Abstract
There are very few root genes that have been described in rice as a monocotyledonous model plant so far. Here, the OsRAA1 (Oryza sativa Root Architecture Associated 1) gene has been characterized molecularly. OsRAA1 encodes a 12.0-kD protein that has 58% homology to the AtFPF1 (Flowering Promoting Factor 1) in Arabidopsis, which has not been reported as modulating root development yet. Data of in situ hybridization and OsRAA1∷GUS transgenic plant showed that OsRAA1 expressed specifically in the apical meristem, the elongation zone of root tip, steles of the branch zone, and the young lateral root. Constitutive expression of OsRAA1 under the control of maize (Zea mays) ubiquitin promoter resulted in phenotypes of reduced growth of primary root, increased number of adventitious roots and helix primary root, and delayed gravitropic response of roots in seedlings of rice (Oryza sativa), which are similar to the phenotypes of the wild-type plant treated with auxin. With overexpression of OsRAA1, initiation and growth of adventitious root were more sensitive to treatment of auxin than those of the control plants, while their responses to 9-hydroxyfluorene-9-carboxylic acid in both transgenic line and wild type showed similar results. OsRAA1 constitutive expression also caused longer leaves and sterile florets at the last stage of plant development. Analysis of northern blot and GUS activity staining of OsRAA1∷GUS transgenic plants demonstrated that the OsRAA1 expression was induced by auxin. At the same time, overexpression of OsRAA1 also caused endogenous indole-3-acetic acid to increase. These data suggested that OsRAA1 as a new gene functions in the development of rice root systems, which are mediated by auxin. A positive feedback regulation mechanism of OsRAA1 to indole-3-acetic acid metabolism may be involved in rice root development in nature.
The growth and development of crops depend on their roots to take up water and nutrient material from soil. The root system of rice (Oryza sativa) consists of embryonic and postembryonic roots. The rice embryonic roots, originating from the radical, emerge after germination. They develop into two forms, one primary root and several seminal roots. Postembryonic roots include adventitious roots that are formed from nodes of the plant and lateral roots that develop on all root types.
As is known, plant hormone auxin (indole-3-acetic acid [IAA]) plays an important role in controlling root development, such as inhibiting elongation of a primary root and promoting formation of adventitious roots, lateral roots, and root hairs. In Arabidopsis, it has been reported that genes involved in the auxin signal transduction pathway are able to control the development of roots. For example, mutants of the AUX/IAA gene family, such as SHY2/IAA3, SLR1/IAA14, IAA28, and MSG2/IAA19, show reduced lateral roots or no lateral roots (for review, see Reed, 2001). Overexpression of AUX1, TIR1, or NAC1 can promote lateral root development (Gray et al., 1999; Xie et al., 2000; Marchant et al., 2002). Overexpression of SINAT5 produces fewer lateral roots, whereas overexpression of a dominant-negative C49S (Cys-49 to Ser) mutant of SINAT5 develops more lateral roots. The lateral root phenotypes correlate with the expression of NAC1 (Xie et al., 2002). As a central regulator, auxin regulates cell division, expansion, and differentiation. It also functions in many other aspects of plant growth and development, such as promoting hypocotyl and stem elongation, mediating root and shoot gravitropism, maintaining apical dominance, and promoting vascular pattern formation (Thimann, 1977; Sachs, 1991; Estelle and Klee, 1994; Hobbie, 1998).
Auxin is possibly synthesized in young leaves and root meristem (Thimann, 1977) and can be transported from young leaves to the roots. In the root tip, it is transported from down to up in the cortex (Doerner, 2000). The polarity in transportation causes the gradient of auxin concentration, which then contributes not only to embryonic patterning and vascular differentiation (Goldsmith, 1977; Thimann, 1977; Sachs, 1991; Liu et al., 1993; Przemeck et al., 1996; Uggla et al., 1998; Sabatini et al., 1999), but also to the cell-patterning process within the root meristem (Sabatini et al., 1999).
In Arabidopsis, many genes, such as the AUX/IAA family, the SAUR (small auxin up-regulated RNA) family, and the GH3 family (Hagen et al., 1984; McClure and Guilfoyle, 1989; Abel and Theologis, 1996), are regulated by auxin. Some auxin-regulated genes show tissue-specific expression patterns (Key, 1989; Crowell and Amasino, 1994; Gray et al., 1998). A sequence motif of 5′-TGTCTC-3′, which is necessary for auxin response factor binding, is found in promoters of many auxin-regulated genes (Liu et al., 1994; Ulmasov et al., 1997a, 1997b, 1999a, 1999b; Guilfoyle et al., 1998). Previous studies have confirmed that auxin can give auxin response a feedback regulation by inducing SCFTIR1-dependent degradation of AUX/IAA proteins (del Pozo and Estelle, 1999; Gray et al., 1999, 2001). Moreover, the auxin signal transduction pathway may be conserved between monocot and dicot plants. Eleven rice genes that encode the homologous protein sequences to the auxin response factors of Arabidopsis were isolated (Sato et al., 2001). OsIAA1, a monocot member of the AUX/IAA family, was demonstrated to be regulated by auxin and light in rice (Thakur et al., 2001).
In monocot plants, the molecular mechanism that establishes the morphology of root systems is blurry so far. Only a small number of mutants related to root formation were found in monocots. For example, rtcs mutant is completely devoid of all adventitious roots (Hetz et al., 1996); rt1 mutant forms few or no crowns and brace roots (Jenkins, 1930); asr1 mutant forms defective seminal roots (De Miranda, 1980); and slr1 mutant and slr2 mutant display short lateral roots (Hochholdinger et al., 2001). Although the genes corresponding to these mutants have not been cloned, studies of the characters of mutants are useful to understand the mechanism of grass root development. Concerning effects of ethylene on the development of adventitious roots, it has been proposed that submergence or treatment with ethylene can induce the growth of adventitious roots of deepwater rice (Vergera et al., 1976; Suge, 1985; Bleecker et al., 1986, 1987). So far, genes related to root development in rice have not yet been studied thoroughly.
In this paper, we used a reverse genetics approach to study the functions of a novel auxin-induced gene regulating root development in rice. Based on the database of expression sequence tag (EST) and genomic sequences, a cDNA and its promoter sequence were cloned. Its expression patterns and phenotypes of overexpression in transgenic plants were analyzed.
RESULTS
OsRAA1 Gene Encodes a Small Protein and Is Conserved in Plants
Based on the EST database, an unknown small protein gene corresponding to an EST (AU071162) was identified in rice (BAB07982). A PAC clone (P0462H08) containing the EST was screened from the database of DDBJ/GenBank/EMBL. The RACE strategy was utilized to amplify the full-length cDNA in rice.
The full-length cDNA consists of 725 nucleotides containing a 327-nucleotide open reading frame (ORF). The 5′-upstream untranslated region is 82-bp long, and the 3′-downstream untranslated region is 339-bp long. The ORF was predicted to encode a 109-amino acid polypeptide with a calculated molecular mass of 12 kD and a pI of 9.45 (Fig. 1A). There are three amino acids that are slightly rich in protein: Ser, Val, and Leu. It was named OsRAA1 (Oryza sativa Root Architecture Associated 1) since our data suggested that its functions are related to root morphology development in rice. Comparative analysis between sequences of the cDNA and those of the PAC clone (P0462H08) at chromosome 1 in rice suggested that the OsRAA1 gene has no intron.
Figure 1.
The analysis of the OsRAA1 gene and its protein sequence. A, Nucleotide and putative protein sequence of OsRAA1 cDNA. B, Protein sequence multiple alignment of OsRAA1. BaLeP1 (BE421936), BaSpP1 (BG342901), and BaSpP2 (BE196402) are from barley leaf and spike; MuFPF1 (Y11987) is from mustard; WhRoP1 (BE428690) and WhRoP2 (BE428819) are from wheat root; ChrFPF1 (BE034410) and ChrFPF2 (BE033961) are from chrysanthemum; MossFPF1 (AW699964) is from moss (Physcomitrella patens); AtFPF1 (Y11988) is from Arabidopsis; ZeaTaP2 (BE552830) is from maize. C, Protein multialignment of rice putative OsRAA1/FPF1 family genes. OsRAAL1 (OsRAA like) is from rice contig AAAA01001788.1, which contains EST AU070455; OsRAAL2 is from rice chromosome 7 (AP003982). D, The phylogenetic tree of the FPF1/OsRAA1 gene family processed by software DNAMAN. In the phylogenetic tree at bottom, the numbers of amino acid substitutions per alignment site are indicated on the branches. E, OsRAA1 promoter structure. TAACAAA/TTTGTTA is a gibberellin response element; TGTCTC is an AuRE. Bar = 100 bp.
The results of BLAST in the GenBank showed that OsRAA1 shared a 58% sequence homology of amino acids with AtFPF1 (Flowering Promoting Factor 1; Y11988). A series of ESTs, including MossFPF1 (AW699964) from moss (Physcomitrella patens); ChrFPF1 (BE034410) and ChrFPF2 (BE033961) from chrysanthemum root; MuFPF1 (Y11987) from mustard; ZeTaP1 (BE552830) from the cDNA library of maize tassel primordia; WhRoP1 (BE428690) and WhRoP2 (BE428819) from the wheat root library; and BaLeP1 (BE421936), BaSpP1 (BG342901), and BaSpP2 (BE196402) from the barley leaf library and spike library, were also obtained from the database (Fig. 1, B and D). The analysis showed that homologous genes of OsRAA1 exist ubiquitously from the lower plant (such as moss) to the higher plant (such as chrysanthemum). It indicates that this gene family may play important roles in plant development.
Comparative alignments analysis of the OsRAA1 sequence suggested that four conserved domains presented in these proteins. The first motif is -GVWV/IF- in the N-terminal part; the second one is -GWERYY- in the middle part; and the third and fourth ones are -DLIS/ALP- and -H/YMYDI/VVV/I- in the near C-terminal part, respectively (Fig. 1C). Interestingly, the conservative domains except for the second one appear in the hydrophobic region on the hydrophobicity plot (data not shown). Besides that, there were seven Leu conserved.
There are at least three members of this gene family in Arabidopsis (Y11988, T04505, and T49976). In rice, there is another EST (AU070455) shared sequence with OsRAA1. The complete sequence corresponding to the EST was obtained from the GenBank (contig AAAA01001788.1). At rice chromosome 7 (AP003982), there is a fragment of genomic sequence, which can also be deduced to a protein with high homology with OsRAA1 (Fig. 1C). These three sequences in rice belong to three different subfamilies (data not shown).
Phylogenetic analysis of the AtFPF1/OsRAA1 gene family indicated that homologs were divided into three branches. It also showed there are different members expressed even in the same organ (Fig. 1D). For example, there are two ESTs (WhRoP1 and WhRoP2) from the wheat root library, and they are located at different branches of the phylogenetic tree. Another two ESTs (BaSpP1 and BaSpP2) are in the barley spike library, which belong to a different subfamily. OsRAA1 is located in a branch that was different from those of AtFPF1.
The OsRAA1 promoter sequence of 1,987 bp was isolated and analyzed. Elements of GA and auxin response were involved in the promoter region. The conservative GA response complex consists of pyrimidine box (C/TCTTTTC/T), GA response element (TAACAAA), and element of TATCCA. Those elements appeared in the promoter region (Fig. 1E), but it was a very weak response to GA3 treatment (data not shown). There is an auxin response element (AuRE) core sequence 5′-TGTCTC-3′ located at −150 to −145 (Fig. 1E). In reverse orientation, another AuRE (5′-CTCTGT-3′) is located at −158 to −153. The presence of two AuRE motifs suggested that the OsRAA1 gene is probably regulated by auxin.
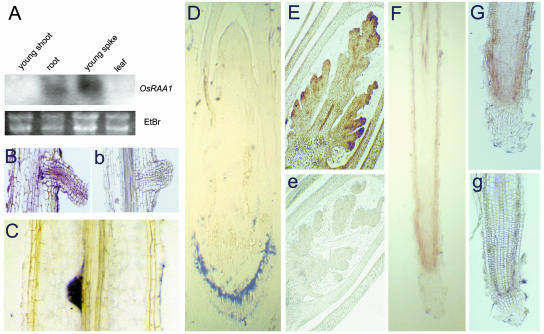
The expression pattern of OsRAA1 was investigated as to its localization. Northern analysis showed that the OsRAA1 mRNA was specifically transcripted in the organs of roots and spikes (Fig. 2A). No signal was detected in either young shoots or leaves. Data of RNA in situ hybridization showed a more distinct expression pattern. As shown in Figure 2, F and G, signals of OsRAA1 were detected in the root apex, including quiescent center and dividing cells. Stronger signals were shown in the cortex of root apical meristem and pericycle of root apex. On the contrary, no distinct signal could be detected in the cortex of the mature zone. A transcript of OsRAA1 was also present in lateral roots, especially in the lateral root primordia and the pericycles of the branch zones (Fig. 2, B and C). In situ hybridization also showed that OsRAA1 mRNA is expressed in the apical meristem of young spikes (Fig. 2E). Besides that, there is a distinct signal in the collenchyma cells of margin vascular bundles between shoot and roots (Fig. 2D). As is known, the adventitious roots are differentiated from these collenchyma cells (Fujii, 1959).
Figure 2.
OsRAA1 expression patterns. A, OsRAA1 northern blot of RNA from tissues of young shoots, roots, young spikes, and leaves in wild-type plants. Young spikes were harvested from 70-d-old plants grown in a growth chamber. The remaining materials were from plants grown for 14 d. Ethidium bromide (EtBr) staining shows equal RNA loading. B to G, In situ localization of OsRAA1 transcript in wild-type rice with an antisense probe. B, Longitudinal section of young root with lateral root. C, Longitudinal section of young root with lateral root primordium. D, Longitudinal section of young shoot, including the junction part between shoot and roots. E, Longitudinal section of young spike. F, Longitudinal sections of root tip. G, Details of young root meristem zone. b, e, and g are the control sections with a sense probe of OsRAA1 corresponding to B, E, and G sections, respectively. C and D photos were taken after mounting of slides using resin, so signals are blue. The remaining photos were taken before mounting of the slide, therefore, signals are brown.
To further confirm the expression patterns, an expression vector of GUS (β-glucuronidase) driven by OsRAA1 promoter was constructed and transformed into rice plants. The transgenic plants showed the patterns of GUS staining in the roots, which are similar to the data of in situ hybridization described above. Figure 3A showed that strong signals were detected in lateral roots and their primordia. It was clear that there were staining signals of GUS activity in the division and elongation zones of the root apex, while there was no signal detected in root cap (Fig. 3B). GUS activity appeared in the palea/lower palea vascular strands (Fig. 3C). The conjugated part between anther and filament also showed a strong signal in the GUS transgenic plant driven by the OsRAA1 promoter (Fig. 3E). Although a few signals appeared at some parts of young leaves, they were irregular and unstable (Fig. 3D).
Figure 3.
The localization of OsRAA1∷GUS gene expression in the transgenic rice. A, A primary root with lateral root. B, Apical region of a primary root. C, Mature spikelet. D, Young shoot with some young leaves. E, Floral organs of a spikelet. Arrow is a morphogenesis site of lateral root with GUS activity.
Therefore, mRNA of OsRAA1 is present in the rapidly growing zones, which indicates its possible roles in the cell growth and/or division. Interestingly, its expression pattern in the root was similar to that of AUX1 (Marchant et al., 2002) and TIR1 and NAC1 in Arabidopsis (Gray et al., 1999; Xie et al., 2000), which are involved in auxin transport or signal transduction pathway.
OsRAA1 Overexpression in the Root Results in Reduced Growth of Primary Root, Increased Formation of Adventitious Roots, and Helix Roots
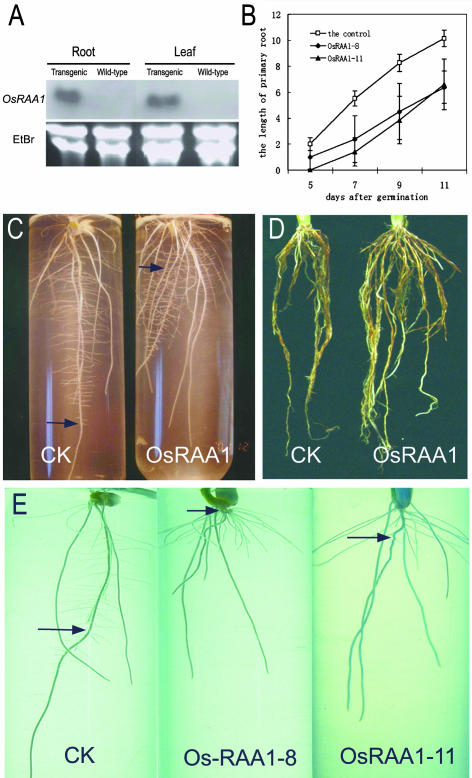
There were 20 independent Ubi∷OsRAA1 transgenic lines with characters of a tolerance to hygromycin and a positive staining of GUS marker to be obtained from the different transformed rice callus in the experiments. Longer flag leaves of the transgenic plants were a clear phenotype in comparison with the wild-type control without a hygromycin treatment. To investigate its role in root development, the seeds of 12 independent transgenic lines with the longest flag leaves were harvested. More than 50 seeds of these lines were germinated in the half-strength MS medium with 75 mg L−1 hygromycin, respectively. Leaves of these seedlings were cut and stained with 5-bromo-4-chloro-3-indolyl-β-d-GlcUA (X-Gluc). Seedlings with positive staining signals were chosen for analysis of phenotypes (data not shown). The transgenic plants of Ubi∷OsRAA1 with positive GUS staining also showed a stronger hybridization signal either in leaf or root before any signal appeared in the untransformed control in the northern-blot analysis (Fig. 4A). Results of the northern blot suggested that OsRAA1 displayed constitutive overexpression patterns in transgenic plants. As shown in Table I and Figure 4, C to E, the transgenic plants had more adventitious roots than the untransformed control plants under the same growth conditions. When the seedling grew for 9 d, a transgenic plant had two more roots than the wild-type control on the average. Moreover, roots of the transgenic plants were almost longer by 0.5 cm. At the same time, primary roots in transgenic rice were shorter than that of the wild-type control (Table I). It was also observed that although the lateral roots of primary roots in the transgenic plants were shorter than those of the wild-type control plant, the lateral roots of the remaining roots were much longer (Fig. 4C). This suggests that OsRAA1 may promote formation of the lateral roots. When the plants developed to the heading stage, the transgenic plants of Ubi∷OsRAA1 had grown more roots than the wild-type control (Fig. 4D). Besides that, another clear root phenotype with a helix appeared in the transgenic plant. In representative lines, such as OsRAA1-8 and OsRAA1-11, the primary roots formed a helix to various extents, while the wild-type plants had a normal primary root under the same conditions (Fig. 4E). The phenotypes, such as reduced growth of primary root and increased adventitious roots and lateral roots, are very similar to that of overexpression of TIR1 and NAC1 in Arabidopsis (Gray et al., 1999; Xie et al., 2000).
Figure 4.
The expression pattern of OsRAA1 and the phenotype of root in the Ubi∷OsRAA1 overexpression transgenic rice. A, Northern-blot analysis for a representative Ubi∷OsRAA1 transgenic plant to show an overexpression. Ethidium bromide (EtBr) staining shows equal RNA loading. B, Statistical analysis of primary root growth in Ubi∷OsRAA1 transgenic plant. More than 10 seedlings were examined for each point. C, Roots of 12-d-old seedlings grown in the half-strength MS medium. CK, control. OsRAA1, Representative of Ubi∷OsRAA1 transgenic line. D, Mature roots at flowering stage. E, The wild-type plant roots (CK), transgenic line 8 (OsRAA1-8), and transgenic line 11 (OsRAA1-11) at 9 d after germination in the half-strength MS medium. Arrow is a primary root.
Table I.
The comparative analysis of root numbers between the transgenic plants and the wild-type control
| Wild-Type Control | Ubi∷OsRAA1 | P | |
|---|---|---|---|
| Root numbers | 6.08 ± 1.56 | 7.82 ± 2.48* | 0.0064 |
| Root numbers (>0.5 cm) | 4.17 ± 1.71 | 6.91 ± 2.67* | 1.35E–4 |
Asterisks indicate the significance of difference between the control and the Ubi∷OsRAA1 transgenic plant populations as determined by repeated-measures analysis of variance (two samples t test proceeded by Origin 6.0; P < 0.05). The numbers are the mean ± se.
Constitutive Expression of OsRAA1 Results in Longer Leaves and Filaments
When the transgenic plants of Ubi∷OsRAA1 grew for 2 weeks after germinating from a solution of hygromycin (75 mg L−1), they were transferred into the soil and then cultured in a greenhouse. The OsRAA1 gene expressed constitutively in leaf and root in the transgenic plants. During the first month after transfer, the transgenic plants grew more slowly than the wild-type controls without a hygromycin treatment. But just before the booting stage, the leaves of the transgenic plant were longer than those of the wild-type control. Especially at the heading stage, the flag leaves of the transgenic plants were distinguished as longer than those of the control plants. As shown in Table II and Figure 5A, the flag leaves of the transgenic plants were about 44 cm (up to 60 cm in some independent lines) in length on the average, while those of the wild-type controls were about 31 cm. The leaf length of transgenic plants was 1.4 times longer than the control. The results of a scan electronic microscope showed that silica cell length of the transgenic plant was longer than that of the control, which was supported by statistical tests (P < 0.05; Fig. 5D). There were five silica cells in the limited area in the flag leaf of the transgenic plants on the average, while six cells appeared at the same size area in the wild-type control (Fig. 5, B and C). In other words, the cell length of transgenic plants was 1.2 times larger than the wild-type control based on the scan electronic microscope data. The difference between the wild-type control and the transgenic plant reached a significant level in statistics (P < 0.05; Table II). It was suggested that the length increase of flag leaves in the transgenic plant might be caused by both extension of cells and an increase in cell numbers. Extension of cells, however, contributed more than increase of cell numbers to leaf extension in transgenic plants.
Table II.
The comparative analysis of leaf length between the transgenic plants and the wild-type control
| Wild-Type Control | Ubi∷OsRAA1 | P | |
|---|---|---|---|
| Length of a flag leaf (cm) | 31.64 ± 4.79 | 44.53 ± 8.13* | 4.76E–10 |
| Length of a last second leaf (cm) | 49.38 ± 8.41 | 58.96 ± 5.03* | 2.63E−5 |
Asterisks indicate the significance of difference between the control and the Ubi∷OsRAA1 transgenic plant populations as determined by repeated-measures analysis of variance (two samples t test proceeded by Origin 6.0; P < 0.05). The numbers are the mean ± se.
Figure 5.
The phenotypes of Ubi∷OsRAA1 constitutive expression transgenic rice plant. A, Flag leaf of wild-type control plant (left) and the Ubi∷OsRAA1 transgenic plant (right). B, Silica cells of flag leaf of wild-type control plant. C, Silica cells of flag leaf of the transgenic plant. D, Statistical analysis of the length of silica cells in the flag leaves (P < 0.05; two samples t test proceeded by Origin 6.0). More than 50 cells were examined for each bar. E, Flowers of wild-type control plant. F and G, The transgenic plant with different lines (D–F) to show longer filaments.
Abnormal florets appeared in the spikes of the transgenic plant (Fig. 5, F and G). The yellow and plump anthers of the wild-type control plant were just above penniform carpels at that stage (Fig. 5E). Conversely, stamens of the transgenic plants with OsRAA1 constitutive expression resulted in abnormally longer filaments with white and shrunken anthers (Fig. 5, F and G). Actually, some filaments were too long to be peeled off from the palea; they curved and attached tightly to the palea.
Auxin Regulates the Transcription of the OsRAA1 Gene
Based on the AuRE sequence in OsRAA1 promoter, the expression patterns in roots, and the phenotypes of transgenic plants with constitutive expression of OsRAA1, we hypothesize that expression of OsRAA1 gene is regulated by auxin. Northern blot showed that the OsRAA1 mRNA was increased from treatment of 10−5 m IAA. More than double expression was induced by the treatment of 10−4 m IAA (Fig. 6A). The GUS staining data of the OsRAA1∷GUS transgenic plant showed that auxin induced a stronger signal in the transgenic plant, while a weaker blue signal appeared in the untreated control (Fig. 6B). Suppression of the inhibitor of auxin polar transport on GUS staining clearly occurred as the treatment time went on (Fig. 6B). The inhibitor experiment may support an explanation that decrease of endogenous auxin weakened GUS expression-driven OsRAA1 promoter. The results suggested that expression of the OsRAA1 gene in nature is induced by auxin.
Figure 6.
OsRAA1 expression is regulated by auxin. A, Northern-blot analysis for OsRAA1 expression treated with IAA of different concentration. Ethidium bromide staining (EtBr) shows equal RNA loading in the blot. The bar shows a relative amount of the gene expression at the bottom. B, Effect of GUS staining on treatment of 1 μm NAA (no. 2) and water as a control (no. 1) and on treatment of the auxin polar transport inhibitor (HFCA, 0.5 μm) for 0, 1, 2, and 4 h (corresponding to nos. 3, 4, 5, and 6, respectively). Data of auxin (nos.1 and 2) and HFCA (nos. 3–6) were from different experiments. C, Roots of wild-type plant (14 d). D, Effect of NAA (1 μm) on root growth of the wild-type plant. The seedling was incubated for 14 d after germination. Arrows show primary roots.
The effect of auxin on root development was observed (Fig. 6). Interesting phenotypes occurred in the wild-type roots treated with 1-napthaleneacetic acid (NAA). Growth of the roots was strongly inhibited by auxin as compared with untreated wild-type control (Fig. 6, C and D; Table III). The numbers of adventitious roots, however, increased up to 11 in a treated seedling plant. In the untreated control, there were five adventitious roots under the same conditions (Table III). All of the treated plants had helix-like primary roots (Fig. 6D). The phenotypes were similar to those status in tomato that auxin stimulated lateral root growth at a concentration at which primary root growth was inhibited (Muday and Haworth, 1994).
Table III.
Effect of auxin (1 μm NAA) on root development in rice
| Control | Treatment | P | |
|---|---|---|---|
| Number of adventitious roots | 6.3 ± 1.2 | 11.4 ± 1.8* | 3.86E–6 |
| Length of a primary root (cm) | 5.6 ± 0.8 | 0.9 ± 0.3* | 5.35E–10 |
Asterisks indicate the significance of difference between treatment group and the control as determined by repeated-measures analysis of variance (two samples t test proceeded by Origin 6.0; P < 0.05). The numbers are the mean ± se.
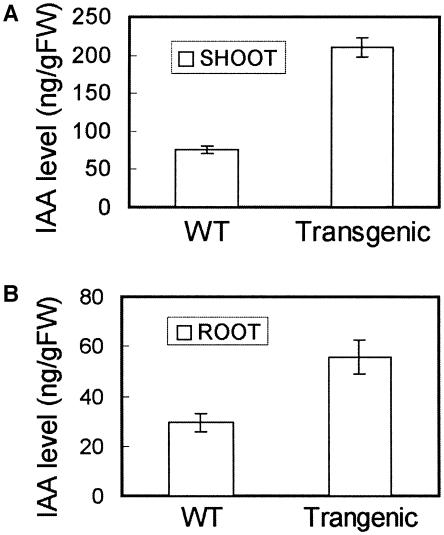
In the Ubi∷OsRAA1 transgenic plant, results of auxin treatment showed that a promotion role on the adventitious root initiation induced by IAA was more distinguished than that in the wild-type control (Fig. 7A). However, the adventitious root initiation responses to 9-hydroxyfluorene-9-carboxylic acid (HFCA), a polar auxin transport inhibitor, between the transgenic plants and the wild-type control exhibited similar phenotype (Fig. 7B). Results of auxin level determined showed that IAA level either in shoot or root was obviously higher in the transgenic plant of Ubi∷OsRAA1 than that in the wild- type control (Fig. 8). And IAA of the transgenic shoot was 2.8 times greater than the wild-type control, while those of the root were 1.9 times greater. It suggested overexpression of OsRAA1 resulted in accumulating a higher level of IAA in the transgenic plant.
Figure 7.
Effects of auxin and HFCA on adventitious root in the OsRAA1 transgenic rice plants. A, Treatment of IAA. B, Treatment of HFCA, auxin polar transport inhibitor.
Figure 8.
Auxin levels of the OsRAA1 overexpression transgenic plant and wild type. A, IAA level in shoot. B, IAA level in root. The roots and shoots used were selected 9 d after germination.
It is well known that gravitropism is a phenomenon mainly regulated by auxin. The seedlings were reoriented horizontally in the darkness to check their gravitropic response. Until reoriented for 3 h, any gravitropic response of the overexpression line was not detected, while the wild-type control showed a distinct response (Fig. 9, A, B, and D). In fact, the transgenic lines with extreme overexpression completely lost gravitropic response until reoriented for 20 h. The kinetics of gravitropic curvature showed a vigorous graviresponse from 3 h after gravistimulation in the transgenic plant (Fig. 9D). Results in Figure 9, C and D, showed that treatment of IAA obviously deferred gravitropic response in the wild-type control, which was similar to that in maize and Arabidopsis (Ishikawa and Evans, 1993; Yamamoto and Yamamoto, 1998; Kim et al., 2000; Ottenschlager et al., 2003). The response phenotype of the overexpression line was identical to those of treatment of IAA in the wild type. These results suggest that the constitutive expression of OsRAA1 might disturb the auxin distribution and reduce gravitropic response in rice roots.
Figure 9.
Overexpression of OsRAA1 inhibited gravitropism. Root gravitropic response treated with horizontal reorientation for 0 h, left, and 3 h, right (A–C). A, Wild-type control. B, Overexpression Ubi∷OsRAA1 transgenic seedling. C, Wild-type seedling treated with IAA (0.4 μm). D, Time course of gravitropic response (more than 20 plants were examined for each point).
DISCUSSION
The OsRAA1 gene belongs to a novel gene family that appears to be conserved in plants from lower to higher. Members of this gene family in both Arabidopsis and rice have no introns in their genomic sequences (Fig. 1; Kania et al., 1997). These characters are similar to those of the SAUR gene family. SAUR family genes have no introns and form gene clusters in their genomic sequence. Most of them encode about 10 kD, a small protein. Their expression can be regulated by auxin (McClure and Guilfoyle, 1989). The OsRAA1, however, has no obvious homology to the members of the SAUR family. So the OsRAA1 may belong to a new small protein family, which can also be regulated by auxin.
FPF1 protein was firstly studied as a flowering promoting factor in mustard (Melzer et al., 1990; Kania et al., 1997). It was indicated that FPF1 is involved in the GA-dependent signaling pathway (Kania et al., 1997), and it may work synergistically with AP1 and LFY to modulate the competence to flowering in the shoot apical meristem (Melzer et al., 1999). It can promote flowering in Arabidopsis (Kania et al., 1997). OsRAA1 did not obviously modulate flowering time in rice (data not shown), although it shared sequence homologous with AtFPF1 (Fig. 1), which promotes flowering in Arabidopsis. There are a few reports concerning function on root development of flowering regulated gene. Known MADS box genes were involved in the control network of flower development. It may be a report only by Zhang and Forde (1998) that the MADS gene functions in signal transduction in root development. Constitutive expression of OsRAA1 could result in the longer leaves and filaments that appeared at the last stage of rice development (Fig. 5), but the young leaves of transgenic plants had no obvious changes at the earlier stage. Unlike AtFPF1, OsRAA1 expression was not obviously induced by GA treatment (data not shown), although GA-response elements appeared in the promoter sequence region (Fig. 1).
The expression pattern shows that OsRAA1 transcript was always in rapidly growing cells, such as primordia of the lateral roots, the steles of young adventitious roots and lateral roots, the meristem, and the division zone of the root apex (Figs. 2 and 3). Cells of these tissues generally undergo rapid division and elongation, and auxin plays important roles in these processes (Boerjan et al., 1995; Celenza et al., 1995; Casimiro et al., 2001). OsRAA1 may contribute to cell extension and division, which are mediated by microtubule in the cell (Fig. 5; Konishi and Sugiyama, 2003). Moreover, the expression pattern of OsRAA1 in roots was similar to the pattern of GUS expression driven by DR5 promoter when treated with exogenously applied auxin (Ulmasov et al., 1997b). Actually, the expression pattern of OsRAA1 (Figs. 2 and 3) also showed auxin distribution in the root (Marchant et al., 2002).
OsRAA1 gene expressed mainly in root and spike (Fig. 2). The putative auxin influx carrier AUX1 modifies root architecture to promote lateral root formation (Marchant et al., 2002). TIR1, the component of the SCF ubiquitin-ligase complex responsible for ubiquitin-mediated protein degradation induced by auxin, is also expressed with a similar pattern in the root (Gray et al., 1999). And NAC1, which mediates auxin signal transduction downstream of TIR1 and promotes lateral root development, also shows the same expression pattern in the Arabidopsis root (Xie et al., 2000). Actually, the OsRAA1 expression pattern has a few differences to AUX1, which has been approved as putative auxin influx carrier. Expression signal of AUX1 gene can be detected in a subset of columella, lateral root cap, and stele tissues of root apex (Marchant et al., 2002; Swarup and Bennett, 2003), while there was no detectable signal of OsRAA1 gene expression in root cap (Fig. 2). As is known, root formation regulated by auxin is particularly sensitive to a variety of changing growth conditions. Adventitious root initiation and growth of the plants with overexpression of OsRAA1 were more sensitive to treatment of auxin than the control plants, while their responses to HFCA in both transgenic line and the wild type showed few differences (Fig. 7). So it is possible that OsRAA1 is dependent on auxin polar transport in nature.
Our data in northern blot (Fig. 6A) and OsRAA1 promoter-GUS transgenic plants (Fig. 6B) suggested that the expression of OsRAA1 can be induced by auxin. The OsRAA1 gene expressed constitutively with higher efficiency in the transgenic plants (Fig. 4). Auxin regulates initiation and growth of root in diverse plants (Muday and Haworth, 1994; for review, see Bernasconi, 1998; Oono et al., 2003; Tatematsu et al., 2004). The wild-type plants treated with auxin exhibited similar phenotypes to those expressing OsRAA1 constitutively in root development (Figs. 4 and 6). The phenotypes of reduced primary root elongation and increased adventitious roots in transgenic lines (Fig. 4) are similar to auxin effects (for review, see Bernasconi, 1998). Response of the Ubi∷OsRAA1 transgenic plants to treatment of IAA was more sensitive than the wild-type plants. Higher level of IAA occurred in the transgenic line (Fig. 8). So it is likely that a positive feedback of OsRAA1 was involved in IAA metabolism.
Constitutive expression of OsRAA1 could also delay the gravitropic response of roots. Some extreme transgenic plants showed no response at all. In Arabidopsis, gravitropic curvature of IAA-treated roots was close to zero, despite a slight but significant downward curvature. Roots on 2,4-D showed no or even tenuous upward curvature (Ottenschlager et al., 2003). The reports have described the relative effectiveness of auxins IAA, NAA, and 2,4-D on inhibition of root gravitropism in Arabidopsis plants (Yamamoto and Yamamoto, 1998). As is known, several models were proposed for the mechanism of gravitropism, such as the statolith hypothesis and the plasmalemma central control model (for review, see Bernasconi, 1998). Although the mechanisms to perceive gravity are different, there is a common central principle that gravity triggers polar transport of IAA to the low side of the root in the cortex of elongation zone and inhibits cell elongation on that side. OsRAA1 may be involved in the cell elongation since OsRAA1 is indeed regulated by auxin. Different distribution of auxin triggered by gravity in the cortex cells may cause the different expression level of OsRAA1. Root formation and development is particularly sensitive to a variety of changing auxin levels. Enhanced and constitutive expression of OsRAA1, however, disturbs regulation of endogenous IAA level and delays the gravitropic response in transgenic plant (Figs. 8 and 9). Both auxin and overexpression of OsRAA1 delay initial gravitropic response in rice root but do not inhibit gravitropism per se.
OsRAA1 shows tissue-specific expression in roots and spikes, and it was induced by auxin. Constitutive overexpression of the gene resulted in the increased number of adventitious roots and reduced growth of primary roots. At the same time, other phenotypes controlled by auxin, such as root helix and gravitropic response, appeared in the roots of the transgenic plants. Overexpression of OsRAA1 increased adventitious root initiation and reduced gravitropic response in rice, which was similar to the responses of the wild-type plants to auxin. At the same time, overexpression of OsRAA1 also caused endogenous IAA to increase. Our data supported that OsRAA1 as a new gene functions in the development of rice root systems, which are mediated by auxin. A positive feedback regulation mechanism of OsRAA1 to IAA metabolism may be involved in rice root development in nature. The biochemical mechanism of the protein functioning in regulation of root development will be addressed by an approach of yeast two-hybridization and immunology.
MATERIALS AND METHODS
Isolation of OsRAA1 and Construction of Vectors
Total RNA of rice (Oryza sativa L. cv Zhonghua 10) root was isolated using the QIAGEN RNeasy plant mini kit (Qiagen, Valencia, CA). The mRNA was purified from total RNA using an Oligotex Poly(A) mRNA purification kit (Qiagen). Then two fragments of OsRAA1 cDNA were amplified by 5′ and 3′ RACE using a Marathon cDNA amplification kit (Clontech, Palo Alto, CA). The double-strand cDNA was synthesized and ligated to the Marathon cDNA adaptor. The 5′- and 3′-end cDNA sequences were amplified using Adaptor primer 1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′) and the ORF forward and reverse primers (5′-AGGGGTTTGGGTGTTGAAG-3′ and 5′-CTAGCTCTGATTGCAAGAAGAATGAAG-3′), respectively. The whole ORF was cloned into pBluescript II SK+ (Stratagene, La Jolla, CA) and sequenced.
The cassette of UbiPro+OsRAA1+Noster was ligated into the multicloning sites of the binary vector pCAMBIA1301 to construct a vector of Ubi∷OsRAA1, which carried a gene of GUS as a marker (Cambia, Canberra, Australia; Roberts et al., 1997). A promoter of about 2.0-kb upstream of OsRAA1 gene was amplified using a PCR approach based on the sequence information of PAC clone (P0462H08) of rice. It was inserted into 5′ end of the GUS gene (gusA) in pCAMBIA1301 to create a vector of GUS driven by OsRAA1 promoter, OsRAA1∷GUS.
RNA in Situ Hybridization
In situ hybridization was performed as described by Schwarzacher and Heslop-Harrison (2000) and Xu et al. (2001). The roots and the shoot apical meristem sections for in situ hybridization were cut from 8-d-old seedlings. Young spikes of about 2 mm in length were harvested from rice plants at the stooling stage. All tissues were fixed in a fixative solution containing 50% ethanol, 5% acetic acid, and 3.7% formaldehyde (37%). Samples were dehydrated by a series of graded ethanol and gradually incubated in a mixture of 90% xylenes and 10% chloroform. Paraffin Plus chips (Sigma, St. Louis) were gradually added into that mixture. Subsequently, the xylene/paraplast solution was changed into molten paraplast. Samples were sectioned at 7 μm on a rotary microtome (Leica, Wetzlar, Germany) and affixed onto poly-Lys coated slides (Sigma). The digoxigenin-labeled antisense and sense riboprobes were synthesized using linear plasmid according to the manual of the DIG RNA labeling kit (Roche, Basel). These riboprobes were hydrolyzed into small fragments of about 150 nucleotides in length. The sections were hybridized in the hybridization mixture with a probe concentration of 0.4 ng μL−1 after being dewaxed gradually, hydrated gradually, and treated with proteinase K of 1 mg L−1 for 30 min.
Gene Transformation
Rice embryonic calli were induced on scutella from germinated seeds and transformed with strain EHA105 of Agrobacterium tumefaciens containing the desired binary vector, as described by Hiei et al. (1994) and Huang et al. (2000). Transgenic plants were selected in half-strength MS medium containing 75 mg L−1 hygromycin (Sigma). Hygromycin-resistant plants from calli, defined as transgenic plants of the T0 generation, were transplanted into soil and grown at a greenhouse at 28°C. For analysis of root phenotypes of transgenic plants, seeds of the T1 generation were germinated in half-strength MS medium containing 75 mg L−1 hygromycin and confirmed by GUS staining. The OsRAA1∷GUS transgenic plants were selected the same way. T1 generation of transgenic plants were used in the analysis experiments.
RNA Gel-Blot Analysis
Total RNA electrophoresis and the programs of the RNA transferred and cross-linked onto a nylon membrane (Hybrid N+; Amersham, Buckinghamshire, UK) were performed as described by Sambrook et al. (1989) and Ge et al. (2000). Total RNA of 15 μg was loaded on each lane. The probe of OsRAA1 cDNA labeled with [32P]dCTP (China Isotope, Beijing) was synthesized for hybridization. After hybridization for 20 h at 68°C, the membrane was washed once with 2× SSC plus 0.1% SDS at 68°C for 20 min, then washed with 1× SSC plus 0.1% SDS at 37°C for 30 min. The membrane was exposed to the x-ray film (Kodak, Rochester, NY) at −70°C for 3 to 7 d.
Histochemical Localization of GUS Activity
GUS staining was performed according to the method described by Jefferson (1989). Different organs of OsRAA1∷GUS transgenic seedling were incubated in a solution containing 50 mm NaP buffer at pH 7.0, 5 mm K3 Fe(CN)6, 5 mm K4 Fe(CN)6, 0.1% Triton X-100, and 1 mm X-Gluc, and incubated at 37°C for 12 h. All samples were vacuum treated for 5 min before the staining.
Auxin Treatment
Treatment of IAA for Analysis of Northern Blot
Surface-sterilized rice seeds were germinated in water and grown in the half-strength MS liquid medium in the chamber (25°C). Seedlings of 14 d were transferred into half-strength MS liquid medium with a different concentration of auxin. The samples were harvested and frozen in liquid nitrogen immediately for isolation of total RNA.
Treatment of OsRAA1∷GUS Transgenic Plant with NAA for Histochemical Display of GUS Activity
The seedlings of the T1 generation of OsRAA1∷GUS transgenic plant were grown in the green house for 2 months. After cleaning the roots, the seedlings were transferred into a liquid medium of half-strength MS and grown for 3 d. Then the seedlings were moved into half-strength MS liquid medium containing NAA of 2.5 × 10−6 m. Fresh roots were cut down after treatment for various times (0, 1, 2, and 4 h) and stained in the X-Gluc solution for 1 h. The roots after staining were stored in 95% ethanol.
Observation of the Leaf Surface Cells using Scanning Electron Microscope
When the rice plants accomplished heading, the flag leaves of the Ubi∷OsRAA1 transgenic plants and the untransformed control were harvested. Small blade fragments from the absolute middle part of the flag leaves were immediately fixed in FAA (3.7% formaldehyde, 50% ethanol, 5% acetic acid) for 12 h and dehydrated in a graded ethanol series. The dehydrated materials were critical point dried in liquid CO2 and mounted on metallic stubs. The mounted material shadowed with gold before viewing with the S-800 scanning electron microscope (Hitachi, Haramachi, Fukushima, Japan).
Determination of IAA Level in Plant
IAA level in seedling was determined by a method of ELISA, (Li and Zhou, 1996).
Chemicals
All chemicals in the experiments were from Beijing Chemicals, Beijing except for those labeled otherwise above.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY659938.
Acknowledgments
We thank Dr. Z.Y. Wang and Dr. J.X. He (the Carnegie Institution of Washington and Stanford) and Dr. R.J. Chen (Institute of the Nobel Foundation) for their critical reading and comments on the manuscript. We thank Mrs. Wei-Min Teng for the plasmid of KSP9. and Dr. H.W. Xue (Shanghai Institute of Plant Physiology, CAS) for his help on establishment of gene transformation of rice.
This work was supported by the Major State Basic Research Program of China (grant no. G19990116), by the Innovation Grand of CAS, National Nature Science Foundation of China (NSFC; grant no. 30270143), and by the State Project of Transgenic Plant (J99–A–024) as well as the State High-Tech Project (2001AA222281).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041996.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi P (1998) Auxin. In L Taiz, E Zeiger, eds, Plant Physiology. Sinauer Associates, Sunderland, MA, pp 543–590
- Bleecker A, Rose-John S, Kende H (1987) Evaluation of 2,5-norbornadiene as a reversible inhibitor of ethylene action in deepwater rice. Plant Physiol 84: 395–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A, Schuette JL, Kende H (1986) Anatomical analysis of growth and developmental patterns in the internode of deepwater rice. Planta 169: 490–497 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Onckelen HV, Montagu MV, Inze D (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7: 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR (1995) A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev 9: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Amasino RM (1994) Cytokinins and plant gene regulation. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL, pp 233–241
- De Miranda LT (1980) Inheritance and linkage of root characteristic from Pueblo maize. Maize Genet Coop News Lett 54: 18–19 [Google Scholar]
- del Pozo JC, Estelle M (1999) Function of the ubiquitin-proteosome pathway in auxin response. Trends Plant Sci 4: 107–112 [DOI] [PubMed] [Google Scholar]
- Doerner P (2000) Root patterning: Does auxin provide positional cues? Curr Biol 10: 201–203 [DOI] [PubMed] [Google Scholar]
- Estelle M, Klee HJ (1994) Auxin and cytokinin in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 555–578
- Fujii Y (1959) Studies on the correlation between number of the vascular bundles in leaf and arrangement of the roots at the node in rice plants. Crop Sci Soc Japan Proc 27: 67–70 [Google Scholar]
- Ge L, Liu JZ, Wong WS, Hsiao WLW, Chong K, Xu ZK, Yang SF, Li N (2000) Identification of a novel multiple environmental factor responsive 1-aminocyclopropane-1-carboxylate synthase gene, NT-ACS2, from tobacco. Plant Cell Environ 23: 1169–1182 [Google Scholar]
- Goldsmith MHM (1977) The polar transport of auxin. Annu Rev Plant Physiol 28: 439–478 [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF (TIR1) -dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998) How does auxin turn on genes? Plant Physiol 118: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt AJ, Guilfoyle TJ (1984) Auxin regulated gene expression in intact soybean hypocotyl and excised sections. Planta 162: 147–153 [DOI] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hobbie LJ (1998) Auxin: molecular genetic approaches in Arabidopsis. Plant Physiol Biochem 36: 91–102 [Google Scholar]
- Hochholdinger F, Park WJ, Feix GH (2001) Cooperative action of SLR1 and SLR2 is required for lateral root-specific cell elongation in maize. Plant Physiol 125: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JQ, Wei ZM, An HL, Xu SP, Zhang B (2000) High efficiency of genetic transformation of rice using Agrobacterium mediated procedure. Acta Bot Sin 42: 1172–1178 [Google Scholar]
- Ishikawa H, Evans ML (1993) The role of the distal elongation zone in the response of maize roots to auxin and gravity. Plant Physiol 102: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jenkins MT (1930) Heritable characters of maize XXXIV-rootless. J Hered 21: 79–80 [Google Scholar]
- Kania T, Russenberger D, Peng S, Apel K, Melzer S (1997) FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key JL (1989) Modulation of gene expression by auxin. Bioessays 11: 52–58 [DOI] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS (2000) Involvement of Brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol 123: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Sugiyama M (2003) Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130: 5637–5647 [DOI] [PubMed] [Google Scholar]
- Li ZT, Zhou X (1996) Plant Hormones and Their Enzyme Linked Immuno-Sorbent Assay. Jiangsu Sci. & Tech. Press, Nanjing, Jiangsu, China, pp 124–132
- Liu CM, Xu ZH, Chua NH (1993) Auxin polar transport is essential for the establishment of bilateral symmetry during early plant development. Plant Cell 5: 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle TJ (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91–93 [DOI] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Kampmann G, Chandler J, Apel K (1999) FPF1 modulates the competence to flowering in Arabidopsis. Plant J 18: 395–405 [DOI] [PubMed] [Google Scholar]
- Melzer S, Majewski DM, Apel K (1990) Early changes in gene expression during the transition from vegetative to generative growth in the long-day plant Sinapis alba. Plant Cell 2: 953–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Haworth P (1994) Tomato root growth, gravitropism, and lateral development: correlation with auxin transport. Plant Physiol Biochem 32: 193–203 [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi KI, Tanaka A, Uchimiya H (2003) p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg GR, Ishikawa H, Evans M, Palme K (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck GKH, Mattsson J, Hardtke CS, Sung ZR, Berleth T (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200: 229–237 [DOI] [PubMed] [Google Scholar]
- Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Roberts CS, Rajagopal S, Yang W, Nugroho S, Smith L, Nguyent T, Ravi KS, Dransfield L, Harcourt R, Vijayachandra K, et al (1997) A comprehensive new set of modular vectors to allow both routine and advanced manipulations and efficient transformation of rice by both Agrobacterium and direct gene-transfer methods. Rockefeller Foundation Meeting of the International Program on Rice Biotechnology, September 15–19, 1997, Malacca, Malaysia
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sachs T (1991) Pattern Formation in Plant Tissues. Cambridge University Press, Cambridge, UK
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sato Y, Nishimura A, Ito M, Ashikari M, Hirano HY, Matsuoka M (2001) Auxin response factor family in rice. Genes Genet Syst 76: 373–380 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison P (2000) Practical in situ Hybridization. BIOs Scientific Publishers, New York
- Suge H (1985) Ethylene and gibberellin: regulation of internodal elongation and nodal root development in floating rice. Plant Cell Physiol 26: 607–614 [Google Scholar]
- Swarup R, Bennett M (2003) Auxin transport: the fountain of life in plants? Dev Cell 5: 824–826 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur JK, Tyagi AK, Khurana JP (2001) OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res 8: 193–203 [DOI] [PubMed] [Google Scholar]
- Thimann KV (1977) Hormone Action in the Life of the Whole Plant. University of Massachusetts Press, Amherst, MA
- Uggla C, Mellerowicz EJ, Sundberg B (1998) Indole-3-acetic acid controls cambial growth in Scots pine by positional signaling. Plant Physiol 117: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997. a) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999. a) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999. b) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997. b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara BS, Jackson B, Datta SK (1976) Deep water rice and its response to deep water stress. In Climate and Rice. International Rice Research Institute, Los Banos, Philippines, pp 301–319
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang SY, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu YY, Chong K, Xu ZH, Tan KH (2001) Expression Patterns of a Vernalization-related Genes Responding to Jasmonate. Acta Bot Sin 43: 871–873 [Google Scholar]
- Yamamoto M, Yamamoto KT (1998) Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol 39: 660–664 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]