C), although the activator potential was repressed (data not shown). Evidently, formation of high Mr aggregates is only observed with the full-length HsfA2, and the interaction of its C-terminal domain with Hsp17.4-CII is only one aspect of this phenomenon. Clearly, these interesting aspects need further studies including HsfA2 of other plants in combination with their potential sHsp corepressors.
Our results show for the first time important elements of a protein network controlling the intracellular distribution, stability, and activator function of tomato HsfA2. Previously, we showed that HsfA1 as master regulator of the hs response in tomato is responsible for hs-induced new synthesis of HsfA2 (Mishra et al., 2002) and for its nuclear retention (Scharf et al., 1998a; Heerklotz et al., 2001). The new elements added to the protein network are the nucleocytoplasmic sHsps of classes CI and CII accumulating together with HsfA2 as a result of the hs-induced gene expression. Both types of sHsps play completely different roles in the HsfA2 network. Hsp17.4-CII functions as a putative repressor of HsfA2 activity and most probably represents the specific anchor protein for the incorporation of HsfA2 into HSG complexes formed only under hs conditions. In contrast to this, class CI sHsps are essential for resolubilization of the HSG complexes in the recovery period and/or for keeping HsfA2/Hsp17.4-CII complexes in soluble state at control temperatures. This leads to the interesting hypothesis that in thermotolerant tomato cells HsfA2 is stabilized in a transcriptional inactive form by interaction with soluble Hsp17.4-CII in presence of Hsp17-CI and is kept competent for a rapid activation under repeating hs conditions. In addition to our results reported in this paper, two earlier observations support this hypothesis: (1) In preinduced tomato culture cells a transient increase in the nuclear localization of HsfA2 was found at the beginning of a second hs treatment (Scharf et al., 1998b), and (2) the detection of Hsp17-CI in nuclei purified from heat stressed tomato cells was reported by Wollgiehn et al. (1994).
Although we have probably identified the four major components of the HsfA2 network, we cannot exclude that in the reality of an hs-induced cell, additional proteins might be important for the control of the activity state and stability of HsfA2 as well. Our analysis was mainly based on the coexpression of the partner proteins in tobacco protoplasts. Although probably very similar, this cannot completely simulate the situation in native tomato cells. Further experiments are required, e.g. an RNAi approach in tomato protoplasts that would allow selectively knocking-down expression of these and other components with putative functional interactions to the HsfA2 network.
MATERIALS AND METHODS
General Materials and Methods
The use of tobacco (Nicotiana plumbaginifolia) mesophyll protoplasts for polyethylene glycol-mediated transformation and transient gene expression was published (Treuter et al., 1993; Scharf et al., 1998a; Döring et al., 2000) and polyclonal antisera from rabbit against tomato (Lycopersicon esculentum) HsfA2, Hsp17-CI and Hsp17-CII were described before (Lyck et al., 1997; Scharf et al., 1998a; Kirschner et al., 2000). Peptide-specific antisera against tomato Hsp17 class CI and class CII were generated in rabbits by immunization with synthetic, class-specific peptides (Eurogentec, Seraing, Belgium) corresponding to nonhomologous amino acid sequence motifs in the N-terminal and C-terminal parts of LpHsp17.7-CI and LpHsp17.4-CII, respectively. If not indicated otherwise, the peptide-specific antisera were used for immunoblot detections of class CI and class CII sHsps (see below). For the requirements of double-immunostaining class-specific antibodies were raised in guinea-pigs (Eurogentec) by immunization with purified GST fusion proteins of tomato Hsp17.7-CI and Hsp17.4-CII expresssed in Escherichia coli BL21DE3. Monoclonal antibodies, clone 16B12 and 9E10 (Hiss Diagnostics, Freiburg, Germany), were used for immunodetection of 3HA- and Myc-tagged proteins, respectively. Secondary antibodies conjugated with horseradish peroxidase or fluorescent dyes CY2 or CY3 were obtained from Sigma-Aldrich (Deisenhofen, Germany) and Dianova (Hamburg, Germany), respectively.
LMB was kindly provided by Minoru Yoshida, Tokyo, and used as specific repressor of the nuclear export receptor exportin 1 as described before (Kudo et al., 1999; Heerklotz et al., 2001).
For indirect immunofluorescence of protoplasts we followed the procedures described by Scharf et al. (1998a) and Heerklotz et al. (2001). Fixation of protoplasts was done with 3.7% (w/v) paraformaldehyde in microtubule stabilizing buffer (100 mm PIPES buffer, pH 6.8, 2 mm EGTA, 1 mm MgSO4).
For microscopic analysis a Zeiss Axiophot microscope (Zeiss, Oberkochen, Germany) combined with a Color View F12 System (Olympus, Hamburg, Germany) was used. Captured images were resized and combined using Photoshop 5.5 Software (Adobe Systems, La Jolla, CA). Confocal laser scan micrographs were captured using a Leica CLSM (Leica, Bensheim, Germany) and Imaris Software (Bitplane, Zurich).
Standard procedures were used for cloning (Ausubel et al., 1993; Sambrook and Russell, 2001). Plant expression vectors are derivatives of pRT101 (Töpfer et al., 1988) and modified versions for expression of triple HA- or Myc-tagged proteins were described before (Kirschner et al., 2000; Siddique et al., 2003). The expression vector encoding HsfA2M4, a mutant form of HsfA2 with disrupted NES, was described (Heerklotz et al., 2001). For deletions or combinations of functional parts of HsfA1 and HsfA2, we used unique SalI sites in different regions of the cDNA introduced by site-directed mutagenesis (Treuter et al., 1993; Boscheinen et al., 1997; Döring et al., 2000). The principle modular structures of the chimeric proteins of Hsfs A1 and A2 as well as fusions of their C-terminal domains (CTD) to the DNA-binding domain (DBD) of the yeast transcription factor Gal4 are given in the block diagrams in Figure 3C and Figure 7B, respectively.
To minimize possible interferences of the endogenous set of sHsps in tobacco protoplasts, two inactive HsfA2 variants were used in those experiments where the intracellular distribution or the aggregation behavior of HsfA2 and coexpressed sHsps was analyzed. The corresponding expression plasmids are pRTHsfA2M2 encoding a mutant form of HsfA2 impaired in DNA-binding (Lyck et al., 1997) and pRT3HA-HsfA2 encoding HsfA2 with a triple HA-tag preceeding the N-terminal DBD.
For Hsf-dependent GUS reporter assays, we used the phsp17gus vector containing the promoter region of the soybean hsp17.3-B gene upstream of the TATA box (Treuter et al., 1993) and the Gal4DBDgus plasmid as reporter for monitoring the activator potential of Gal4DBD-HsfCTD fusion constructs (Döring et al., 2000).
Two-Hybrid Protein Interaction Assay
The yeast two-hybrid screening of a tomato cDNA library derived from heat stressed Lycopersicon peruvianum cell cultures and protein interaction tests were done with the pBDGal4 bait and pADGal4 prey vector system (Stratagene, Amsterdam) as described before (Scharf et al., 1998a; Bharti et al., 2000; Siddique et al., 2003). As bait vector, pBDGal4-HsfA2.11.16B encoding the Gal4 DNA binding domain fused to the C-terminal part of tomato HsfA2 (amino acids 98–351) with the two activator motifs inactivated by mutation (W297>A, W337>E, and L341>A) was used (Döring et al., 2000).
Hsp Expression Vectors for Yeast and Plant Cells
The prey vectors pADGal4xLpHsp17.4-CII and pADGal4xLpHsp17.3-CII encoding fusion proteins of the Gal4 activator domain with the two formes of cytosolic Hsp17 class CII proteins known in tomato. The exchange of individual amino acid residues that are distinct in both sequences (see also Fig. 1) was done by PCR using the Taq Plus Precision system (Stratagene) and forward and reverse primer complementary to plasmid sequences flanking the Hsp17 cDNA inserts in combinations with the following mutagenesis primers:
- Hsp17.4-CII:
- Pr0839R (K35I) 5′-GCATCACGAACATAGATCTTTGATGGTGC-3′
- Pr1089R (V48A) 5′-GGATACTCTTTCACGTCAGCTGGTGTCGCAGCC-3′
- Pr0841R (D55N) 5′-CGAAAACATATGAATTCGGATACTCTTTCACG-3′
- Pr1407F (V96A) 5′-GAGAAAGAAGGTGCAAAGTTTATCCGGATGGAG-3′
- Hsp17.3-CII:
- Pr1071R (A48V) 5′-GCATCACGAACGTACTTCTTTGATGGTGC-3′
Amplified DNA fragments were cut at the appropriate restriction sites and ligated into EcoRI-XbaI linearized pADGal4. The mutant Hsp17.4-CII Q151E was generated by subcloning a PagI-XbaI cDNA fragment derived from pADGal4xLpHsp17.3-CII into PagI-XbaI linearized pADGal4xLpHsp17.4-CII. The corresponding plant expression constructs were generated by ligation of the mutagenized cDNA fragments into pRT104 linearized with EcoRI and XbaI.
Prey constructs and plant expression vectors encoding class CI sHsps of tomato (LpHsp17.7-CI) or pea (PsHsp18.1-CI) and class CII sHsps of pea (Pisum sativum; PsHsp17.1-CII) or Arabidopsis (AtHsp17.6-CII) were described before (Kirschner et al., 2000; Siddique et al., 2003).
Genomic DNA was used as template for PCR amplification of DNA fragments encoding class CI and class CII sHsps from the tomato cv L. esculentum var Moneymaker (Mishra et al., 2002). Forward and reverse primer corresponding to gene-specific sequences in the 5′- and 3′-UTR were synthesized on the basis of sequence informations available from The Institute for Genomic Research tomato EST database (http://www.tigr.org/tdb/tgi/lgi) for LeHsp17.6-CI (TC98617), LeHsp17.8-CI (TC98618), and LeHsp18.1-CI (TC101691) or from the National Center for Biotechnology Information sequence database (http://www.ncbi.nlm.nih.gov) for LeHsp17.4-CII (AF090115) and LeHsp17.6-CII (U72396). Flanking restriction sites were introduced for subcloning the amplified DNA fragments into XhoI-XbaI linearized pRT101. Following the same procedure, the plant expression vector encoding AtHsp17.7-CII (At5g12030) was generated.
Protein Extraction, Polyacrylamide Gel Electrophoresis, and Immunoblot Analysis
For protein extraction harvested protoplasts were lysed by three cycles of freeze-thawing in NEB500 buffer containing 25 mm HEPES, pH 7.5, 500 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 10 mm NaF, 0.2% (w/v) NP40, and 10% (w/v) glycerol. For all protein extraction buffers used in this study, Complete protease inhibitor cocktail tablets were added as recommended by the manufacturer (Roche Diagnostics, Mannheim, Germany). Cellular debris was removed by centrifugation for 5 min at 10,000 rpm at 4°C. Aliquots corresponding to approximately 15,000 protoplasts were heated with 1 vol. of 2× SDS sample buffer and separated on 14% SDS-polyacrylamide gels (SDS-PAGE).
Protein extracts for separation on native 3% to 20% polyacrylamide pore exclusion gels were prepared in nondenaturing buffer as described before (Kirschner et al., 2000).
For immunoblot analysis, proteins were transferred to 45-μm nitrocellulose membranes (PROTRAN BA85, Schleicher and Schuell, Dassel, Germany) and processed for chemiluminescence detection following the manufacturer's protocol (Perkin Elmer Life Sciences, Rodgau-Jügesheim, Germany).
Ultracentrifugation
For separation of soluble and insoluble protein fractions, protoplasts were extracted with high salt and detergent buffer (50 mm Tris-HCl, pH 7.8, 500 mm NaCl, 25 mm KCl, 5 mm MgCl2, 30 mm EDTA, 0.5% [w/v] NP40, 0.2% [w/v] sarcosyl, 5% [w/v] saccharose, 5% [w/v] glycerol, 14.2 mm β-mercaptoethanol, and protease inhibitors). After removal of cellular debris as described above, insoluble proteins were sedimented by centrifugation of WPE for 60 min at 100,000g at 4°C (Sorvall Micro-Ultracentrifuge RC M120, rotor RP100-AT4, Newtown, CT). Pellets were dissolved in high salt extraction buffer equal to the volume of the supernatant. After precipitation with acetone, proteins of both fractions were further processed equally for separation on SDS-PAGE and immunoblot detection as described above.
Corresponding chemiluminescence signals from the supernatant (S100) and pellet (P100) fractions were quantified by using the Image 1D software (Amersham Biosciences, Freiburg, Germany). Relative amounts of a given protein in the insoluble P100 fraction were calculated on the basis that the signal densities of both fractions (S100 + P100) correspond to 100%.
Size Exclusion Chromatography
Protein extracts in NEB500 corresponding to approximately 500,000 protoplasts were injected on a Superdex 200 HR30/10 filtration column (Amersham Biosciences). Size exclusion chromatography (SEC) was performed at 4°C with elution buffer (25 mm HEPES, pH 7.5, 500 mm NaCl, 1 mm EDTA, 0.2% [w/v] NP40, and 1 μg mL−1 pefabloc) at a flow rate of 0.4 mL min−1. Fractions of 0.8 mL were collected and after acetone precipitation the distribution of indicated proteins was analyzed by SDS-PAGE and immunoblot detection. As molecular mass standards thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), lactate dehydrogenase (140 kD), and bovine serum albumin (67 kD) were used.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY608694.
Acknowledgments
We thank Gisela English for the excellent technical assistance and Drs. Lutz Nover, Markus Fauth, and Pascal von Koskull-Döring for their helpful and critical discussions during preparation of this manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SCHA 577/6 to K.-D.S.) and Fonds der Chemischen Industrie (to K.-D.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042820.
References
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N (1998) HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol 18: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors (1993) Current Protocols in Molecular Biology. John Wiley & Sons, Indianapolis
- Baler R, Zou J, Voellmy R (1996) Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones 1: 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj S, Ali A, Ovsenek N (1999) Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol Cell Biol 19: 8033–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Schmidt E, Lyck R, Bublak D, Scharf KD (2000) Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J 22: 355–365 [DOI] [PubMed] [Google Scholar]
- Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Carlson T, Fackenthal DL, Paddock D, Storey K, Lea K (2000) Complex regulation of the yeast heat shock transcription factor. Mol Biol Cell 11: 1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf K-D (1997) Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet 255: 322–331 [DOI] [PubMed] [Google Scholar]
- Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L (2000) Role of AHA motifs for the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12: 265–278 [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Bukau B (2002) Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol Life Sci 59: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ (2000) Chaperone substrates inside the cell. Trends Biochem Sci 25: 210–212 [DOI] [PubMed] [Google Scholar]
- Fu S, Meeley R, Scanlon MJ (2002) Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R (2001) Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem 276: 45791–45799 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Protein folding: molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Haslbeck M (2002) SHsps and their role in the chaperone network. Cell Mol Life Sci 59: 1649–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L (2001) The balance of nuclear import and export determines the intracellular distribution of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Schöffl F (2002) Interaction between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J Exp Bot 53: 371–375 [DOI] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder J, Nover L (2000) Transient expression and heat stress induced aggregation of endogenous and heterologous small heat stress proteins in tobacco protoplasts. Plant J 24: 397–412 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification of a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 96: 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Schöffl F (1996) An hsp70 antisense gene affects the expression of Hsp70/Hsc70, the regulation of Hsf, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Mol Gen Genet 252: 11–19 [DOI] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Treuter E, Scharf KD, Nover L (1997) Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202: 117–125 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nair SC, Toran EJ, Rimerman RA, Hyermstad S, Smithgall TE, Smith DF (1996) A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf 1, and the aryl hydrocarbon receptor. Cell Stress Chaperones 1: 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L (1987) Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb Technol 9: 130–144 [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D (1989) Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol 9: 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB (1982) A regulatory upstream promoter element in the Drosophila hsp70 heat-shock gene. Cell 30: 517–528 [DOI] [PubMed] [Google Scholar]
- Pelham HRB, Bienz M (1982) A synthetic heat-shock promoter element confers heat-inducibility on the Herpes simplex virus thymidine kinase gene. EMBO J 1: 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI (1998) Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev 12: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L (1998. a) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Höhfeld I, Nover L (1998. b) Heat stress response and heat stress transcription factors. J Biosci 23: 313–329 [Google Scholar]
- Scharf KD, Rose S, Zott W, Schöffl F, Nover L (1990) Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J 9: 4495–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI (1998) Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Port M, Tripp J, Weber C, Zielinski D, Calligaris R, Winkelhaus S, Scharf KD (2003) Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones 8: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai LJ, McFall SM, Huang K, Demeler B, Fox SG, Brubaker K, Radhakrishnan I, Morimoto RI (2002) Structure-function analysis of the heat shock factor-binding protein reveals a protein composed solely of a highly conserved and dynamic coiled-coil trimerization domain. J Biol Chem 277: 735–745 [DOI] [PubMed] [Google Scholar]
- Töpfer R, Schell J, Steinbiss HH (1988) Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res 16: 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Nover L, Ohme K, Scharf KD (1993) Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol Gen Genet 240: 113–125 [DOI] [PubMed] [Google Scholar]
- Walter S, Buchner J (2002) Molecular chaperones: cellular machines for protein folding. Angew Chem Int Ed Engl 41: 1098–1113 [DOI] [PubMed] [Google Scholar]
- Wollgiehn R, Neumann D, zur Nieden U, Müsch A, Scharf KD, Nover L (1994) Intracellular distribution of small heat stress proteins in cultured cells of Lycopersicon peruvianum. J Plant Physiol 144: 491–499 [Google Scholar]
- Wu C (1995) Heat stress transcription factors. Annu Rev Cell Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Young JC, Barral JM, Hartl UF (2003) More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci 28: 541–547 [DOI] [PubMed] [Google Scholar]