Abstract
To study the early steps of flower initiation and development in grapevine (Vitis vinifera), we have isolated two MADS-box genes, VFUL-L and VAP1, the putative FUL-like and AP1 grapevine orthologs, and analyzed their expression patterns during vegetative and reproductive development. Both genes are expressed in lateral meristems that, in grapevine, can give rise to either inflorescences or tendrils. They are also coexpressed in inflorescence and flower meristems. During flower development, VFUL-L transcripts are restricted to the central part of young flower meristems and, later, to the prospective carpel-forming region, which is consistent with a role of this gene in floral transition and carpel and fruit development. Expression pattern of VAP1 suggests that it may play a role in flowering transition and flower development. However, its lack of expression in sepal primordia, does not support its role as an A-function gene in grapevine. Neither VFUL-L nor VAP1 expression was detected in vegetative organs such as leaves or roots. In contrast, they are expressed throughout tendril development. Transcription of both genes in tendrils of very young plants that have not undergone flowering transition indicates that this expression is independent of the flowering process. These unique expression patterns of genes typically involved in reproductive development have implications on our understanding of flower induction and initiation in grapevine, on the origin of grapevine tendrils and on the functional roles of AP1-and FUL-like genes in plant development. These results also provide molecular support to the hypothesis that Vitis tendrils are modified reproductive organs adapted to climb.
The early phases of reproductive development and flower formation have been well characterized in the herbaceous model plants Arabidopsis and Antirrhinum majus (Egea-Cortines and Davies, 2000; Theissen, 2001; Mouradov et al., 2002; Simpson and Dean, 2002). In Arabidopsis, transition to flowering is dependent on the activity of floral meristem identity genes such as LEAFY (LFY), FRUITFULL (FUL), APETALA1 (AP1), and CAULIFLOWER (CAL; for review, see Pidkowich et al., 1999; Simpson et al., 1999; Kieffer and Davies, 2001; Zik and Irish, 2003). LFY expression in lateral meristems seems to be responsible for their acquisition of flower meristem identity (Blázquez et al., 1997). FUL, CAL, and AP1 are closely related MADS-box genes that belong to the SQUAMOSA (SQUA) gene subfamily, also called AP1/FUL gene lineage (Theissen et al., 2000; Becker and Theissen, 2003; Litt and Irish, 2003). Together with LFY, they play functionally redundant roles in the control of flower meristem identity (Mandel et al., 1992; Weigel et al., 1992; Mandel and Yanofsky, 1995; Liljegren et al., 1999; Ratcliffe et al. 1999; Ferrándiz et al., 2000). In Arabidopsis, the patterns of expression of these genes relate well with their proposed function. FUL is up-regulated in the shoot apical meristem (SAM) around the transition to flowering and before the initiation of flower meristems. LFY and AP1 are expressed in emerging flower meristems, while FUL is soon excluded from this region (Weigel et al., 1992; Mandel and Yanofsky, 1995). Apart from their function as floral meristem identity genes, AP1 and FUL also seem to play roles as organ identity genes at later stages of flower development. AP1 was initially defined as a class A gene involved in sepal and petal identity (Irish and Sussex, 1990; Coen and Meyerowitz, 1991; Theissen, 2001). FUL has been shown to play a role in carpel and fruit development that affects valve, replum, and style morphology (Gu et al., 1998; Ferrándiz et al., 2000).
Genetic and molecular characterization of the flowering process in different species reveals a conservation of the basic genetic mechanisms controlling the early stages of flower formation (Theissen and Saedler, 1999; Ng and Yanofsky, 2001). However, when homologous genes from species other than Arabidopsis have been analyzed, differences—regarding expression patterns and mutant phenotypes—have been found that may reflect distinct roles from those described for the Arabidopsis genes. For instance, the lack of examples of A-function mutants in most angiosperms analyzed questions a general role of AP1-like genes as A-function genes (Huijser et al., 1992; Yu et al., 1999; Litt and Irish, 2003). Moreover, in monocots, SQUA-like genes do not seem to be always functional orthologs of their Arabidopsis counterparts, based on their relatively large number and expression patterns (Schmitz et al., 2000; Theissen et al., 2000; Yu and Goh, 2000; Gocal et al., 2001). More recent studies have even questioned the existence of the AP1 gene lineage outside core eudicots (Litt and Irish, 2003). All these data suggest that flower initiation and development may involve common regulatory mechanisms to all the angiosperms as well as species-specific mechanisms whose genetic and molecular bases are yet unknown.
We are studying the reproductive biology of grapevine (Vitis vinifera), a woody perennial vine with a pattern of organ formation and development quite distinct to those previously described for annual herbaceous plants (Mullins et al., 1992). Wild grapevines are vigorous climbing plants with pressure-sensitive tendrils that allow them to climb into the forest canopy to a height of 20 to 30 m, supported by the forest trees. When grown from seeds, grapevine seedlings undergo a short-lived juvenile phase during which they produce leaves with a spiral phyllotaxis for the first 6 to 10 nodes. Later on, phyllotaxis changes and this event marks the transition to the adult phase of the plant. In adult plants phyllotaxis is alternate, with the SAM producing a characteristic sequence of leaves and lateral meristems, historically called uncommitted primordia (Tucker and Hoefert, 1968; Pratt, 1974; Gerrath and Posluszny, 1988a; Gerrath et al., 1998). Due to unequal internode elongation, lateral meristems that arise alternate to leaf primordia become opposed to leaves in the expanded shoot. Lateral meristems give rise to tendrils for a long period of time (2–5 years) before the plant initiates flowering. Upon flowering induction, inflorescences are formed in place of tendrils from the same lateral meristems (Pratt, 1971; Srinivasan and Mullins, 1981; Posluszny and Gerrath, 1986; Gerrath and Posluszny, 1988a; Morrison, 1991). As they develop from the same meristematic structures, tendrils and inflorescences have been proposed to be homologous organs. Furthermore, inflorescences and tendrils can substitute for each other depending on environmental conditions or hormonal treatments, and occasionally tendrils can give rise to flowers and fruits (Darwin, 1875; Bugnon, 1953; Pratt, 1971, 1974; Srinivasan and Mullins, 1981; Boss and Thomas, 2000, 2002). The presence of lateral meristems that give rise to tendrils, inflorescences, or intermediate organs and the production by the SAM of both leaves and lateral meristems that become opposed at maturity are a special feature of the Vitaceae. Attempts to explain their unusual organs position and their origin at the SAM have generated equally plausible interpretations on shoot formation, either sympodial or monopodial (Gerrath and Posluszny, 1988a; Gerrath et al., 1998).
In temperate regions, grapevine requires two consecutive growing seasons to flower. A rise in light intensity and temperature seems to be required to promote flowering (Butrosse, 1974; Mullins et al., 1992) that is induced in latent summer buds. Upon flowering induction, the SAM produces a few (2–3) lateral meristems that will give rise to inflorescence meristems. The inflorescence meristems form several inflorescence branch meristems before the bud enters dormancy at the end of the summer. The next spring, additional inflorescence branch meristems are formed before each one divides into a cluster of 3 to 4 flower meristems that develop into flowers (Srinivasan and Mullins, 1981; Gerrath and Posluszny, 1988b; Gerrath, 1993; May, 2000; Carmona et al., 2002).
Because of its particular developmental features, we are interested in the process of flower initiation in grapevine. Furthermore, grapevine belongs to the Vitaceae, a basal family of core eudicots (Judd et al. 1999), and understanding the genetic and molecular control of flower initiation and development in this family can shed light on the evolution of the regulation of flower development in angiosperms. Previously, we isolated VFL, the grapevine ortholog of FLO/LFY, and described its characteristic expression patterns in meristematic regions (Carmona et al., 2002). We have now investigated the role of other meristem identity genes during flower initiation and development. We have identified two genes within the SQUA subfamily, VFUL-L and VAP1, which could correspond to the grapevine FUL-like and AP1 orthologs. Our results are consistent with VFUL-L having a role in floral transition and carpel and fruit development. In contrast, the expression patterns of VAP1 do not support its role as an A-function gene. Unexpectedly, both genes are highly expressed during tendril development. These unique expression patterns of genes traditionally involved in reproductive development are discussed in relation to the special features of grapevine development and the ontogenetic relationship between tendrils and inflorescences.
RESULTS
Isolation of Grapevine FUL- and AP1-Like Genes
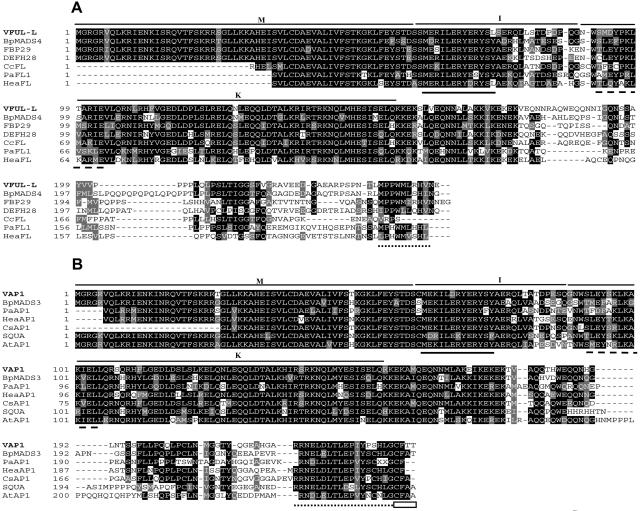
Genes belonging to the SQUA subfamily of MADS-box transcription factors were isolated using a 3′/5′RACE strategy. Two different cDNA types were identified. One of them (GenBank accession no. AY538747) contained an ORF of 741 bp preceded by a 5′-untranslated region of 83 bp. The 3′-untranslated region showed length heterogeneity, with four polyadenylation sites at positions 910, 997, 1012, and 1029 bp. The other cDNA type (GenBank accession no. AY538746) contained an ORF of 723 bp and the 5′-untranslated region was 166 bp in length. Two polyadenylation sites at positions 1058 and 1075 were detected in its 3′-untranslated region. The encoded MADS-box proteins (247 and 241 amino acids, respectively) aligned well with those from the SQUA subfamily (Fig. 1, A and B) based on sequence conservation in the I-region, which is typical for each MADS-box subfamily (Fig. 1, A and B, continuous line; Elo et al., 2001). Furthermore, differences at the beginning of the K-box and especially at the C terminus, where a prenylation motif (CaaX) is typical of AP1/SQUA-like proteins (Rodriguez-Concepción et al., 1999) but is absent in FUL-like proteins, allowed us to identify the grapevine MADS-box proteins as belonging to the FUL and AP1/SQUA clades (Fig. 1, A and B, dashed lines, dotted line, and white box; Theissen et al., 2000; Berbel et al., 2001; Elo et al., 2001; Becker and Theissen, 2003).
Figure 1.
Sequence analysis of VFUL-L, VAP1, and related SQUA-like proteins. A, VFUL-L protein and sequence comparison to other FUL-like proteins. B, VAP1 protein and sequence comparison to AP1/SQUA-like proteins. In both alignments, the MADS-box (M), I-region (I), and K-box (K) are indicated. In the I-region, the sequence highly conserved between both proteins is underlined with a continuous line. Dashed lines indicate distinctive sequences in the K-box. The highly divergent C-terminals show some conserved motives in each group (dotted line), mainly in the AP1 group that includes the prenylation motif (white box). C, Phylogenetic tree of the predicted SQUA-like proteins from different eudicots plant species (accession no. in parentheses): Antirrhinum SQUA, DEFH28, AmFUL (X63701, AY040247, AY306139); Arabidopsis AP1, CAL, FUL, AtFL (Z16421, L36925, AY072463, Q9LI72); Apple MdMADS2, MdMADS5 (U78948, AJ000759); Betula pendula BpMADS3, BpMADS4, BpMADS5 (X99653, X99654, X99655); Cauliflower BOAP1 (Z37968); Clarkia concinna CcFL (AY306143); Corylopsis sinensis CsAP1, CsFUL (AY306146, AY306147); Eucalyptus EAP1 (AF305076); Heuchera americana HeaAP1, HeaFL, HeaFUL (AY306148, AY306149, AY306150); Nicotiana sylvestris NsMADS2 (AF068726); Nicotiana tabacum NAP1-1, NAP1-2, NTMADS5 (AF009126, AF009127, AF068724); Petunia PFG, FBP26, FBP29 (AF176782, AY370517, AF335245); Pea PEAM4 (AF461740); Phytolacca americana PaAP1, PaFL1, PaFL2, PaFUL (AY306160, AY306161, AY306162, AY306163); Potato POTM1-1, SCM1 (U23757, AF002666); Silene latifolia SLM5 (X80492); Sinapis alba SaMADSC, SaMADSB (Q41276, Q41274); Tomato TRD4 (AY098732). Bootstrap support values are indicated when over 50. In order to root the SQUA subfamily, Arabidopsis SEP1 (M55551), SEP2 (M55552), and SEP3 (AF015552) were used as outgroup.
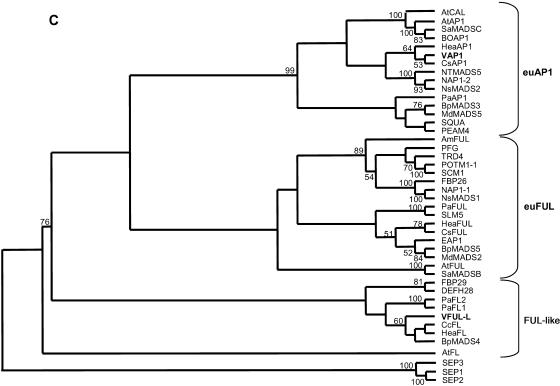
Recent phylogenetic analyses have identified two clades of FUL-like genes in core eudicots, euFUL and FUL-like (Litt and Irish, 2003). To determine the evolutionary relationships between proteins encoded by the isolated grapevine genes and other core eudicots MADS-box proteins of the SQUA subfamily, a phylogenetic tree was constructed using the complete predicted protein sequences (Fig. 1C). This analysis grouped the grapevine genes within the FUL-like and the euAP1 clades, respectively (Fig. 1C), for what we refer to as VFUL-L and VAP1, respectively. The existence of a putative euFUL gene cannot be ruled out with our experimental approach.
VFUL-L and VAP1 Are Expressed throughout Grapevine Reproductive Development
The temporal and spatial expression patterns of VFUL-L and VAP1 were analyzed in buds collected during two consecutive years and in developing and mature reproductive structures (Figs. 2 and 3, see “Materials and Methods” and Carmona et al., 2002 for description of developmental stages).
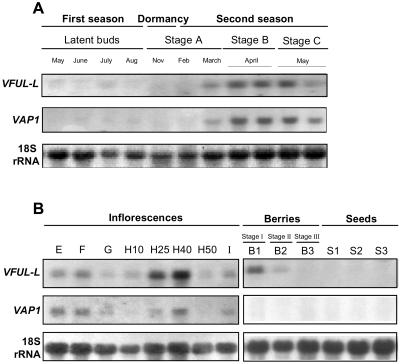
Figure 2.
Expression of VFUL-L and VAP1 during reproductive development studied by RNA-blot. A, Expression of VFUL-L and VAP1 in buds during two consecutive growing seasons. Latent buds in the first growing season, winter buds (stage A) and spring buds (stages B and C) from the second growing season. B, VFUL-L and VAP1 expression in developing inflorescences (phenological stages E, F, G, H10, H25, H40, H50, and I), berries, and seeds. 18S RNA was used as a quantitative control of loading.
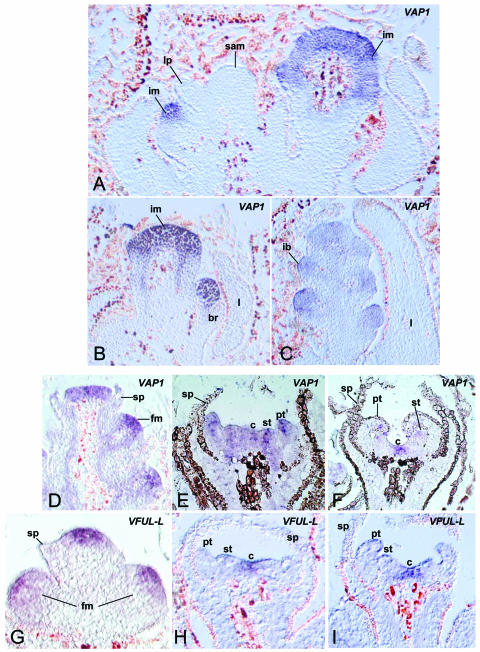
Figure 3.
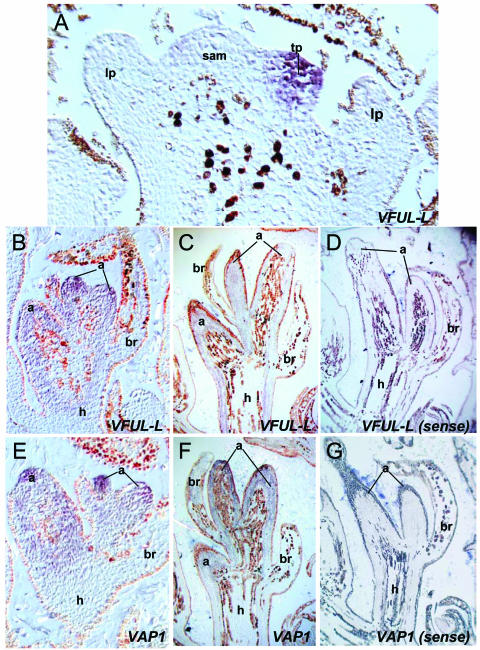
VFUL-L and VAP1 spatial expression patterns during reproductive development. A to C, First season, VAP1 expression during inflorescence development. D to I, Second season, VAP1 and VFUL-L expression patterns during early flower development. A, VAP1 expression in young inflorescence meristems. VAP1 is not expressed in the SAM and leaf primordia. B, Expression of VAP1 in inflorescence meristems. C, Expression of VAP1 in inflorescence branch meristems at a later stage of development. D, Close-up of an inflorescence branch where flower meristems have just formed. VAP1 mRNA accumulates in young flower meristems and becomes excluded from the sepal-forming region. E, VAP1 mRNA preferentially accumulates at the tip of petal, stamen, and carpel primordia during organogenesis. F, VAP1 expression at a slightly later stage of flower development when transcripts are mainly detectable at the prospective carpel-forming region. G, Expression of VFUL-L in young flower meristems during the second season. Notice that this expression is more spatially restricted than that of VAP1 (D). H, VFUL-L expression in flower meristems of a stage similar to E. I, VFUL-L expression in the carpel-forming region of slightly more advanced flowers comparable to F. im, inflorecence meristem; br, bract; lp, leaf primordium; l, leaf; ib, inflorescence branch meristem; fm, flower meristem; sp, sepals; pt, petals; st, stamens; c, carpel.
In buds, the RNA levels of VFUL-L and VAP1 were analyzed by RNA-blot hybridization experiments (Fig. 2A). Transcripts of VFUL-L and VAP1 were barely detected in buds of the first season (Fig. 2A, latent buds) and in dormant buds (Fig. 2A, dormancy). During the first season, the SAM produces lateral meristems that generate inflorescence meristems. During the second season, expression of VFUL-L and VAP1 was already detectable in winter buds (end of stage A). The expression of the two transcripts increased significantly in swelling buds (stage B) and decreased in sprouting buds when shoots were beginning to grow out (stage C, Fig. 2A, second season). During stages A, B, and C of the second season, inflorescence meristems divide to generate additional branch inflorescence meristems, which give rise to flower meristems and flowers.
Expression of both genes was also analyzed in developing inflorescences, berries, and seeds of the growing shoots (Fig. 2B). During inflorescence development, the expression patterns of VFUL-L and VAP1 were very similar, but the levels of VFUL-L transcripts were higher than those of VAP1. Transcripts were detected during flower development (Fig. 2B, stages E-H40). VFUL-L levels were particularly high during stage H when carpel development takes place (Fig. 2B, H25 and H40). The expression of the two genes decreased in mature flowers (Fig. 2B, stages H50 and I). VAP1 expression was not detected during fruit formation and maturation (Fig. 2B, berries and seeds). However, VFUL-L transcripts were detected during stage I of berry development. This expression decreased at stage II and disappeared as the ripening of the berry progressed (stage III). VFUL-L transcripts were not detected in seeds.
To determine the spatial distribution of VAP1 and VFUL-L transcripts at different stages of reproductive development, we carried out in situ hybridization experiments (Fig. 3). The expression patterns of both genes were very similar during the first season when inflorescence meristems are forming (VAP1, Fig. 3, A–C, VFUL-L, not shown), but they diverged during the second season when flower initiation and development takes place (Fig. 3, D–I).
In the first season, VAP1 and VFUL-L were detected from the earliest stages of inflorescence development (Fig. 3, A and B). At later stages, VAP1 and VFUL-L were strongly expressed in the inflorescence branch meristems but not in their subtending bracts (Fig. 3C). In the second season, when flower meristems were initiated, VAP1 and VFUL-L expression patterns diverged. VAP1 was broadly expressed in the newly formed flower meristems (Fig. 3D). As soon as sepal primordia began to grow, VAP1 was excluded from the sepal-forming region and became restricted to the inner part of the meristem that forms the petals, stamens, and carpels (Fig. 3D). Later, when petals and stamen primordia were visible, VAP1 mRNA preferentially accumulated at the tips of these growing primordia (Fig. 3, E and F) and then became mainly restricted to the carpel-forming region (Fig. 3F). On the other hand, VFUL-L was expressed in a small area of the central part of the meristem already in very young floral meristems, (Fig. 3G). During floral organ formation, VFUL-L was not detected in sepal, petal, or stamen primordia but was confined to the prospective carpel-forming region (Fig. 3, H and I).
VFUL-L and VAP1 Are Expressed during Tendril Initiation and Development
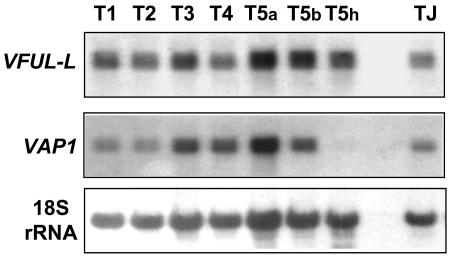
Gene expression was also analyzed during vegetative development by RNA-blot hybridization. VFUL-L and VAP1 transcripts were not detected in the vegetative apices, leaves, or shoots at different stages of cane development (E, G, H50, and I) nor in roots of in vitro grown plants (data not shown). However, high levels of both transcripts were observed in tendrils. To further investigate this expression, three regions were distinguished in tendrils: the hypoclade (h) or basal zone, the branching zone (b) where the hypoclade splits in two arms, and the inner and outer arms (a). First, we analyzed the transcription of both genes in the arms of five consecutive developing tendrils nearest to the shoot apex (Fig. 4, T1–T5a). VFUL-L and VAP1 mRNAs were detected in all five tendrils with highest levels of expression in the most developed one (T5a). Then, we studied the spatial expression of VFUL-L and VAP1 in the three regions of tendril 5 (T5): arms (T5a), branching zone (T5b), and hypoclade (T5h). Expression of VFUL-L was high in the three tendril zones, while that of VAP1 was high in the arms, low in the branching zone, and undetectable in the hypoclade.
Figure 4.
VFUL-L and VAP1 expression during tendril development studied by RNA blot. Expression in the arms of five consecutive tendrils numbered from the shoot apex (T1, T2, T3, T4, and T5a) of an elongated shoot. Expression in three regions of tendril T5: the arms (T5a), the branching zone (T5b), and the hypoclade zone (T5h). Expression in the first formed tendril of a young plant (TJ). 18S RNA was used as a quantitative control of loading.
In situ hybridization experiments confirmed the results obtained by RNA-blot hybridizations. VFUL-L and VAP1 genes were expressed all along tendril development (Fig. 5). VFUL-L and VAP1 were expressed in the lateral meristem as soon as it became distinguishable in the periphery of the SAM (Fig. 5A for VFUL-L, VAP1 not shown). Both genes continued to be expressed throughout tendril development in different spatial patterns. VFUL-L was detected in all the regions of the developing tendrils (Fig. 5, B and C), whereas VAP1 expression was restricted to the tendril arms (Fig. 5, E and F). Transcripts accumulated in the parenchyma but not in the vascular tissues.
Figure 5.
VFUL-L and VAP1 expression in tendrils studied by in situ hybridization. A to C, VFUL-L expression. E to F, VAP1 expression. A, VFUL-L is transcribed in the lateral meristems formed in the second season that will give rise to a tendril but not in the leaf primordia. B to C, VFUL-L is expressed throughout developing tendrils. D, Tendrils of a similar stage hybridized with a sense VFUL-L probe. E to F, VAP1 expression is restricted to the apical region (arms) of the developing tendril. G, Tendrils of a similar stage hybridized with a sense VAP1 probe lp, leaf primordium; tp, tendril primordium; br, bract; a, tendril arm; h, tendril hypoclade.
In Arabidopsis and in other herbaceous and woody species, expression of AP1-and FUL-like genes has been associated with developing reproductive structures. Detection of their grapevine homologs in tendrils, considered to be vegetative climbing structures, raised the question of whether this was a consequence of the maintenance, in tendrils, of an expression induced in inflorescences upon flowering. This could reflect the homology between tendrils and inflorescences, but it might not have a functional relevance. Alternatively, this expression could be independent of the flowering process and have a role on its own. To test whether VFUL-L and VAP1 expression in tendrils was related to the reproductive state of the plant, we analyzed their expression in the earliest formed tendrils of young plants that had not undergone flowering induction. Expression of both genes was already detected in the first formed tendril of plants grown from seeds that would still grow vegetatively for 2 to 5 more years before initiating flowering (Fig. 4, TJ). This indicates that this expression is not dependent on flowering induction but is specifically associated to tendril development. Other genes involved in the flowering process such as VFL (Carmona et al., 2002) or VSOC1, the putative ortholog of SOC1 (Samach et al., 2000), were not expressed at this early stage (data not shown), indicating that these expression patterns were not common to all genes involved in flowering initiation but typical of VFUL-L and VAP1.
DISCUSSION
We have isolated and characterized two grapevine MADS-box genes belonging to the SQUA subfamily or AP1/FUL gene lineage (Theissen et al., 2000; Becker and Theissen, 2003; Litt and Irish, 2003). Conservation of typical protein motifs in the deduced proteins grouped them as members of the FUL-like and AP1/SQUA (euAP1) gene clades, and consequently they were named as VFUL-L and VAP1. In spite of some peculiarities, their expression patterns also support that they are the putative orthologs of previously described FUL-like and AP1-like genes in other plant species.
VFUL-L and VAP1 expression in grapevine is related to reproductive development and spans two growing seasons as previously described for VFL (Carmona et al., 2002). Their expression coincides with the time of flowering induction and inflorescence development during the first season and flower initiation and development during the second season (Srinivasan and Mullins, 1981; Gerrath and Posluszny, 1988b; Gerrath, 1993; May, 2000; Carmona et al., 2002). Such a bimodal pattern of expression has also been observed in other temperate woody perennials like kiwifruit (Walton et al., 2001) and apple trees (Sung et al., 1999; Kotoda et al., 2000). In contrast to the expression of VFL, VFUL-L and VAP1 transcripts were not detected in the SAM or in leaf primordia.
VFUL-L and VAP1 are initially coexpressed in lateral meristems and are maintained at high levels in their derived structures. Coexpression of VFUL-L and VAP1 is in contrast to what has been found in herbaceous species like Arabidopsis and Antirrhinum where AP1-and FUL-like genes display mutually excluding spatial patterns of expression. AP1 and SQUA are specifically expressed in floral meristems (Huijser et al., 1992; Mandel et al., 1992), whereas FUL and DEFH2 are expressed in the SAM after flowering transition (Gu et al.,1998; Müller et al., 2001). Coexpression of both genes in reproductive meristems has also been described in Silene latifolia and Malus domestica (Hardenack et al., 1994; Sung et al., 1999). Since AP1- and FUL-like genes were originated by duplication from a common ancestral AP1/FUL gene (Becker and Theissen, 2003; Litt and Irish, 2003), their similar expression patterns might reflect an ancestral condition related to their common phylogenetic origin.
During flower development, expression patterns of VFUL-L and VAP1 are not coincident. Very early, expression of VFUL-L becomes restricted to the carpel-forming region at the central part of the flower meristem and continues to be expressed at high levels through the early stages of fruit development. This expression pattern suggests that VFUL-L may play a role in carpel and fruit development in a similar way to what has been described for euFUL in Arabidopsis (Mandel and Yanofsky, 1995; Gu et al., 1998). A similar role has been pointed out for DEFH28, another FUL-like gene from Antirrhinum, based on sequence and expression pattern similarities as well as on functional analyses in transgenic Arabidopsis (Müller et al., 2001). These results support the suggestion that euFUL and FUL-like genes might be involved in similar functional roles (Litt and Irish, 2003). On the other hand, expression of VAP1 during grapevine flower development differs substantially from that of AP1 (Mandel et al., 1992). Grapevine VAP1, initially expressed in the young flower meristem, soon becomes excluded from the sepal-forming region. Later, its expression is preferentially detected in the tip of the growing petals and stamens and in the developing carpel. Carpel expression has also been reported for the Antirrhinum SQUA gene (Huijser et al., 1992) and Silene AP1-like gene (Hardenack et al., 1994). In contrast, Arabidopsis AP1 is only expressed in sepal and petal primordia and the phenotype of ap1 mutants suggested that it could be responsible for the A-function in the ABC model (Irish and Sussex, 1990; Coen and Meyerowitz, 1991; Theissen, 2001). The lack of expression of VAP1 in the sepal whorl is not consistent with a function of this gene in the specification of sepal identity. These results are in agreement with previous observations which questioned a role for the Antirrhinum SQUA gene (Huijser et al., 1992) and the Gerbera hybrida AP1 ortholog (Yu et al., 1999) in the specification of sepal identity and provide additional arguments to revise the concept of the A-function in flower organ identity (Litt and Irish, 2003).
The strong and distinctive expression of VFUL-L and VAP1 in developing tendrils and the fact that their expression is independent of the flowering induction suggest a relevant role of these genes in tendril development. This could represent a novel role for these genes that would have been recruited for the development of tendrils in Vitis. Examples of gene recruitment to carry out different developmental functions have been previously described. For instance, UNIFOLIATA (UNI) and PEA FIMBRIATA (PEAFIM), the pea orthologs of Arabidopsis LEAFY and UNUSUAL FLORAL ORGANS, do not only participate in flower initiation and development in pea but are also required for the development of its compound leaves (Gourlay et al., 2000; Taylor et al., 2001). The legume-Rhizobium symbiosis is another example of recruitment of genes for the evolution of a new developmental pathway (Szczyglowski and Amyot, 2003). Among them, several MADS-box genes expressed in flowers, such as Alfalfa nmh7 and ngl9 genes, are also expressed in infected nodule cells (Zucchero et al., 2001) and could have a role in the development of nodules. The expression of VFUL-L and VAP1 in the grapevine lateral meristems and later in tendrils and inflorescences could, on the other hand, reflect an ancestral role of both genes in the specification and proliferation of these meristems and their derivatives, as has been previously proposed for the ancestral AP1 and AG-related gene lineages (Irish, 2003). Moreover, we cannot completely rule out that tendril expression represents a residual expression without functional significance in a structure that has a common origin with reproductive organs.
Transition from juvenile to adult phase in grapevine is marked by the initiation of lateral meristems at the flanks of the SAM. In vegetative growing plants, these structures differentiate as tendrils. However, upon flowering induction, several consecutive lateral meristems generate inflorescence meristems. Which are the genetic bases of this pattern of growth in which equivalent primordia give rise to either reproductive (inflorescence) or vegetative (tendril) structures? We have shown that VFUL-L and VAP1 as well as VFL (Carmona et al., 2002) are expressed in the lateral meristems independently of their fate. However, VFL, VFUL-L, and VAP1 show differential expression in the derived structures. The three genes are highly expressed throughout inflorescence development. However, in tendril primordia, VFL is only transiently expressed, whereas VFUL-L and VAP1 are maintained until late developmental stages. This differential expression of VFL in the two structures might suggest that a threshold level of VFL expression is required for the development of inflorescence and flower meristems instead of tendrils. A threshold level of LFY has also been shown to be necessary to induce flower initiation in Arabidopsis (Blázquez et al., 1997). This could be responsible in part for the generation of two different types of structures from a common primordium. The plasticity of grapevine lateral meristem development could represent an adaptation to a climbing habit based on the conversion of a reproductive organ (the inflorescence) into a climbing organ (the tendril). Additional functional experiments will be required to confirm this hypothesis.
MATERIALS AND METHODS
Plant Material
Grapevine (Vitis vinifera L. var Tempranillo) samples were collected in the fields of Instituto Madrileño de Investigaciones Agrarias (Alcalá de Henares, Madrid). RNA-blot and in situ hybridization analyses were performed on plant materials collected and fixed at different developmental stages during two growing seasons.
Cloning of VFUL-L and VAP1 and Sequence Analyses
Cloning of VFUL-L and VAP1 was performed using a 3′/5′-RACE strategy (Frohman et al., 1988), following the instructions of a commercial RACE kit (Roche Diagnostics GmbH, Mannhein, Germany). 3′-RACE was performed with oligo(dT)-primed single-stranded cDNA synthesized from total RNA of developing buds from phenological stage B (Baggiolini, 1952). Further PCR amplification was performed with anchored primers to the 3′-end and degenerate primers from highly conserved regions of SQUA-like genes. These primers were 5′-AAGAAAGCTCAT/CGAGATCTCT/CGT-3′ from the MADS-box for the first amplification and 5′-ATGGAA/GAGGATA/TCTTGAAC/AGGTATGA-3′ from the highly conserved motif of the I-region (Fig. 1, A and B) for the reamplification. Amplified fragments from two independent experiments were cloned in pGEM-T easy vector (Promega, Madison, WI). Thirty clones corresponding to the 3′ region were sequenced to analyze their sequence diversity. Two different cDNA sequences were found, which, based on sequence similarities, were designated as VFUL-L and VAP1. For 5′-RACE, specific single-stranded cDNAs were synthesized using a VFUL-L or VAP1 specific-primers (5′-ATCTTCTCCCACAAAGTGCCTCAGGTTCCT-3′ and 5′-ATGTTTAGACAGGGAAGCTGCTGTCGCAGT-3′, respectively). Then, terminal transferase was used to add a homopolymeric A-tail to the 3′ end of the cDNAs. Tailed cDNAs were amplified with a VFUL-L specific primer (5′-AGCCAGTTTCCCTGTGAGTCAGGAT-3) or a VAP1 specific primer (5′-ATCTTCCCCCAAAAAGTGCCTTTGGCTTCT-3′), respectively, and an oligo(dT)-anchor primer. The resulting products were reamplified in a second PCR using nested VFUL-L (5′-AGAGAAGCTGCTTCTCTCCGAAAGTGAG-3′) or VAP1 (5′-CAGTCAGCTGCCTCTCTGCATAAGAA-3′) specific primers and an anchor primer. The gene sequences were completed after two successive 5′-RACE experiments, which allowed the isolation of the middle part and the 5′end of the genes. The complete coding region of the cDNAs were obtained by reverse transcriptase-PCR with primers flanking the VFUL-L sequence (5′-AGCAAACCACTCTTCCCACACATA-3′ and 5′-TCGCGTAAAAAATTCTCACCAGG-3′) or the VAP1 sequence (5′-CGCAAAACCAAAAGATGGGAAGAGGT-3′ and 5′-CTATAAGTCCATATTCACATGGAACT-3′), respectively, using Pfu DNA Polymerase (Stratagene, La Jolla, CA). These cDNAs were cloned in pGEM-T easy vector. Six clones of each cDNA were completely sequenced and compared. For sequencing, the Big Dye Terminator Cycle sequencing kit and a sequencer (Prism 377, ABI, Sunnyvale, CA) were used.
Phylogenetic Analyses
To generate a phylogenetic tree, predicted proteins were aligned with ClustalW. Using this original data set, 100 data sets were generated by bootstrap resampling using SEQBOOT program. Distance matrices were made for each bootstrap data set using the PRODIST program-Dayhoff PAM matrix algorithm. The distance matrices obtained were used to construct 100 unrooted trees by the neighbor-joining method using the NEIGHBOR program. A consensus tree was obtained using CONSENSE. SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs belong to the PHYLIP program (Phylogeny Inference Package, version 3.57c, Department of Genetics, University of Washington, Seattle). Arabidopsis SEP1, SEP2, and SEP3 (Pélaz et al., 2000) were used as outgroup members.
RNA-Blot Hybridization Analyses
Plant material was collected from organs and tissues at different developmental stages (phenological stages A–I; Baggiolini, 1952). In the first growing season, young buds in the axils of the leaves (latent buds) were collected at equivalent branch positions from May to August (Fig. 2A, latent buds). For phenological stage A (winter buds), samples were collected in November (dormancy period), February, and March (Fig. 2A, stage A). In the second growing season, swelling buds were collected during early and advanced phenological stages B and C in April and May (Fig. 2A, stages B and C). As the shoot began to elongate, expression was analyzed in inflorescences taken at the stages E, F, G, H, and I. Four samples were collected from inflorescences during stage H (H10, H25, H40, and H50), the number indicating the length of the shoot in centimeters (Fig. 2B, inflorescences). Stages E and F correspond to inflorescences separated from leaves in the shoot and with developing flowers that begin to grow out from beneath protective bracts. Along stage E, sepals develop and petals and stamens primordia become visible. At stages G and H10, the inflorescences are well developed, but not the flowers, which are still present in compact groups. Later in stage H (H25–H50), the inflorescences posses separated flowers which are still undergoing maturation. Development of flower organs span stages E, F, G, and H, being gynoecium initiated last and developing along stage H. Stage I correspond to the beginning of anthesis. During fruit setting and maturation, berries and seeds from stage I to III (Fig. 2B, berries and seeds) were separately analyzed. Stage I correspond to a rapid growth of the berry, stage II to a slow growth and maturation of the seeds, and stage III to the ripening, when the color change or veraison takes place (Mullins et al., 1992). Different organs of the plant from phenological stages E to I were also independently analyzed: shoot apex, young leaves, and shoots. Leaves, shoots, and roots from in vitro grown plants were also tested (data not shown). To analyze expression in tendrils, samples were collected from the arms of the first five tendrils of advanced stage H shoots (Fig. 4, T1–T5). Tendril number 1 corresponds to the last formed by the shoot apex. Tendrils number 5 were dissected in three regions: the inner and outer arms (a), the branching zone (b), and hypoclade zone (h; Fig. 4, T5a, b, and h). In young plants coming from seeds, tendril samples correspond to the first formed tendril (Fig. 4, TJ). Total RNA extraction was performed following the protocol of Chang et al. (1993). For RNA-blot hybridization analyses, 15 μg of total RNA was loaded per lane of agarose/formaldehyde gels, electrophoretically separated, and transferred to Hybond-N+ membranes. To avoid cross hybridization with other MADS-box genes, 3′-end probes excluding MADS-box sequences were used. Filters where hybridized with a 32P-radiolabeled VFUL-L probe (734 bp) or VAP1 probe (720 bp), corresponding in both cases to the 3′end of the gene and the 3′ untranslated region, obtained with specific primers for VFUL-L (5′-TGGAAAGGATCCTTGAACGATATGA-3′ and 5′-AGAAGGCACATGTGCCAAAATA-3′) and for VAP1 (5′-TGGAGAAGATCCTTGATCGCTATGA-3′ and 5′-AATATAAGGGGAAACATCTTAA-3′). Hybridization was performed overnight at 65°C as described by Church and Gilbert (1984). Filters were washed several times in 2× SSC and 0.1% (w/v) SDS and once in 0.1× SSC and 0.1% (w/v) SDS for 20 min at 65°C.
RNA In Situ Hybridization
In situ hybridizations were carried out on plant tissue collected and fixed during two growing seasons at the developmental stages described above. Late stages of flower and fruit development could not be analyzed by this method due to the high levels of background obtained in this plant material. Digoxigenin labeling of RNA probes, tissue preparation, and hybridization were performed as described by Coen et al. (1990). The templates for the VFUL-L and VAP1 riboprobes were the 734-bp and 720-bp fragments, respectively, containing the 3′region of the genes as described above, cloned in pBlueScript KS vector. The hybridized sections were visualized with Nomarski optics in a Leica DMR microscope (DMR, Leica, Wetzlar, Germany).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY538747 and AY538746.
Acknowledgments
We thank Félix Cabello and the Instituto Madrileño de Investigaciones Agrarias (Alcalá de Henares, Madrid) for providing plant material for this research, Gemma Bravo and Elena González for excellent technical assistance, Leonor Ruíz-Garcia for providing material from young grapevine plants, and two anonymous reviewers for their suggestions and constructive criticism.
This work was supported by the Comunidad Autónoma de Madrid (grant no. 07G–0046–2000 and a postdoctoral fellowship to M.C.) and by Ministerio de Ciencia y Tecnología (grant no. BIO2001–3891–C02–02). Support to research activity at Centro Nacional de Biotecnología is provided through a specific agreement of Consejo Superior de Investigaciones Científicas-Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040832.
References
- Baggiolini M (1952) Les stades repères dans le developpement annuel de la vigne et leur utilisation pratique. Rev Romande Agric Vitic Arbor 8: 4–6 [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrándiz C, Cañas LA, Madueño F, Beltrán JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Boss PK, Thomas MR (2000) Tendrils, inflorescences and fruitfulness: a molecular perspective. Aust J Grape Wine Res 6: 168–174 [Google Scholar]
- Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape “green revolution” mutation. Nature 416: 847–850 [DOI] [PubMed] [Google Scholar]
- Bugnon (1953) Recherches sur la Ramification des Ampelidacées. Presses Universitaires de France, Paris
- Butrosse MS (1974) Climatic factors and fruitfulness in grapevines. Hort Abstr 44: 319–325 [Google Scholar]
- Carmona MJ, Cubas P, Martinez-Zapater JM (2002) VFL, the grapevine. FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiol 130: 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Church JM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R (1990) Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Darwin C (1875) The Movement and Habits of Climbing Plants, Ed 2. D. Appleton and Co., New York
- Egea-Cortines M, Davies B (2000) Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci 5: 471–476 [DOI] [PubMed] [Google Scholar]
- Elo A, Lemmetyinen J, Turunen ML, Thika L, Sopanen T (2001) Three MADS-box genes similar to APETALA1 and FRUITFULL from silver birch (Betula pendula). Physiol Plant 112: 95–103 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNA from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrath JM (1993) Developmental morphology and anatomy of grape flowers. Hortic Rev 13: 315–337 [Google Scholar]
- Gerrath JM, Lacroix CR, Posluszny U (1998) Phyllotaxis in the vitaceae. In RV Jean, D Barabe, eds, Symmetry in Plants. World Scientific, Singapore, pp 89–107
- Gerrath JM, Posluszny U (1988. a) Morphological and anatomical development in the Vitaceae. I. Vegetative development in Vitis riparia. Can J Bot 66: 209–224 [Google Scholar]
- Gerrath JM, Posluszny U (1988. b) Comparative floral development in some members of the Vitaceae. In P Leins, SC Tucker, PK Endress, eds, Aspects of Floral Development. J Cramer, Berlin, pp 121–131
- Gocal FW, King RW, Blundell CA, Schwartz OM, Andersen CH, Weigel D (2001) Evolution of floral meristem identity genes: analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol 125: 1788–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay CW, Hofer JMI, Ellis THN (2000) Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA and TENDRIL-LESS. Plant Cell 12: 1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R (1998) The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hardenack S, Ye D, Saedler H, Grant S (1994) Comparison of MADS-box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 6: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lonnig WE, Meijer H, Saedler H, Sommer H (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF (2003) The evolution of floral homeotic gene function. Bioessays 25: 637–646 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd WS, Campbell CS, Kellogg E, Stevens PF (1999) Plant Systematics. A Phylogenetic Approach. Sinauer Associates, Sunderland, MA
- Kieffer M, Davies B (2001) Developmental programmes in floral organ formation. Semin Cell Dev Biol 12: 373–380 [DOI] [PubMed] [Google Scholar]
- Kotoda N, Wada M, Komori S, Kidou S, Abe K, Masuda T, Soejima J (2000) Expression pattern of homologues of floral meristem identity genes LFY and AP1 during flower development in apple. J Am Soc Hortic Sci 125: 398–403 [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1995) The Arabidopsis AGL8 MADS-box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7: 1763–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P (2000) From bud to berry, with special reference to inflorescence and bunch morphology in Vitis vinifera L. Aust J Grape Wine Res 6: 82–98 [Google Scholar]
- Morrison JC (1991) Bud development in Vitis vinifera L. Bot Gaz 152: 304–315 [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller BM, Saedler H, Zachgo S (2001) The MADS-box gene DEFH28 from Antirrhinum is involved in the regulation of floral meristem identity and fruit development. Plant J 28: 169–179 [DOI] [PubMed] [Google Scholar]
- Mullins MG, Bouquet A, Williams LE (1992) Biology of the Grapevine. Cambridge University Press, Cambridge, UK
- Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Pélaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pidkowich MS, Klenz JE, Haughn GW (1999) The making of a flower: control of floral meristem identity in Arabidopsis. Trends Plant Sci 4: 64–70 [DOI] [PubMed] [Google Scholar]
- Posluszny U, Gerrath JM (1986) The vegetative and floral development in the hybrid grape cultivar Ventura. Can J Bot 64: 1620–1631 [Google Scholar]
- Pratt C (1971) Reproductive anatomy of cultivated grapes—a Review. Am J Enol Viticult 22: 92–109 [Google Scholar]
- Pratt C (1974) Vegetative anatomy of cultivated grapes—a Review. Am J Enol Viticult 25: 131–150 [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepción M, Yalovsky S, Gruissem W (1999) Protein prenylation in plants: old friends and new target. Plant Mol Biol 39: 865–870 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42: 899–913 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C (1999) When to switch to flowering. Annu Rev Cell Dev Biol 15: 519–550 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Mullins G (1981) Physiology of flowering in the grapevine—a Review. Am J Enol Viticult 32: 47–63 [Google Scholar]
- Sung SK, Yu GH, An G (1999) Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiol 120: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Amyot L (2003) Symbiosis, inventiveness by recruitment? Plant Physiol 131: 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Hofer J, Murfet I (2001) Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13: 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H (2000) A short history of MADS-box genes. Plant Mol Biol 42: 115–149 [PubMed] [Google Scholar]
- Theissen G, Saedler H (1999) The golden decade of molecular floral development (1990–1999): a cheerful obituary. Dev Genet 25: 181–193 [DOI] [PubMed] [Google Scholar]
- Tucker SC, Hoefert L (1968) Ontogeny of the tendril in Vitis vinifera. Am J Bot 55: 1110–1119 [Google Scholar]
- Walton EF, Podivinsky E, Wu RM (2001) Bimodal patterns of floral gene expression over the two seasons that kiwifruit flowers develop. Physiol Plant 111: 396–404 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 29: 843–859 [DOI] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pöllänen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH (1999) Organ identity genes and modified pattern of flower development in Gerbera hybrida (Asteraceae). Plant J 17: 51–62 [DOI] [PubMed] [Google Scholar]
- Yu H, Goh CJ (2000) Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol 123: 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003) Flower development: initiation, differentiation, and diversification. Annu Rev Cell Dev Biol 19: 119–140 [DOI] [PubMed] [Google Scholar]
- Zucchero JC, Caspi M, Dunn K (2001) ngl9: a third MADS-box gene expressed in alfalfa root nodules. Mol Plant Microbe Interact 14: 1463–1467 [DOI] [PubMed] [Google Scholar]