Abstract
The screening of the Versailles collection of Arabidopsis T-DNA transformants allowed us to identify several male gametophytic mutants, including poky pollen tube (pok). The pok mutant, which could only be isolated as a hemizygous line, exhibits very short pollen tubes, explaining the male-specific transmission defect observed in this line. We show that the POK gene is duplicated in the Arabidopsis genome and that the predicted POK protein sequence is highly conserved from lower to higher eukaryotes. The putative POK homolog in yeast (Saccharomyces cerevisiae), referred to as Vps52p/SAC2, has been shown to be located at the late Golgi and to function in a complex with other proteins, Vps53p, Vps54p, and Vps51p. This complex is involved in retrograde trafficking of vesicles between the early endosomal compartment and the trans-Golgi network. We present the expression patterns of the POK gene and its duplicate P2 in Arabidopsis, and of the putative Arabidopsis homologs of VPS53 and VPS54 of yeast. We show that a POK::GFP fusion protein localizes to Golgi in plant cells, supporting the possibility of a conserved function for Vps52p and POK proteins. These results, together with the expression pattern of the POK::GUS fusion and the lack of plants homozygous for the pok mutation, suggest a more general role for POK in polar growth beyond the pollen tube elongation process.
Pollen tube growth is a vital process during male gametophyte development, since it allows male gametes to reach the ovules and achieve fertilization (Preuss, 1995). The elongation of the pollen tube, as well as that of animal axons, plant root hairs, fungal hyphae, and moss protonema, is achieved by a polarized mode of growth, termed tip growth (Franklin-Tong, 1999; Palanivelu and Preuss, 2000; Hepler et al., 2001), which involves the tip-localized exocytosis of Golgi-derived vesicles containing cell wall precursors (Franklin-Tong, 1999). The tip of the pollen tube is devoid of organelles, but contains almost exclusively Golgi-derived vesicles (Geitmann and Emons, 2000). In the shank of the tubes, an inverse fountain pattern of cytoplasmic streaming is observed; organelles and vesicles are transported toward the apex in the cortical region and basipetally in the central cytoplasm (Pierson et al., 1990). Actin filaments and microtubules, which are organized in longitudinal arrays more or less parallel to the elongation axis, act as tracks for cytoplasmic streaming and allow delivery of vesicles to the tip (Pierson et al., 1990; Vidali and Hepler, 2001). The use of membrane structure dyes, such as FM1-43 or FM4-64 (Parton et al., 2001; Camacho and Malhó, 2003), has revealed dynamic endo/exocytosis processes at the tip of the pollen tube, but the molecular events underlying these processes are still poorly understood. The recent identification of the Golgi-associated tobacco (Nicotiana tabacum), Rab GTPase NtRab2, predominantly expressed in pollen, suggests that tip growth and vesicle trafficking could be tightly linked (Cheung et al., 2002). Moreover, a GTP-bound dominant negative form of NtRab2 blocks endoplasmic reticulum to Golgi trafficking and inhibits pollen tube growth (Cheung et al., 2002).
The functions of numerous vesicle-trafficking proteins and several different protein-sorting pathways are well described in yeast (Saccharomyces cerevisiae; for review, see Kucharczyk and Rytka, 2001). Although many homologs of these proteins have been identified in plants (e.g. Ahmed et al., 1997; Conceição et al., 1997; Bassham and Raikhel, 1998; Sanderfoot et al., 1998; Zheng et al., 1999), little is known about their precise roles. However, knowledge from yeast has accelerated progress in identifying plant vacuolar protein complexes. For example, the isolation of an Arabidopsis insertion mutant for the VACUOLELESS 1 gene (Rojo et al., 2001), which encodes a protein homolog of the yeast Vps16p, recently lead, through comparison with the yeast model, to the characterization of three components of the first vacuolar complex identified in plants (Rojo et al., 2003). Considering the differences in cellular organization and the higher number of potential secretory pathway proteins in plant versus yeast (Sanderfoot et al., 2000; Jürgens and Geldner, 2002), it seems likely that the vesicle-trafficking machinery has undergone specific modifications in plants (Bassham and Raikhel, 2000; Jürgens and Geldner, 2002).
Using insertional mutagenesis, we identified several putative male gametophytic mutants (Bonhomme et al., 1998a, 1998b; Procissi et al., 2001), which included four affecting the specific step of pollen tube development. The poky pollen tube (pok) mutant, previously named A9 or Ttd8, exhibits very reduced pollen tubes when grown in vitro (Bonhomme et al., 1998b). In this article, we show that the POKY POLLEN TUBE (POK) gene is duplicated in the Arabidopsis genome and is ubiquitously expressed in plant tissues. However, the expression pattern of a POK::GUS fusion protein suggests higher protein expression in roots and reproductive tissues. The predicted POK protein shares significant homology with the yeast Vps52p/SAC2 protein, which has been shown to be involved in the retrograde transport of Golgi-derived vesicles toward the late Golgi (Conibear and Stevens, 2000). We report the cloning of Arabidopsis genes encoding putative homologs of the Vps52p protein partners and comparison of their expression patterns with that of the POK gene. Furthermore, a POK::GFP fusion protein localizes to Golgi structures in living plant cells, supporting the hypothesis of conservation of the function of both POK and Vps52p proteins in Arabidopsis.
RESULTS
The pok Phenotype Affects Pollen Tube Tip Growth
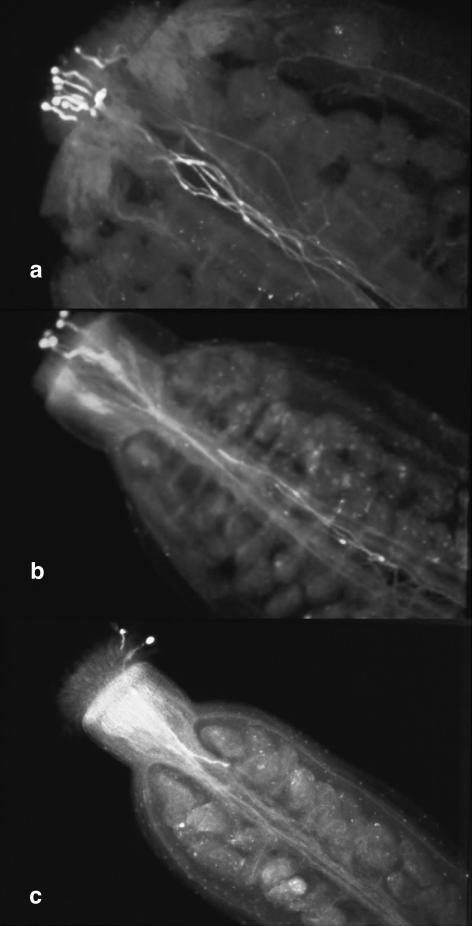
In order to define the pok phenotype in situ, aniline blue staining was performed after limited pollination of wild-type pistils with pollen grains, either from wild-type or from hemizygous pok plants. Sixteen hours after pollination, all wild-type pollen grains had germinated and completely elongated their pollen tubes, reaching the base of the ovary (Fig. 1a). The number of pollen tubes counted in the style was similar to the number of pollen grains deposited (Fig. 1a, n = 7). On the contrary, pollen from pok plants produced both elongated and short pollen tubes (Fig. 1b, five pollen grains on stigma, three tubes in the ovary). When only one or two grains were deposited, we commonly observed short, poky pollen tubes 16 h after pollination, which are presumed to carry the T-DNA construct (Fig. 1c). This observation is consistent with a role for the POK gene in pollen tube growth in vivo.
Figure 1.
The pok phenotype. In situ limited pollinations (after 16 h), followed by aniline blue staining, of wild-type pistils with pollen grains from wild-type (a) or hemizygous pok plants (b and c).
Nonetheless, some pollen tubes were still able to elongate sufficiently to reach ovules located in the upper part of the ovary (Fig. 1c). This would explain the residual male gametophytic T-DNA transmission (0.46%) in the pok mutant line (Bonhomme et al., 1998a, 1998b). Despite this residual transmission, numerous screens to isolate homozygous pok plants remained unsuccessful, suggesting that homozygous pok mutations lead to precocious lethality.
POK Is Duplicated in the Arabidopsis Genome, but the pok Phenotype Can Be Assigned to the POK Gene Alone
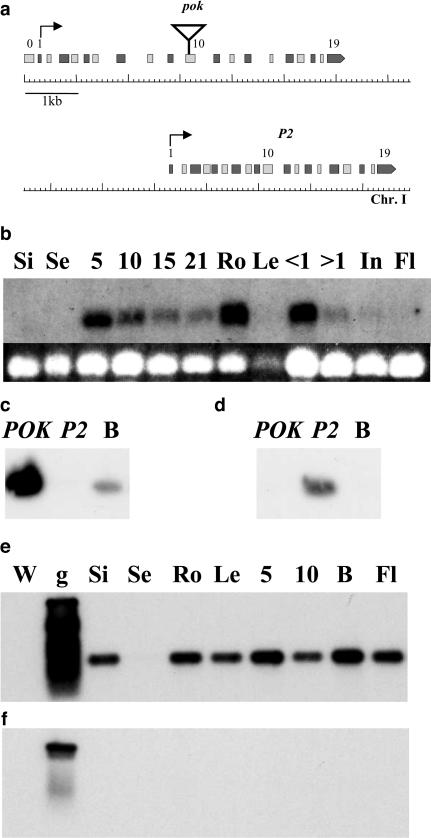
Genomic regions flanking the single T-DNA construct inserted in the pok mutant line were cloned (Bonhomme et al., 1998a) and sequenced, leading to the identification of a pair of duplicated genes located 4 kb apart on the bottom of chromosome I (Fig. 2a): the POK gene, in which the T-DNA insertion occurred (At1g71270), and the P2 gene (At1g71300).
Figure 2.
The POK and P2 genes. a, Structure of the POK and P2 genes on chromosome I. Even exons are in light gray and uneven exons are in dark gray to facilitate comparison between duplicated gene structures. Triangle, T-DNA insertion in pok line. Black arrows, Predicted translation start. b to f, Expression of the POK and P2 genes. b (top), Northern-blot hybridization of total RNA from various wild-type tissues (see below) with a probe overlapping POK exon 10. Autoradiography exposure was 1 week. b (bottom), Loading control showing EtBr stained gel image. c and d, Specificity test for POK (c) and P2 (d) primers used for RT-PCR. POK, cloned POK cDNA. P2, cloned P2 cDNA. B, flower bud cDNAs. e and f, Autoradiography of blotted PCR products obtained with either POK (e) or P2 (f) specific primers on cDNA from various wild-type tissues. Si, siliques; Se, seeds; Ro, roots; Le, leaves; 5, 5-d; 10, 10-d; 15, 15-d; 21, 21-d-old plant; B, flower buds; <1, flower buds less than 1 mm in size; >1, flower buds more than 1 mm in size; In, inflorescences; Fl, mature flowers; W, water control; g, genomic wild-type DNA.
The POK and P2 genes (Fig. 2a) are composed of 20 and 19 exons, respectively (with an additional 5′ exon in POK). The exon-intron structure (Fig. 2a) is strictly conserved and exon sequences share 90% identity, whereas intron sequences are largely divergent and shorter in P2 than in POK.
Pok hemizygous mutant plants were complemented with an 11.2-kb genomic clone overlapping the POK gene, but excluding P2. The complementation construct was linked to the hpt gene, whereas the pok T-DNA insertion was marked with the nptII gene, conferring hygromycin (H) and kanamycin (K) resistance, respectively, to the transformed plants. Among the progeny of infiltrated mutant plants, five plants were selected that, when selfed, showed 2:1 segregation for kanamycin resistance, instead of 1:1 observed in the pok mutant. Furthermore, these five plants showed 5:1 segregation for hygromycin resistance, as expected in the case of independent segregation of both T-DNA insertions. The self progeny of one of these potentially complemented plants was analyzed. Descendants homozygous for hygromycin resistance either were also homozygous for kanamycin resistance (H/H, K/K), or showed a 3:1 ratio of kanamycin-resistant to kanamycin-sensitive progeny (H/H, K/-). Crossing these latter plants as male in with wild type confirmed that T-DNA transmission was restored. Moreover, descendants that had lost the complementing construct (showing a fully hygromycin-sensitive progeny) showed 1:1 segregation for kanamycin resistance. Finally, when germinating pollen from a complemented descendant (K/-, H/H) in vitro, the same proportion of elongated pollen tubes (78%, out of 348 counted pollen grains) was obtained as for wild-type pollen (77% of elongated tubes, out of 340 pollen grains), showing phenotypic restoration for this plant. In the control hemizygous pok line, 45% of elongated pollen tubes were counted out of 325 pollen grains.
To rule out potential lesions in the linked P2 gene, the P2 genomic region was completely sequenced in the pok mutant and no mutations were observed.
Taken together, these data confirm that the T-DNA insertion in the POK gene alone is responsible for the observed tip growth defect in pok plants.
Expression of the POK and P2 Genes
Northern-blotting experiments using exon 10 of the POK gene as a probe (Fig. 2b) revealed a transcript of the expected size (2.2 kb) at every developmental stage tested and in almost all plant tissues. However, there was some variation in transcript abundance; very low levels of transcripts were detected in seeds, whole inflorescence, and mature flowers, while higher levels were observed in roots and flower buds less than 1 mm in size (i.e. before pollen mitosis [PM] I).
Due to their strong identity, the POK and P2 transcripts could not be distinguished in hybridization experiments. However, oligonucleotides specific for each cDNA were designed (Fig. 2, c and d) and reverse transcription (RT)-PCR analysis was used to characterize the expression pattern of both transcripts independently. For the POK gene, RT-PCR results, although not quantitative, were qualitatively similar to those obtained by northern blotting, with the POK cDNA detected in all tissues, although at very low levels in seeds (Fig. 2e). For the P2 cDNA, no RT-PCR products were detected in any sample (Fig. 2f), even after two rounds of nested amplification (not shown).
Taken together, the weak, or absent, expression of P2 versus POK and the presence of the POK cDNA in all RT-PCR samples argue that the northern-blot analysis shown in Figure 2b faithfully reflects POK mRNA distribution only.
The POK and P2 Transcripts
Pollen and root cDNA libraries were screened with a probe corresponding to the T-DNA insertion region and overlapping the predicted exon 10 of the POK gene. Two types of clones were obtained from the pollen cDNA library: a POK partial cDNA clone (1,793 nt long) and a P2 cDNA clone (2,096 nt long). The screening of the root cDNA library allowed the isolation of a longer POK cDNA clone (2,217 nt); however, no cDNA clone for the P2 gene was found.
Putative initiator ATGs for both transcripts (NetStart prediction software: http://www.cbs.dtu.dk/services/NetStart/), located in exon 1 (Fig. 2a), predict a total length for POK and P2 coding sequences of 2,088 and 2,106 nt, respectively. 5′RACE experiments using POK-specific primers indicated a 5′UTR of 156 to 158 nt, located 31 to 33 nt upstream of the 5′ terminus of the root cDNA.
POK and P2 Are Highly Conserved among Eukaryotes
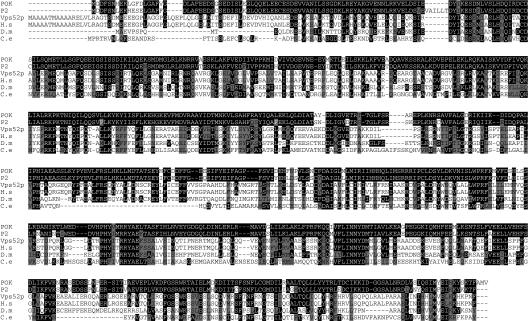
The POK and P2 predicted proteins are 696 and 702 amino acids, respectively, with a predicted molecular mass of approximately 80 kD. Both proteins share 88% identity and 93% similarity. According to BLAST software, POK shows 23% identity and 43% similarity with the yeast SAC2/Vps52p protein (Kölling et al., 1994; Conibear and Stevens, 2000). Predicted homologous proteins are also found in other species, such as Homo sapiens, Drosophila melanogaster, and Caenorhabditis elegans, with 23% to 35% identity and 43% to 59% similarity over the entire sequences (Fig. 3). Although some amino acids appear well conserved among the different species (Fig. 3), no particular structure, signal peptide, or known motifs are predicted in the POK sequence.
Figure 3.
POK is conserved among eukaryotes. Alignment of POK and P2 protein sequences with their putative orthologs in S. cerevisiae (Vps52p, accession, AAB64912), H. sapiens (H.s, accession, AAH32108), D. melanogaster (D.m, accession, AAF52254), and C. elegans (C.e, accession, AAA68727), according to ClustalW software (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html). Black boxes, Amino acids identical to POK amino acids. Gray boxes, Amino acids strongly similar to POK amino acids.
The AtVPS53 and AtVPS54 Genes
Conibear and Stevens (2000) showed that the yeast Vps52p protein functions in complex with two other proteins called Vps53p and Vps54p. We therefore searched for Arabidopsis genes encoding potential homologs of Vps53p and Vps54p. Performing BLAST analysis, we identified two candidate genes that we called AtVps53 and AtVps54 (At1g50500 and At4g19490, respectively). The corresponding predicted proteins share, respectively, 19% and 22% identity and 38% and 44% similarity with their yeast counterparts.
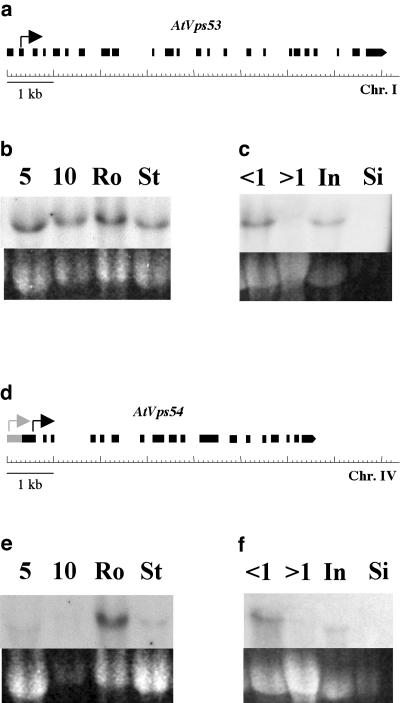
AtVps53 and AtVps54 cDNA clones were isolated from the pollen cDNA library. They are both 2.7 kb long and contain 24 and 18 exons, respectively (Fig. 4, a and d). The predicted proteins contain 829 and 1,001 amino acids, respectively, compared to 822 and 889 amino acids for Vps53p and Vps54p. A predicted translation start for the AtVps53 transcript is located in the middle of exon 2 (Fig. 4a, black arrow). For AtVps54, GENSCAN software (Chris Burge, Stanford University) predicts a long exon 1 (Fig. 4d, gray arrow), but RT-PCR experiments did not allow amplification of this region (not shown). The first in frame ATG of the isolated AtVps54 pollen cDNA is located at the end of exon 1 (Fig. 4d, black arrow), which would give rise to a 784-amino acid protein.
Figure 4.
The AtVps53 and AtVps54 genes. a and d, Structure of AtVps53 (a) and AtVps54 (d) genes on chromosomes I and IV, respectively. Black boxes, Exons identified by sequencing the pollen cDNAs. Gray box, Predicted additional region of AtVps54 first exon (GENSCAN software, http://genes.mit.edu/GENSCAN.html). Arrows indicate predicted translation start (for AtVps54, gray arrow, first ATG of the predicted mRNA; black arrow, first in frame ATG in the available pollen cDNA). b, c, e, and f, Expression of the AtVps53 and AtVps54 genes. b and e, Northern blots performed on total RNA from wild-type vegetative tissues, with a probe corresponding to AtVps53 (b) or AtVps54 (e) cDNAs. Top, Autoradiographs after 1 week exposure. Bottom, Corresponding loading controls (EtBr staining). c and f, Northern blots performed on total RNA from wild-type reproductive tissues, with a probe corresponding to AtVps53 (c) or AtVps54 (f) cDNAs. Top, Autoradiographs after 4-d exposure. Bottom, Corresponding loading controls (EtBr staining). Abbreviations same as for Figure 2.
Expression of these two vps-like genes was studied by northern-blot analysis (Fig. 4, b, c, e, and f). As observed for POK, both AtVps53 and AtVps54 are ubiquitously expressed. Their basal expression level is low, although higher for AtVps53 compared to AtVps54 in all vegetative tissues (Fig. 4, b and e), and higher for AtVps54 in roots versus other tissues (Fig. 4e). In reproductive tissues, both transcripts are more abundant in buds less than 1 mm in size compared with later stages (Fig. 4, c and f).
Distribution of the POK::GUS Fusion Protein
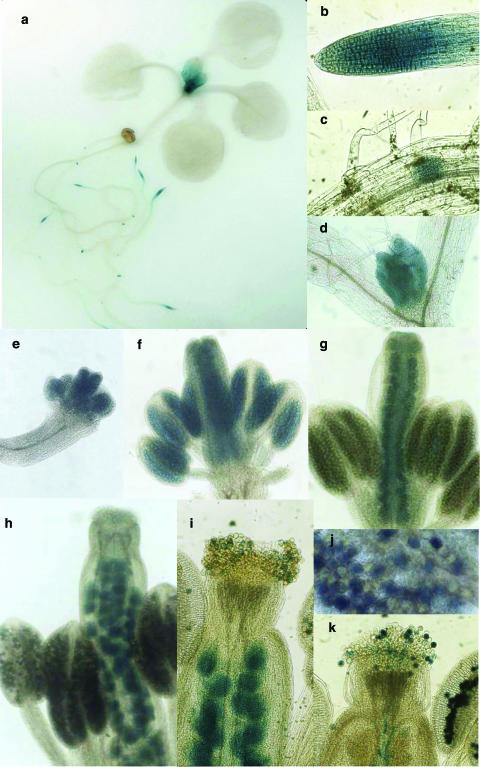
Since an active translational fusion between the first 10 exons of the POK gene and the uidA gene carried by the T-DNA occurred in the pok mutant, GUS staining allowed us to determine a preliminary expression pattern for the POK protein. In 8- to 10-d-old plantlets, strong blue staining was detected in the root apex (Fig. 5a), with enhanced intensity in the elongation zone (Fig. 5b), in emerging lateral root primordia (Fig. 5c), in very young leaves (Fig. 5d), and sometimes in stipules (Fig. 5a).
Figure 5.
GUS staining assays on hemizygous pok mutants. GUS staining on 10-d-old plantlets (a and d), 8-d-old roots (b and c), and successive stages of floral development (e–j). e, premeiotic stage, pistil size, 0.1 mm; f, tetrad stage, pistil size, 0.5 mm; g, microspore/PMI stage, pistil size, 0.8 mm; h, trinucleate pollen stage, pistil size, 2 mm; i, postpollination stage; j, trinucleate pollen stage; k, control transgenic (hemizygous) line, expressing GUS in pollen grains and pollen tubes, postpollination stage.
In reproductive tissues, GUS staining was observed at different stages of floral development (Fig. 5, e–j). Blue coloration was detected from the earliest stages of flower development, before meiosis and gametogenesis (bud size, 0.1 mm; Fig. 5e), and maintained later (Fig. 5, f–j). As flower bud size reached 1.6 to 1.7 mm (i.e. after PMII), expression of the fusion protein became strictly gametophytic in the male reproductive tissues, whereas it was clearly sporophytic in the female tissues, since all ovules were stained (Fig. 5h). At maturity, half of the pollen grains within the anthers were blue (Fig. 5j), as expected for hemizygous plants. At later postpollination stages, all developing seeds were stained (Fig. 5i). However, no blue staining was observed in the pistil below the stigma surface (Fig. 5i), compared to a control line (T-DNA insertion line; A. Guyon, A. Lécureuil, P. Guerche, and S. Bonhomme, unpublished data) expressing GUS in pollen and for which a blue coloration was observed in the style, corresponding to elongated pollen tubes (Fig. 5k; note that developing seeds are not stained). The absence of staining in the style of the pok mutant could be the result of lower GUS expression in this line compared to our control line, as well as of lack of effective pok pollen tube growth.
POK Is Localized to Golgi in Living Plant Cells
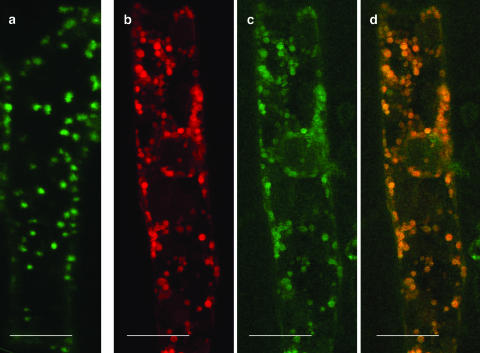
The subcellular localization of the POK protein was investigated using a POK::GFP fusion construct containing the complete open reading frame of the POK gene fused with the GFP coding sequence. Both cDNAs were cloned downstream of 2.2 kb of the predicted POK promoter region. Onion (Allium sativum) epidermal cells were bombarded with this POK::GFP construct. The collected signal exhibited a cytoplasmic dispersed punctate pattern (Fig. 6a) characteristic of the previously described Golgi distribution pattern in plant cells (Boevink et al., 1998; Batoko et al., 2000). Onion cells were also cobombarded with both the POK::GFP construct and a ST::DsRed construct (provided by F. Brandizzi, Oxford Brookes University, Oxford, UK; Saint-Jore et al., 2002), where the signal anchor sequence of the rat α-2,6-sialyltransferase gene was used as a late Golgi marker. Both GFP (Fig. 6c) and DsRed (Fig. 6b) signals exhibited a very similar distribution within the cell. The overlay and coincidence of both signals (Fig. 6d) strongly suggests that the POK::GFP fusion protein localizes to Golgi in onion epidermal cells.
Figure 6.
Localization of the POK::GFP fusion protein in onion epidermal cells. Onion epidermal cells were cobombarded with the POK::GFP and the ST-DsRed constructs. Resulting fluorescent signals have been collected independently. a and c, GFP signal; b, DsRed signal; d, overlay of b and c. Scale bar represents 80 μm.
DISCUSSION
POK and Pollen Tube Elongation: Slowly to the Goal
pok can be classed a late gametophytic mutant, since microsporogenesis and gametogenesis show no phenotypic differences compared to wild type. Moreover, we showed that pollen tube elongation is impaired in pok. Contrary to the raring-to-go Arabidopsis mutant (Johnson and McCormick, 2001), initial steps of pollen germination do not seem to be affected in pok. Indeed, in vitro germination (Bonhomme et al., 1998b) and in situ pollination experiments both show that at least some pok pollen grains are able to germinate, but pollen tubes subsequently fail to elongate. The limited transmission of the pok T-DNA insertion after pollinating wild-type pistils with pollen from the pok mutant suggests that, although reduced, elongation is still possible in pok pollen tubes. We previously showed (Bonhomme et al., 1998b) that the percentage of male transmission obtained after such crosses is 10 times higher in the upper third of the ovary (i.e. the ovary fraction located beneath the style). This observation leads to the hypothesis that the T-DNA transmission defect observed in pok line selfing progeny likely results from a competition between the pok pollen tubes and the wild-type pollen tubes. We also conclude that the pok mutation strictly affects the tip growth processes in pollen, since pok pollen tubes that are able to reach the upper ovules subsequently accomplish fertilization.
POK Function May Not Be Restricted to Tip Growth
Although weak T-DNA transmission through the male gametophyte has been observed, suggesting incomplete penetrance of the mutation, no plants homozygous for the pok mutation were isolated. This suggests that the POK function is not restricted to reproduction, but could also be essential in early developmental events. This hypothesis is reinforced by the expression pattern of the POK gene, which is clearly not restricted to the reproductive organs, but extends to other plant tissues. Indeed, northern-blot experiments showed that, apart from reproductive tissues (see below), POK expression is high in elongating tissues, such as roots or young seedlings. Moreover, intense GUS staining is detected in the elongation zone of the root apex and in the emerging lateral roots of the pok mutant.
POK Expression in Reproductive Tissues
Even though the POK gene basal expression level is quite low, we detected higher levels of POK transcript in young flower buds (i.e. before the uninucleate microspore stage) compared with later stages of floral development. POK gene expression thus seems to be restricted to early steps of microgametogenesis, allowing us to classify it with the early class of pollen-expressed genes, which are transcribed soon after meiosis and whose transcripts accumulate until PMII and then decrease (Mascarenhas, 1990). A similar expression pattern has been reported for the GRP2 mRNA of tobacco (Ylstra and McCormick, 1999) and for some pollen-expressed Petunia genes (Guyon et al., 2000). In addition, GUS assays on pok inflorescences revealed precocious blue staining, which was maintained throughout flower development. In male reproductive organs, the staining appears strictly gametophytic, from PMII until maturity. Nonetheless, the mutant phenotype suggests that the POK protein is only required for the later steps of male gametophyte development (i.e. correct elongation of the pollen tubes). As the POK transcript seems to be unstable in the developing pollen grain, one can hypothesize that it would be rapidly translated and that the POK protein, rather than the POK mRNA, would accumulate in maturing pollen grains. It has been clearly shown that 5′-UTR are involved in both transcriptional and translational regulations of many plant genes (Bate et al., 1996; Jun Hua et al., 2001; Hulzink et al., 2002); investigation of the precise role of the POK gene 5′-UTR may provide new insights into the complex regulation of the early class of pollen-expressed genes.
The transcript level of putative homologs for VPS53 and VPS54 in Arabidopsis, named AtVps53 and AtVps54, was studied. Expression levels of both genes were revealed to be quite weak (particularly for AtVps54); however, expression was higher in roots (mainly for AtVps54). For both AtVps53 and AtVps54, an expression pattern comparable to that of POK was observed in reproductive tissues (i.e. higher expression in young flower buds), suggesting that they also could be part of the early class of pollen-expressed genes. It has been shown that, for a given gene, mRNA stability can be different in pollen and in somatic cells (Ylstra and McCormick, 1999). One can thus hypothesize that, at least in male gametophytes, POK, AtVps53, and AtVps54 transcripts would share the same type of regulation, involving rapid translation of unstable transcripts and subsequent protein accumulation.
POK and P2
A second mRNA, sharing 90% identity with POK and named P2, is also expressed in pollen grains, although at a much lower level, as it was undetectable using RT-PCR. Similar to 17% of the full complement of Arabidopsis genes (Blanc et al., 2000), POK and P2 are two tandemly repeated duplicated genes. We can hypothesize that P2 could be a POK-derived pseudogene, although Blanc et al. (2000) suggest that a highly conserved exon-intron structure between duplicated genes is an indication that both genes are expressed. However, examples of young pseudogenes that have conserved their exon-intron structure and are still expressed at very low levels have been described in Drosophila (Ramos-Onsins and Aguade, 1998), mouse (Hirotsune et al., 2003), and Arabidopsis (Zhang et al., 2000). Identification and phenotypic analysis of P2 mutant lines should help us to establish the role of the P2 gene. It is important to note that, despite the high identity between both transcripts and both predicted proteins, POK and P2 genes are not fully functionally redundant, at least in the male gametophyte where they are both expressed.
POK Shows Significant Homology with Vps52p
The deduced protein sequences of both POK and P2 share 23% to 35% identity with several eukaryotic proteins, all of which seem to be orthologous to one another, including the SAC2/Vps52p protein of yeast. The VPS52 gene, as well as VPS53 and VPS54, were identified by Conibear and Stevens (2000) while screening for novel components of the trans-Golgi network-localized vesicle-transport machinery. Vps52 was found to be allelic to the previously identified SAC2 gene (Novick et al., 1989; Kölling et al., 1994). It was shown that Vps52p, Vps53p, and Vps54p form a stable complex that is peripherally associated with the late Golgi compartment and is involved in the recycling of resident Golgi proteins from an early endosomal, or prevacuolar, compartment to the trans-Golgi network (Conibear and Stevens, 2000). This hypothesis was reinforced when Siniossoglou and Pelham (2001) showed that Vps52p could bind both the Ypt/Rab GTPase Rab6 and the t-SNARE Tlg1p. More recently, a fourth subunit of the VPS complex with a potential regulatory function has been identified in yeast and was named Vps51p (Conibear et al., 2003; Reggiori et al., 2003).
Proteins shown to be components of the vesicle-transport machinery are usually well conserved among eukaryotic species from yeast to human. As observed for POK, most of these genes have been shown to be ubiquitously expressed, but to higher levels in roots (Ahmed et al., 1997; Conceição et al., 1997; Bassham and Raikhel, 1998; Rojo et al., 2003). Although many genes homologous to those involved in the yeast Golgi vesicle trafficking have already been identified in Arabidopsis (e.g. Ahmed et al., 1997; Conceição et al., 1997; Bassham and Raikhel, 1998; Sanderfoot et al., 1998; Zheng et al., 1999), to our knowledge POK would be one of the first for which a corresponding mutant line has been identified.
POK Could Be Involved in Plant Vesicle Trafficking
Using POK::GFP and ST::DsRed fusion constructs in transient assays, we demonstrated that POK shares the same localization in plants as Vps52p does in yeast. Since tip-growing cells, such as pollen tubes, ensure rapid and polarized elongation through active apical exocytosis of Golgi vesicles (Hepler et al., 2001), the phenotype observed in pok mutants can be explained if the POK protein has a function comparable to that of Vps52p. Indeed, if POK is involved in the recycling of Golgi resident membrane proteins, alteration of its function would lead to inefficient vesicle recycling and subsequent decrease of the growth rate. Furthermore, it seems that POK could also have a more general role in non-tip-growing cells, where vesicle-recycling deficiency could also lead to slow growth, explaining the proposed zygotic lethality of homozygous pok mutants. The study of the pok mutant should allow us to progress in the understanding of the molecular mechanisms involved in pollen development and pollen tube growth. Since POK seems to belong to the early class of pollen genes and exhibits a complex expression pattern, study of its regulation could be of particular interest. Besides, the further characterization and analyses of the functions of POK should provide new insights regarding the processes governing not only tip growth, but also vesicle trafficking in the male gametophyte and in somatic cells.
MATERIALS AND METHODS
Plant Material
The T-DNA transmission defect lines of Arabidopsis (L.) Heynh. ecotype Wassilewskija were isolated from the Versailles collection of T-DNA insertion mutants as described in Bonhomme et al. (1998b).
Pollen Tube Growth
In vitro growth of pollen tubes was tested as described in Procissi et al. (2001). In situ pollination and aniline blue staining were performed as described in Procissi et al. (2003).
GUS Assays
Histochemical GUS assays were performed as described in Procissi et al. (2003), except that K3Fe(CN)6 and K4Fe(CN)6 concentrations were increased to 2 mm for staining reproductive tissues.
Complementation Experiments
A XbaI/SalI 11.2-kb fragment corresponding to the POK gene was subcloned from a genomic phage clone (Bonhomme et al., 1998a) and inserted into a binary vector (pZP P200; Hajdukiewicz et al., 1994) bearing the hpt gene. The hemizygous pok line was transformed by floral dipping in an Agrobacterium solution as described by Clough and Bent (1998). Transformants were selected in vitro on hygromycin-containing medium (30 mg/L).
Screening of cDNA Libraries
The pollen cDNA library was constructed from poly(A+) RNA isolated from mature pollen of Arabidopsis ecotype Landsberg erecta. Mature pollen was isolated as described previously (Honys and Twell, 2003). Poly(A+) RNA was isolated directly from isolated pollen after grinding pollen in liquid nitrogen using a pestle and mortar and using the Dynabeads mRNADIRECT mRNA kit (Dynal Biotech, Oslo) according to the manufacturer's instructions. Pollen cDNA was primed with oligo(dT) and cloned into the EcoRI/XhoI sites of the lambda HybriZAP vector (Stratagene, La Jolla, CA).
Root cDNA library was cloned into lambda gt10 (kind gift from M. Salanoubat). Both cDNA libraries were screened with a probe corresponding to exon 10 of the POK gene, with probes corresponding to exons 10 and 22 for the AtVps53 gene and with exons 11 and 15 for the AtVps54 gene.
Northern-Blot Analysis
Seed RNA was extracted following Downing (1992). RNA from other plant tissues was isolated as described in Procissi et al. (2003). Northern blots were performed on 20 μg of total RNA as described in Ausubel et al. (1990).
RT-PCR Experiments
cDNA synthesis was performed with Superscript RT (Life Technologies, Carlsbad, CA) from total RNA (3 μg) treated with DNAse I, following the manufacturer's protocol. cDNAs were amplified with 27 cycles of PCR. Primers used were: POK gene, POK-1 (5′-GGTGTTAAGTGCACATTTTCGTG-3′) and POK-2 (5′-TCACCAAAGAAATCATCACAAAA-3′); P2 gene, P2-1 (5′-TGCTAAGTGCACATTTTCAGT-3′) and P2-2 (5′-TCACCAAAGAAATCATCACAAAA-3′); AtVps53 gene, 53 to 1 (5′-AAAATCAGAAAATACATGGGAC-3′) and 53 to 2 (5′-TCAAGAAATATGCCGTGAC-3′); and AtVps54 gene, 54 to 1 (5′-ATGCGTATCTCCATACATGA-3′) and 54 to 2 (5′-AAAAATCAGATTCGAGCGAT-3′).
5′ RACE Experiments
5′ RACE experiments were performed using a Life Technologies system on roots and flower bud RNA. Successively used primers were specific for exon 5 (5′-GTCTGAACTTATAGAACCAATC-3′), the end of exon 4 (5′-CTGAAATCCACTGAGAAGAGT-3′), and the beginning of exon 4 (5′-GTCACTTTCCTTTATGTAATCC-3′). RACE products were cloned into pGEM-T (Promega, Madison, WI).
GFP Constructs
The POK promoter region (2.2 kb) was amplified from Wassilewskija DNA using the primers 5′-TTTCTAGACGGCAGAGCTTCAGT-3′ and 5′-TCCCCCATGGTTTGGCCCTA-3′. The resulting fragment was cloned and sequenced into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). In parallel, the POK cDNA was amplified using the primers 5′-CTAATGGCGGACTCAAACTGATT-3′ and 5′-GCTACCATGGCGAAAGTCTTGGAGTATTTCCT-3′, allowing replacement of the stop codon by an NcoI restriction site at the 3′ end. The resulting PCR product was cloned and sequenced in the pCRBluntII-TOPO (Invitrogen). An XbaI/BglII promoter fragment and a BglII/NcoI cDNA fragment were isolated from the constructs described above and cloned together into the pTrc99A vector (Amann et al., 1988), where the promoter-cDNA junctions were verified. The resulting XbaI/NcoI fragment was cloned into pCambia1302 vector (CAMBIA GPO, Canberra, Australia) to produce the POK::GFP translational fusion.
Onion Epidermal Cell Bombardment
As described by Weigel and Glazebrook (2002), 7 μg of each construct were used to transform onion (Allium sativum) cells. Bombardments were performed with a PDS-1000/HE Biolistic Particle Delivery System (Bio-Rad Laboratories, Hercules, CA), using 900-psi rupture discs (Bio-Rad). Transformed epidermal cells were observed using a LEICA TCS-NT confocal laser scanning microscope (Leica Mycrosystems, Wetzlar, Germany) with an argon/krypton laser (Omnichrome). GFP signal was collected with an FITC filter set (BP520/30) at 488 nm. DsRed was excited at 568 nm with a BP600/30 filter set.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers bankit481797 (POK), bankit481801 (P2), AAB64912, AAH32108, AAF52254, and AAA68727.
Acknowledgments
We thank F. Brandizzi (Oxford Brookes University) for providing us with the ST::DsRed construct, R. Berthomé for help with bombardments, V. Carpentier and A. Lécureuil for the GUS assays, and C. Horlow and F. Nolent for their contribution to this work. We are also very grateful to C. Mézard and F. Nogué for critical reading of the manuscript and stimulating discussions.
This work was supported by INRA-DGAP (doctoral fellowship to E.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037747.
References
- Ahmed SU, Bar-Peled M, Raikhel NV (1997) Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol 114: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E, Ochs B, Abel KJ (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in E. coli. Gene 69: 301–315 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1990) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Bassham DC, Raikhel NV (1998) An Arabidopsis Vps45p homolog implicated in protein transport to the vacuole. Plant Physiol 117: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV (2000) Unique features of the plant sorting machinery. Curr Opin Cell Biol 12: 491–495 [DOI] [PubMed] [Google Scholar]
- Bate N, Spurr C, Foster GD, Twell D (1996) Maturation-specific translational enhancement mediated by the 5′-UTR of a late pollen transcript. Plant J 10: 101–111 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore Y (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and the Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Horlow C, Guyon A, Férault M, Vezon D, Marchand M, de Laissardière S, Bechtold N, Pelletier G (1998. a) Screening for gametophytic mutations in the Versailles collection of Arabidopsis thaliana transformants: first results for two putative male gametophytic mutants. Acta Hortic 459: 173–181 [Google Scholar]
- Bonhomme S, Horlow C, Vezon D, de Laissardière S, Guyon A, Férault M, Marchand M, Bechtold N, Pelletier G (1998. b) T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distorsion alone. Mol Gen Genet 260: 444–452 [DOI] [PubMed] [Google Scholar]
- Camacho L, Malhó R (2003) Endo/exocytosis in the pollen tube apex is differentially regulated by Ca2+ and GTPases. J Exp Bot 54: 83–92 [DOI] [PubMed] [Google Scholar]
- Cheung A, Chen CY, Glaven RH, de Graaf BHJ, Vidali L, Hepler PK, Wu HM (2002) Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conceição AS, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV (1997) The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9: 571–582 [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH (2003) Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell 14: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH (2000) Vps52p, Vps53p and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11: 305–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing WL (1992) A Brassica napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J 2: 685–693 [PubMed] [Google Scholar]
- Franklin-Tong VE (1999) Signaling in pollination. Curr Opin Plant Biol 2: 490–495 [DOI] [PubMed] [Google Scholar]
- Geitmann A, Emons AMC (2000) The cytoskeleton in plant and fungal cell tip growth. J Microsc 198: 218–245 [DOI] [PubMed] [Google Scholar]
- Guyon VN, Astwood JD, Garner EC, Dunker AK, Taylor LP (2000) Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in Petunia. Plant Physiol 123: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A (2003) An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–96 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulzink RJ, de Groot PF, Croes AF, Quaedvlieg W, Twell D, Wullems GJ, van Herpen MM (2002) The 5′ untranslated region of the ntp303 gene strongly enhances translation during pollen tube growth, but not during pollen maturation. Plant Physiol 129: 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, McCormick S (2001) Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol 126: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Hua X, van de Cotte B, Van Montagu M, Verbruggen N (2001) The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J 26: 157–169 [DOI] [PubMed] [Google Scholar]
- Jürgens G, Geldner N (2002) Protein secretion in plants: from the trans-Golgi network to the outer space. Traffic 3: 605–613 [DOI] [PubMed] [Google Scholar]
- Kölling R, Lee A, Chen EY, Botstein D (1994) Nucleotide sequence of the SAC2 gene of S. cerevisiae. Yeast 10: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Kucharczyk R, Rytka J (2001) Saccharomyces cerevisiae—a model organism for the studies on vacuolar transport. Acta Biochim Pol 48: 1025–1042 [PubMed] [Google Scholar]
- Mascarenhas JP (1990) Gene activity during pollen development. Annu Rev Plant Physiol Plant Mol Biol 41: 317–338 [Google Scholar]
- Novick P, Osmond BC, Bolstein D (1989) Suppressors of yeast actin mutations. Genetics 121: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R, Preuss D (2000) Pollen tube targeting and axon guidance parallels in tip-growth mechanisms. Trends Cell Biol 10: 517–524 [DOI] [PubMed] [Google Scholar]
- Parton RM, Fischer-Parton S, Watahiki MK, Trewavas AJ (2001) Dynamics of the apical vesicle accumulation and the rate of growth are related in individual pollen tubes. J Cell Sci 114: 2685–2695 [DOI] [PubMed] [Google Scholar]
- Pierson ES, Lichtscheidl IK, Derksen J (1990) Structure and behaviour of organelles in living pollen tubes of Lilium longiflorum. J Exp Bot 41: 1461–1468 [Google Scholar]
- Preuss D (1995) Being fruitful: genetics of reproduction in Arabidopsis. Trends Genet 11: 147–153 [DOI] [PubMed] [Google Scholar]
- Procissi A, de Laissardière S, Férault M, Vezon D, Pelletier G, Bonhomme S (2001) Five gametophytic mutations affecting pollen tube growth in Arabidopsis thaliana. Genetics 158: 1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi A, Guyon A, Pierson ES, Giritch A, Knuiman B, Grandjean O, Tonelli C, Derksen J, Pelletier G, Bonhomme S (2003) KINKY POLLEN encodes a SABRE-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J 36: 894–904 [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S, Aguade M (1998) Molecular evolution of the Cecropin multigene family in Drosophila: functional genes vs. pseudogenes. Genetics 150: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ (2003) Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem 278: 5009–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV (2003) The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol Biol Cell 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV (1998) A putative vacuolar cargo receptor colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc Natl Acad Sci USA 95: 9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV (2000) The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptator protein receptors. Plant Physiol 124: 1558–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR (2001) An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J 20: 5991–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Hepler PK (2001) Actin and pollen tube growth. Protoplasma 215: 64–76 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Ylstra B, McCormick S (1999) Analysis of mRNA stabilities during pollen development and in BY2 cells. Plant J 20: 101–108 [DOI] [PubMed] [Google Scholar]
- Zhang J, Pontoppidan B, Xue J, Rask L, Meijer J (2000) The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol Plant 115: 25–34 [DOI] [PubMed] [Google Scholar]
- Zheng H, Fischer von Mollard G, Kovaleva V, Stevens TH, Raikhel NV (1999) The plant v-SNARE AtVTI1a likely mediates vesicle transport from the TGN to the prevacuole. Mol Biol Cell 10: 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]