Abstract
Using a two-element iAc/Ds transposon-tagging system, we identified a rice (Oryza sativa L. cv Nipponbare) recessive mutant, anther indehiscence1 (aid1), showing partial to complete spikelet sterility. Spikelets of the aid1 mutant could be classified into three types based on the viability of pollen grains and the extent of anther dehiscence. Type 1 spikelets (approximately 25%) were sterile due to a failure in accumulation of starch in pollen grains. Type 2 spikelets (approximately 55%) had viable pollen grains, but anthers failed to dehisce and/or synchronize with anthesis due to failure in septum degradation and stomium breakage, resulting in sterility. Type 3 spikelets (approximately 20%) had normal fertility. In addition, aid1 mutant plants had fewer tillers and flowered 10 to 15 d later than the wild type. The Ds insertion responsible for the aid1 mutation was mapped within the coding region of the AID1 gene on chromosome 6, which is predicted to encode a novel protein of 426 amino acids with a single MYB domain. The MYB domain of AID1 is closely related to that of the telomere-binding proteins of human, mouse, and Arabidopsis, and of single MYB domain transcriptional regulators in plants such as PcMYB1 and ZmIBP1. AID1 was expressed in both the leaves and panicles of wild-type plants, but not in mutant plants.
Anther dehiscence is the final step of anther development and results in the release of pollen grains to enable pollination, fertilization, and seed set (Goldberg et al., 1993). In anthers, three tissues—endothecium, septum, and stomium—play important roles in anther dehiscence. The endothecium is an anther wall tissue lying between the epidermal layer and the tapetum. The septum is a filling tissue for spaces between vascular bundles and the two adjacent anther locules. The stomium is a single layer of specialized epidermal cells that joins adjacent anther walls and is the final breakage site for anther dehiscence (Keijzer, 1987). In rice (Oryza sativa), at the beginning of anthesis, filaments begin to elongate and pollen grains swell rapidly. The increased pollen pressure causes the locule to bulge, resulting in rupture of the septum that has been weakened by hydrolytic enzyme action. Subsequent lysis of the middle lamellae of the stomium cells, pollen pressure, and the force induced by the inward bending of the locules causes the stomium to split (Keijzer et al., 1996; Matsui et al., 1999). Finally, shrinkage of the U-shaped, thickened endothecium induces the outward bending of locules, which widens the theca to release swollen pollen grains. Meanwhile, for successful anther dehiscence and pollination, a series of developmental events that leads to the breakage of the stomium and release of pollen grains must synchronize with other developmental processes that occur within both the anther and the floret. However, little is know about the mechanisms that control and coordinate many of these developmental events. Identification of mutants defective in any of these developmental activities would shed light on the mechanism of anther dehiscence and anthesis.
Although a number of spontaneous, as well as induced, cytoplasmic and genic male sterile mutants, which have defects at different developmental stages, have been identified, few have been studied in detail at a molecular or genetic level in rice (Kinoshita, 1997). Recently, the MULTIPLE SPOROCYTE1 (MSP1) gene, which encodes a Leu-rich repeat receptor-like protein, has been shown to be necessary for early sporogenic development and initiation of anther wall formation. The msp1 mutation gives rise to an excessive number of both male and female sporocytes and results in complete male sterility (Nonomura et al., 2003). Proper interaction between tapetal cells and the microspore mother cells is crucial for the functional development of pollen grains. A thermosensitive genic male sterile mutation is found to be associated with premature programmed cell death of the tapetum, resulting in the failure to develop functional pollen grains (Ku et al., 2003). A transcription factor, OsGAMYB, has been shown to play an important role not only in the induction of α-amylase in aleurone, but also in pollen development (Kaneko et al., 2004). To date, no gene related to anther dehiscence has been identified in rice. In Arabidopsis, several genes, such as COI1 (Xie et al., 1998), OPR3/DDE1 (Sanders et al., 2000; Stintzi andBrowse, 2000), DAD1 (Ishiguro et al., 2001), AOS/DDE2-2 (Park et al., 2002; von Malek et al., 2002), and AtMYB26 (Steiner-Lange et al., 2003), affecting anther dehiscence have been cloned and characterized. Mutants of these genes display defects in filament elongation and timing of anther dehiscence, as well as reduced pollen viability. All of these genes, except for AtMYB26, are involved in the jasmonic acid (JA) pathway.
Here we report the identification and characterization of a Ds-tagged rice mutant, designated anther indehiscence1 (aid1), having defects in the development of pollen grains, endothecium strength, septum degradation, and/or stomium breakage that result in the failure of anther dehiscence and its synchronization with anthesis, leading to partial to complete spikelet sterility. Other associated phenotypes were reduced tiller numbers and delayed flowering. AID1 encodes a single MYB DNA-binding domain protein. The MYB domain of AID1 is closely related to that of telomere-binding proteins, including TRF1 and TRF2 of human and mouse, TRP1 and TBP1 of Arabidopsis, and of single MYB-repeat transcriptional regulators in plants such as MYB1 of Petroselinum crispum and IBP1 of maize (Zea mays).
RESULTS
Identification of Mutants with Partial to Complete Spikelet Sterile Phenotype
The initial iAc/Ds mutagenesis protocol is described elsewhere (Upadhyaya et al., 2002). Essentially, DsG- and iAc-containing rice lines were produced by Agrobacterium-mediated transformation, followed by genetic crossing and production of subsequent progenies for phenotypic screening. Out of 12 F3 progeny plants from an F2 mutagenic plant (iAc+Ds+), derived from a cross between iAc and DsG transgenic lines (Upadhyaya et al., 2002), 11 were found to be stable (devoid of iAc) Ds insertion plants (transposed Ds linked to the original Ds launching pad). Among these 11 stable Ds insertion plants, 1 showed complete sterility and 5 showed partial sterility, ranging from 79% to 99%. The remaining 5 plants were phenotypically similar to the wild type (Table I). All the progeny of the 5 partially sterile F3 plants (B4-1-1-1-2, -3, -4, -9, and -10) also showed partial to complete sterility (Table I), indicating that they were homozygous mutants. The progeny of 4 phenotypically normal F3 plants (B4-1-1-1-5, -7, -8, and -11) segregated in a ratio of 3:1 (normal to mutant; χ2 = 0.01, P = 0.92), indicating that they were heterozygous mutants. The progeny of the remaining phenotypically normal plant (B4-1-1-1-12) remained unchanged, indicating that it was a segregated Ds null plant (Table I). Subsequent progenies of heterozygous plants showed perfect segregation of mutant phenotype (3:1; χ2 = 0.12, P = 0.73), and all mutant plants (homozygotes) showed partial-to-complete sterility (data not shown). Wild-type pollen grains could fertilize homozygous mutant spikelets, resulting in normal seed setting, which showed a 3:1 segregation of normal to mutant plants in the F2 generation (χ2 = 0.33, P = 0.57; data not shown). These segregation data indicate that aid1 is a single recessive male sporophytic mutation.
Table I.
Cosegregation of Ds insertion in the genomic region corresponding to BAC OSJNBa0035I03 with spikelet sterility
| Line | F3 Generation
|
F4 Generation
|
|||||
|---|---|---|---|---|---|---|---|
| Percentage of Sterility | PCR1a | PCR2b | Genotype of Ds Insertion Locus | Genotype of Ds Insertion Locus | Percentage of Sterility
|
||
| Average | Range | ||||||
| B4-1–1-1-2 | 79.36 | − | + | Ds/Ds | Ds/Ds | 80.31 | 56.55–100.00 |
| B4-1–1-1-3 | 98.73 | − | + | Ds/Ds | Ds/Ds | 91.56 | 54.64–100.00 |
| B4-1–1-1-4 | 92.90 | − | + | Ds/Ds | Ds/Ds | 73.24 | 65.38–100.00 |
| B4-1–1-1-5 | 11.55 | + | + | Ds/+ | Ds/Ds | 75.35 | 58.70–98.32 |
| Ds/+ | 12.97 | 11.51–14.42 | |||||
| +/+ | 17.88 | 13.40–22.36 | |||||
| B4-1–1-1-6 | 100.00 | − | + | Ds/Ds | Ds/Ds | n/ac | n/a |
| B4-1–1-1-7 | 10.37 | + | + | Ds/+ | Ds/Ds | 77.86 | 55.56–100.00 |
| Ds/+ | 17.93 | 0.00–30.86 | |||||
| +/+ | 26.48 | 25.36–27.59 | |||||
| B4-1–1-1-8 | 10.64 | + | + | Ds/+ | Ds/Ds | 73.32 | 60.00–95.01 |
| Ds/+ | 16.78 | 9.04–35.21 | |||||
| +/+ | 16.39 | 9.47–25.00 | |||||
| B4-1–1-1-9 | 98.92 | − | + | Ds/Ds | Ds/Ds | 87.71 | 54.95–100.00 |
| B4-1–1-1-10 | 93.75 | − | + | Ds/Ds | Ds/Ds | 83.20 | 55.26–100.00 |
| B4-1–1-1-11 | 11.27 | + | + | Ds/+ | Ds/Ds | 76.77 | 70.51–88.37 |
| Ds/+ | 14.42 | 7.14–20.02 | |||||
| +/+ | 13.80 | 12.62–15.83 | |||||
| B4-1–1-1-12 | 8.53 | + | − | +/+ | +/+ | 14.87 | 0.00–26.67 |
| Wild type | 6.04 | + | − | +/+ | +/+ | 8.03 | 0.00–19.23 |
Primers used were pm3101_For and pm3101_Rev.
Primers used were pm3101_Rev and Ds3_6587+.
Not applicable.
Characterization of the Spikelet Sterility Mutant
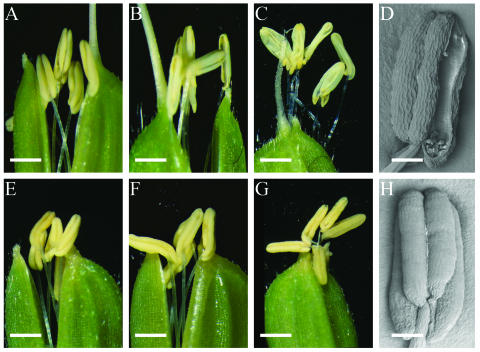
In the wild type, anthers reach the top of the spikelet and begin to dehisce, resulting in release of pollen grains over the stigma of the pistil immediately before spikelet opening (Fig. 1A). After anthesis, the filaments elongate further and the remaining pollen grains are released (Fig. 1B). The spikelet then begins to close, keeping empty anthers outside the spikelet (Fig. 1C). In the mutant, the anthesis of spikelets showed varying levels of abnormalities, resulting in varying levels of spikelet fertility. The predominant mutant phenotype (type 2 spikelet, see below) is shown in Figure 1, E to H. Anthers of the mutant did not dehisce at the time of spikelet opening or even after its closing (Fig. 1, E–G), while other reproductive organs appeared normal. This suggests that the spikelet sterility of the mutant was due mainly to the inability of anthers to dehisce and release pollen grains normally (compare Fig. 1, D and H). Therefore, we have designated this mutant aid1.
Figure 1.
Comparison of anthesis and anther dehiscence between the wild type and the aid1 mutant. A to D, Spikelet and anthers of the wild type. E to H, Spikelet and anthers of the aid1 mutant. A and E, Spikelet at the beginning of anthesis. B and F, Widely opened spikelet. C and G, Closed spikelet after anthesis. D and H, Scanning electron micrograph image of anther from spikelet at its peak stage of anthesis. Bars in A, B, C, E, F, and G are approximately 530 μm. Bars in D and H are 100 μm.
To better understand the variation in spikelet sterility, spikelets of the aid1 mutant were categorized into three types, based on the behavior at anther dehiscence and pollen viability. Type 1 spikelets (approximately 25%) did not show filament elongation, spikelet opening, or anther dehiscence because of abortion of pollen grains; consequently, anthers of type 1 spikelets were smaller and contained no pollen grains or only nonviable pollen grains (no or less starch). Type 2 spikelets (approximately 55%) were normal in filament elongation and spikelet opening, but showed indehiscence (Fig. 1, E–G) or delayed dehiscence, which caused spikelet sterility. There was no significant difference in the percentage of viable pollen grains between type 2 and wild-type spikelets (Table II), but pollen grains of the mutant were slightly smaller (data not shown). Furthermore, pollen grains from manually dehisced type 2 spikelets were able to self-fertilize (data not shown). Type 3 (approximately 20%) spikelets showed normal anther dehiscence and were fertile. The percentage of sterile spikelets (approximately 80%), based on this observation, was consistent with the average percentage of spikelet sterility of the homozygous aid1 mutant plants (73%–92%) shown in Table I.
Table II.
Spikelet types and their pollen viability of the aid1 mutant
| Spikelet Type of the aid1 Mutant | Type 1 | Type 2 | Type 3 | Wild Type |
|---|---|---|---|---|
| Fertility | Pollen sterility | Spikelet sterility | Fertility | |
| Percentage of spikelets | 24.45 | 54.58 | 20.97 | 6.04a |
| Percentage of stained pollen | 0–<10b | 90.01 | 94.79 | 96.83 |
Percentage of spikelet sterility of wild type.
There were two types of anthers showing no filament elongation—one without pollen grains and another with only poorly developed pollen grains.
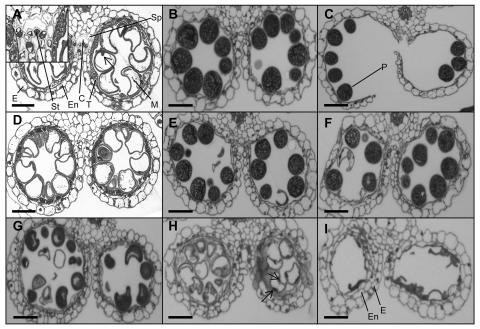
No distinguishable microscopic difference could be observed between cross-sections of anthers of type 3 spikelets of the aid1 mutant and those of wild type in different stages of anther development (compare Fig. 2, B and F). Even for anthers of type 1 and 2 spikelets, no difference in the development of anthers was detected until the nonvacuolate microspore stage (data not shown). In the wild type, at the vacuolate microspore stage, the tapetal cells and the septum began to degenerate, and a cavity was formed due to the stomium dissociation from the enzymatically weakened septum (Fig. 2A). In contrast, at the same developmental stage, breakage of the septum and the formation of the cavity were not observed in anthers of type 2 spikelets (compare Fig. 2, A and D) even after complete maturation of pollen grains (compare Fig. 2, B and E). In anthers of type 1 spikelets, not only were the breakage of the septum and the splitting of the stomium blocked, but the development of pollen grains was also affected. It seemed that the development of pollen grains was arrested at different stages between the bicellular vacuolated stage and the final starch-filling stage. In most anthers, the irregularly shaped nonviable pollen grains were due to incomplete starch accumulation (Fig. 2G); in some anthers, pollen grains failed to accumulate any starch (Fig. 2H), and, in extreme cases, pollen grains degraded completely (Fig. 2I).
Figure 2.
Cross-sections of anther locules from the wild type and the aid1 mutant. A to C, From the wild type. D and E, From type 2 spikelets of the aid1 mutant. F, From type 3 spikelets of the aid1 mutant. G to I, From type 1 spikelets of the aid1 mutant. A and D, Uninucleate vacuolated microspore stage. The nucleus is indicated by the arrow. A, Inset shows the details of the cavity formed after degeneration of the septum. B, E, F, G, and I, Maturation stage of pollen grains. C, Anther dehiscence stage. H, Bicellular vacuolated stage showing the generative and vegetative nuclei (arrows). C, cavity; E, epidermis; En, endothecium; M, microspore; P, pollen grain; Sp, septum; St, stomium; T, tapetum. Bars in A to I are 10 μm.
To determine whether environmental factors contributed to the variation of anther dehiscence and spikelet fertility, the effect of light and humidity was investigated. For dark treatment, panicles of the aid1 mutant were wrapped in black paper bags to achieve a totally dark microenvironment for the panicle. For humidity treatment, panicles of the aid1 mutant were wrapped in a plastic bag and water was injected into the bag daily with a syringe to maintain saturated humidity. Observations were made for anther dehiscence and spikelet sterility as described above. No changes in the percentage of the three types of spikelets were observed (data not shown), suggesting that light and humidity had no effect on the phenotype of the aid1 mutant.
In addition to the partial to complete sterility caused by the defect in pollen development and anther dehiscence, the aid1 mutant showed a delay of 10 to 15 d in heading date and an approximately 40% reduction in tiller numbers; the tiller numbers of the segregated mutant and normal plants in the F4 population were 4.8 and 8.5, respectively.
The aid1 Mutant Could Not Be Corrected by Application of JA
JA, a multifunctional growth regulator, has been shown to be essential for anther dehiscence in Arabidopsis, and exogenous application of JA has been shown to rescue some of the fertility-related mutations (Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002). JA was applied exogenously to the aid1 mutant to see whether the mutation could be corrected. There was no such response with the aid1 mutant. In fact, JA application had a slightly negative effect on spikelet fertility (Table III). In addition, the percentages of the three types of spikelets did not change significantly after the application of JA (data not shown). This suggests, unlike the situation with the OPR3/DDE1 (Sanders et al., 2000; Stintzi and Browse, 2000) and DAD1 (Ishiguro et al., 2001) genes, which control fertility in Arabidopsis, that the AID1 gene is not involved in JA biosynthesis.
Table III.
Effect of JA treatment on spikelet sterility of the aid1mutant
| Wild Type
|
The aid1 Mutant
|
|||||
|---|---|---|---|---|---|---|
| Control | Water | JA | Control | Water | JA | |
| Percentage | 6.80 ± 1.31 | 11.68 ± 3.04 | 26.15 ± 2.97 | 88.77 ± 1.78 | 90.23 ±3.71 | 94.23 ± 0.99 |
The aid1 Mutant Is Ds Tagged
Three different Ds flanking sequences were rescued from mutant plants B4-1-1-1-2, -3, -6, and -8. Database searches indicated that LW602 (accession no. AY429020), LW599 (accession no. AY429019), and LW597 (accession no. AY429018) were rice genomic DNA corresponding to P1-derived artificial chromosome (PAC) P0675A05 (accession no. AP002071), PAC P0015E04 (accession no. AP002069), and bacterial artificial chromosome (BAC) OSJNBa0035I03 (accession no. AP003019), respectively, which all map to the short arm of chromosome 6.
To identify which Ds insertion was associated with the spikelet sterility phenotype, a set of primers were synthesized, based on sequences available in the National Center for Biotechnology Information (NCBI) GenBank database, to amplify the region flanking each of the Ds insertion sites (PCR1). PCR2 and PCR3 amplified Ds3′ and Ds5′ flanking sequence tags (FSTs), respectively (see Table IV for primers used). Assuming that the Ds insertion was in a single-copy gene, using this combination of PCRs, homozygous Ds insertion plants would be PCR1−, PCR2+, and PCR3+; heterozygous Ds insertion plants would be PCR1+, PCR2+, and PCR3+; and Ds null plants would be PCR1+, PCR2−, and PCR3−. These PCR results showed that the Ds insertions mapping to PACs P0675A05 and P0015E04 were present in all 11 F3 plants segregating for spikelet sterility (data not shown), indicating that they were not responsible for the mutant phenotype. The Ds insertion mapping to BAC OSJNBa0035I03 segregated with the mutant phenotype in F3 plants (Table I), indicating that it was responsible for the mutant phenotype.
Table IV.
Primers used in this study
| Primer | Sequence | Target |
|---|---|---|
| Ac_1931+ | 5′-CAGCTCCAAAGACAAAGACAAC-3′ | iAc |
| Ac_2382− | 5′-TGCAGCAGCAATAACAGAGTC-3′ | iAc |
| Ds3_6587+ | 5′-CCGTCCCGCAAGTTAAATATG-3′ | Ds3′ |
| Ds5_112− | 5′-ATCGGTTATACGATAACGGTC-3′ | Ds5′ |
| GPAInt | 5′-TCCAAGTCCACAAGGAAAATTG-3′ | Ds |
| GUS_313- | 5′-TCACTTCCTGATTATTGACCCAC-3′ | Ds |
| Pm3101_For | 5′-AGAGGATTGTTGGGGATGGC-3′ | BAC OSJNBa0035I03 |
| Pm3101_Rev | 5′-GCACAGGTAAATCACAAAGCAAG-3′ | BAC OSJNBa0035I03 |
| OSJN_For | 5′-ATTCGTTCTCGCAGACCGTG-3′ | BAC OSJNBa0035I03 |
| OSJN_Rev | 5′-AGAGGATATAGCAGTTCCAAAGGC-3′ | BAC OSJNBa0035I03 |
| Pm3106_For | 5′-AAGACAAGCACAAAGCTCATCAC-3′ | PAC P0015E04 |
| Pm3106_Rev | 5′-AAGACAAGCACAAAGCTCATCAC-3′ | PAC P0015E04 |
| P0675A05_For | 5′-ACACCTCTAACACCTCTTCC-3′ | PAC 0675A05 |
| P0675A05_Rev | 5′-AAGACGAAACAAGCCTCTCC-3′ | PAC 0675A05 |
| AID1_fg27_For | 5′-ATGCCGCGGCTGATGCCGGAGG-3′ | AID1 |
| AID1_fg27_For2 | 5′-AGAAGAGGAAACATTAAGAAAGG-3′ | AID1 |
| AID1_tigr28_Rev | 5′-CTACCTCATCATGTTTCTCCACTTATCTTTCAAGTC-3′ | AID1 |
| AID1_Rev | 5′-CTTTATCCATCCTTGCAGCCAG-3′ | AID1 |
| RSs1_F | 5′-TGCCTTGATCGAAGCTGAC-3′ | RSs1 |
| RSs1_R | 5′-AGCAAGGGGTAGAGGCTCTC-3′ | RSs1 |
Cosegregation of this Ds insertion and the mutant phenotype was confirmed in the F4 generation by PCR analyses as described above. All 145 F4 progeny plants of five partially (spikelet) sterile Ds insertion plants (B4-1-1-1-2, -3, -4, -9, and -10) showed partial to complete spikelet sterility and were homozygous for Ds (Ds/Ds), with an average percentage of spikelet sterility of 73% to 92% (Table I). Four plants for the heterozygous Ds insertion segregated at a 1:2:1 (32:57:29; χ2 = 0.29, P = 0.87) ratio of homozygous (Ds/Ds), heterozygous (Ds/+), and Ds null (+/+) plants. Observed cosegregation of mutant phenotype and Ds insertion puts the linkage to <0.03 cM, which accounts for less than 11 kb. There were no marked differences in spikelet sterility between Ds/+, +/+ plants, and wild type, while spikelet sterility of Ds/Ds plants ranged from 73% to 78%. The progeny of the Ds null plant B4-1-1-1-12 remained null for Ds and showed a normal phenotype (Table I).
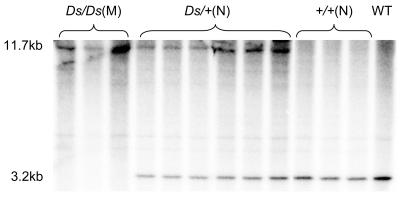
This cosegregation relationship was further confirmed by Southern-blot analysis of F4 plants (Fig. 3). Ds null plants were predicted to contain a hybridizing restriction fragment of 3.2 kb. As there is no recognition site for AvrII within Ds (8.5 kb), homozygous Ds insertion plants would have an 11.7-kb hybridizing restriction fragment (Fig. 4B), while heterozygous Ds insertion plants would have both 3.2- and 11.7-kb hybridizing restriction fragments. Hybridization results were as expected (Fig. 3).
Figure 3.
Cosegregation of Ds insertion in the genomic region corresponding to BAC OSJNBa0035I03 with the mutant phenotype. Genomic DNA (approximately 10 μg) was extracted from progenies of B4-1-1-1-8 (Ds/+) and wild-type plants, digested with AvrII, fractionated on a 0.7% agarose gel, and blotted to a Hybond-N+ membrane. The membrane was hybridized with a 744-bp genomic DNA fragment probe (see Fig. 4B) that covers the region flanking Ds in the genomic region corresponding to BAC OSJNBa0035I03. Homozygous Ds insertion (Ds/Ds) segregants only had an 11.7-kb hybridizing restriction fragment and showed mutant phenotype. Heterozygous Ds insertion (Ds/+) segregants had both 11.7- and 3.2-kb hybridizing restriction fragments and showed normal phenotype. Ds null (+/+) segregants only had a 3.2-kb hybridizing restriction fragment and showed normal phenotype. N, Normal phenotype; M, mutant phenotype; WT, wild type.
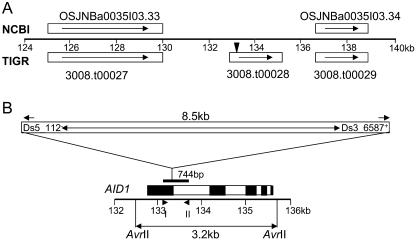
Figure 4.
Schematic structure of the AID1 gene and its position on BAC OSJNBa0035I03. A, Annotation results of NCBI and TIGR for a 16-kb genomic region harboring the Ds insertion responsible for the mutant phenotype. White rectangles represent predicted genes with orientation indicated by arrows. The 3008.t00028 locus is the candidate for the AID1gene. Black triangle indicates the position of Ds insertion. B, Schematic representation of the position of Ds insertion and of the 744-bp probe used for Southern blot. The orientation of Ds insertion is indicated by the Ds border (Ds3′ and Ds5′). Solid arrowheads indicate the positions of primers Ds3_6587+ and Ds5_112−, and triangles indicate the positions of primers OSJN_For (I) and OSJN_Rev (II) used to amplify the 744-bp probe. AvrII restriction sites are shown. The AID1 gene contains five exons (black rectangle) and four introns (white rectangle) with the Ds inserted at the 3′ end of the first exon.
AID1 Encodes a Novel Protein with a Single MYB Domain
The Ds insertion associated with the mutant phenotype is located at a position corresponding to 133381 nt in BAC OSJNBa0035I03 (Fig. 4, A and B). At the time of the initial analysis, this BAC had not been annotated. Based on gene prediction, expressed sequence tags (ESTs) available in the public domain and reverse transcription (RT)-PCR (data not shown), three putative genes were predicted in a 16-kb region spanning the Ds insertion site (124–140 kb of BAC OSJNBa0035I03). This result is consistent with the annotation now available in The Institute for Genomic Research (TIGR) rice database, but differs from that in NCBI (Fig. 4A). The putative AID1 gene corresponds to TIGR's predicted 3008.t00028 locus and contains five exons (Fig. 4B). As the mutant phenotypes were tightly linked to the Ds insertion and loss of-function of the AID1 gene, and the expression of the gene (3008.t00029 locus) downstream of AID1 was not affected in the aid1 mutant (see below), we concluded that AID1 is the gene involved in the developmental events leading to anther dehiscence in rice. Using RT-PCR and RACE approaches, we cloned the AID1 gene (see “Materials and Methods”; accession no. AY429017). The Ds element in the aid1 mutant was inserted within the 206th codon of the AID1 gene (Fig. 5).
Figure 5.
Full-length cDNA of the AID1 gene and its deduced amino acid sequence. The AID1 gene encodes a single MYB domain protein containing 426 amino acids. The triangle indicates the position of the Ds insertion causing the mutant phenotype. Primers used to amplify the cDNA and 3′ RACE products are indicated by italic and arrows. The MYB-like domain with a LKDKW motif (in bold italic) is boxed.
AID1 is predicted to encode a protein of 426 amino acids that contains a single MYB DNA-binding domain at the C terminus. Its MYB domain is closely related to that of telomere-binding proteins, including TRF1 and TRF2 of human and mouse, TRP1 and TBP1 of Arabidopsis, and of single MYB-repeat transcriptional regulators of plants such as MYB1 of P. crispum (Feldbrügge et al., 1997), IBP1 of maize (Lugert and Werr, 1994), and BPF1 of P. crispum and Catharanthus roseus (Costa e Silva et al., 1993; van der Fits et al., 2000) as shown in Figure 6. Alignment of the predicted sequence of AID1 and other telomere-binding proteins revealed 36% to 50% sequence identity in the MYB domains. However, there is no similarity outside the MYB domain, suggesting that AID1 is a novel gene.
Figure 6.
Amino acid alignment of the putative DNA-binding domain of AID1 and other LKDKW-type single MYB domain proteins. Multiple sequence alignment of conserved MYB domains was produced by ClustalW. The core of the LKDKW motif is indicated by bold type. The short dashes represent gaps. Similar and identical amino acids are indicated by dots and asterisks under the amino acids, respectively. OsAID1, Oryza sativa ANTHER INDEHISCENCE1; hTRF1 and hTRF2, human TELOMERIC REPEAT BINDING FACTOR 1 and 2; mTRF1 and mTRF2, mouse TELOMERIC REPEAT BINDING FACTOR 1 and 2; CgTRF1, Cricetulus griseus TELOMERIC REPEAT BINDING FACTOR 1; PcMYB1, P. crispum DNA-binding protein; AtTRP1, Arabidopsis TELOMERIC REPEAT-BINDING PROTEIN 1; AtTBP1, Arabidopsis TELOMERIC DNA-BINDING PROTEIN 1; PcBPF1, P. crispum BOX P-BINDING FACTOR-1; CrBPF1, C. roseus BOX P-BINDING FACTOR-1; ZmIBP1, maize INHIBITOR-BINDING PROTEIN 1. Level of domain sequence identity between AID1 and the other listed sequences ranges from 36% to 50%.
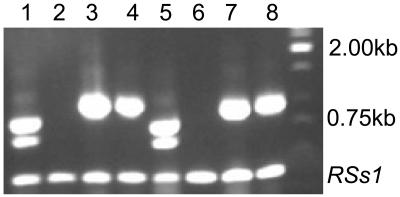
To determine the expression pattern of the AID1 gene, northern-blot analysis was performed with wild-type RNA using a 5′ cDNA probe, but only a very weak signal was detected, indicating that AID1 has low expression. However, AID1 transcripts were detected by RT-PCR in the wild-type panicle as well as leaves, but not in the mutant panicle or leaves. Two transcripts were amplified (Fig. 7). The third exon of the AID1 gene was found to be missing in the smaller transcript, which could result in a premature termination and a nonfunctional protein. RT-PCR also indicated that the expression of the gene downstream of AID1 was not affected in the aid1 mutant (Fig. 7), suggesting that the 3008.t00028 and 3008.t00029 loci are two independent genes.
Figure 7.
Expression analysis of the AID1 gene. RT-PCR was performed by primers OSJN_For and AID1_tigr28_Rev using total RNA extracted from young panicles of the wild type (lane 1) and the aid1 mutant (lane 2), and from leaves of the wild type (lane 5) and the aid1 mutant (lane 6). The bigger and smaller bands are the functional and nonfunctional transcripts of the AID1 gene, respectively. Based on sequence, the third exon of the AID1 gene was found to be missing in the smaller transcript, which could result in a premature transcript. The expression of the gene (3008.t00029) downstream of AID1 was detected both in panicle and leaves of the wild type (lanes 3 and 7) and of the aid1 mutant (lanes 4 and 8). The rice Suc synthase gene RSs1 was used as an internal control for RNA integrity and DNA contamination.
DISCUSSION
The aid1 Mutation Affects Events Leading to Anther Dehiscence
We have identified a mutant, aid1, from a Ds-tagged rice-screening population, in which spikelet sterility is the most obvious phenotype. Based on a tight phenotypic segregation with the Ds tag and associated nonexpression of the tagged AID1 gene in the mutant, we conclude that the AID1 gene is important for fertility as well as increased tillering and early flowering. Sterility in the aid1 mutant was mainly caused by a defect in anther dehiscence (type 2 spikelets), although approximately 25% of the spikelet sterility was due to pollen sterility (type 1 spikelets). The failure of anther dehiscence is associated with a defect in the programmed degradation and breakage of anther wall tissues. In the wild type, from the vacuolated microspore stage of anther development, a cavity begins to form because the stomium dissociates from the broken-down septum (Fig. 2, A and B). Further splitting of the stomium, caused by the pressure of swollen pollen grains and the force induced by the inward bending of the locules, results in the release of pollen grains (Fig. 2C). But this process was not observed in the aid1 mutant (Fig. 2, D and E). Degradation of the septum and breakage of the stomium needs both enzymatic lysis of cell wall and mechanical force (Keijzer et al., 1996). In rice, it has been observed that, prior to dehiscence, pollen grains swell rapidly, and it has been proposed that this rapid swelling generates a significant mechanical force that has a role in rupturing the enzymatically weakened septum (Matsui et al., 1999). It is not clear how the lysis of the cell wall is related to anther dehiscence in the aid1 mutant, but lack of anther dehiscence could be related to insufficient pressure buildup due to the fact that the pollen grains are smaller. Although a defect in programmed tapetum degradation is one of the main reasons for the failure of development of pollen grains (Fei and Sawhney, 2001; Ku et al., 2003), the deposition of starch and other secondary metabolites in the pollen grain is crucial for its viability (Raghavan, 1988). Our results provide further evidence for this, as approximately 25% of spikelets of the aid1 mutant failed to accumulate starch during the late stages of anther development and were completely sterile (Fig. 2, G–I).
The importance of the deposition of lignified cellulose in the secondary wall thickenings of endothecium has been elucidated in Arabidopsis (Steiner-Lange et al., 2003). In the aid1 mutant, the development of the endothecium seemed to be normal. The endothecial cells became highly vacuolate at the early microspore stage, and at the bicellular microspore stage they began to develop fibrous bands and finally became compressed and disorganized (Raghavan, 1988; compare Fig. 2, A and D, B and E). But in those anthers (from type 1 spikelets) that failed to deposit starch, the development of fibrous bands did not appear to be initiated at the bicellular microspore stage and the endothecial cells remained vacuolated (Fig. 2, H and I), suggesting that the deposition of starch in pollen grains and the development of fibrous bands in endothecial cells are related events toward normal maturation of pollen grains and anther dehiscence.
The fact that a small proportion of the anthers did dehisce normally in the aid1 mutant indicates that AID1 is important, but not absolutely essential, for normal anther development. The exact reason for varying levels of defects in anther development in the mutant is yet to be determined, but it is possible that environmental conditions are important for the induction of these events. For example, a thermosensitive genic male sterile mutant has been shown to be associated with premature programmed cell death of the tapetum (Ku et al., 2003). We have shown that humidity and light were not associated with the variable phenotypes of the aid1 mutant, but the effect of temperature needs to be determined. On the other hand, it has been shown that a large number of genes are involved in anther development (for review, see Goldberg et al., 1993) and hence it is logical to assume that other genes provide complementary or supplementary functions in the aid1 mutant to produce approximately 20% normal spikelets in the absence of AID1 (Fig. 7).
The involvement of JA in anther dehiscence has been confirmed by isolating genes involved in JA biosynthesis in Arabidopsis (Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002; von Malek et al., 2002). Our results indicate that AID1 is not involved in JA biosynthesis. It may be a downstream gene in the JA signal transduction pathway or may be involved in a JA-independent pathway responsible for anther development.
AID1 Is a Single MYB Domain DNA-Binding Protein
MYB domain proteins in plants represent the largest gene family of transcription factors, suggesting their participation in a number of processes. They can be classified into three subfamilies, MYB1R, R2R3-type MYB, and MYB3R, depending on the number of adjacent repeats (1–3) in the MYB domain (for review, see Stracke et al., 2001). MYB proteins with one and three MYB repeats are known, but the majority of MYB proteins contain two repeats (i.e. R2R3-type MYB).
The single MYB domain proteins are quite divergent; however, two highly conserved amino acid motifs, SHAQK(Y/F) F (Rose et al., 1999) or LKDKW (Feldbrügge et al., 1997), are present within the DNA-binding domain. Members of the SHAQK(Y/F) F type are known to be transcriptional regulators, such as MYBSt1 (Baranowskij et al., 1994), LeMYB1 (Rose et al., 1999), ZmMRP1 (Gómez et al., 2002), and OsMYBS1-3 (Lu et al., 2002). They also include the circadian clock-associated floral repressor LHY (Schaffer et al., 1998) and CCA1 (Wang et al., 1997). Members of the LKDKW type include telomeric repeat-binding proteins (RTBP1, Yu et al., 2000; AtTRP1, Chen et al., 2001; AtTBP1, Hwang et al., 2001; Smh1, Marian et al., 2003), transcriptional regulators (ZmIBP1, Lugert and Werr, 1994; PcMYB1, Feldbrügge et al., 1997), and elicitor-responsive related DNA-binding proteins (PcBPF1, Costa e Silva et al., 1993; CrBPF1, van der Fits et al., 2000), suggesting that this type of single MYB domain gene has more diverse functions.
AID1 is a novel member of the LKDKW type of single MYB domain proteins (Figs. 5 and 6). The aid1 mutation blocked degradation of the septum and splitting of the stomium. It also affected the deposition of starch in pollen grains and the development of fibrous bands in endothecial cells. These results suggest that AID1 regulates programmed cell death as well as deposition of secondary metabolic materials during maturation of pollen grains. Therefore, AID1 is most likely a transcriptional regulator involved in late developmental events concerned with the maturation of pollen grains and anther dehiscence.
The pleiotropic effects observed, such as fewer tillers and delayed flowering, further suggest that AID1 functions as a transcriptional regulator and has a role to play not only in the reproductive stage, but also in the vegetative stage and in the flowering transition. Consistent with this interpretation, AID1 is expressed in both vegetative and reproductive tissues (Fig. 7). Transcription factors can exhibit multiple functions, and a mutation could lead to pleiotropic effects as observed in the rice GAMYB loss-of-function mutation that shows both impaired α-amylase expression in aleurone and male sterility (Kaneko et al., 2004).
aid1 Is Likely To Be an Allele of the Previously Reported ms1 Mutant
The BAC OSJNBa0035I03 harboring the AID1 gene has been anchored to approximately 13.5 cM on chromosome 6. The genetic distance between AID1 and the well-characterized Waxy (Wx) gene located at 8.3 cM on chromosome 6 (Nagato and Yoshimura, 1998) is approximately 5.2 cM. It is similar to the distance of approximately 5 cM between the Wx gene and the previously reported rice ms1 locus (Nagato and Yoshimura, 1998). ms1 is a spontaneous recessive male sterility mutant with a partial sterility phenotype. We have been unable to obtain the ms1 mutant, which may have been lost, to test the allelism between aid1 and ms1.
In maize, ms2, which maps to chromosome 9, is also closely linked to Wx (Burnham, 1978). Furthermore, five male sterility loci (msg3, msg10, msg13, msg16, and msg19) in barley (Hockett and Eslick, 1971) have been mapped to chromosome 7HS (Korzun et al., 1997). Rice chromosome 6, maize chromosome 9, and barley chromosome 7 have been shown to be syntenous (see Gale et al., 2001). The AID1 orthologs from maize and barley can now be isolated. Any of these male sterile mutants can be sequenced over the AID1 ortholog to pinpoint any mutation. Our initial search for maize or barley ESTs homologous to AID1 has been unsuccessful presumably because the transcript is rare and thus is not represented in the publicly available ESTs.
In conclusion, using a transposon Ds-tagged mutant, we have identified a gene in rice, designated AID1, involved in anther development with pleiotropic effects on tillering and flowering time. AID1 is a single MYB DNA-binding domain protein. The conserved motif LKDKW within the MYB domain of AID1 is identical to those of transcription factors TRP1 and TBP1 of Arabidopsis, MYB1 of P. crispum and IBP1 of maize.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All rice (Oryza sativa) lines used in this study were derived from the japonica cv Nipponbare. The male sterility mutant described in this paper was derived from an F2 generation plant (B4-1-1-1) derived from a cross between iAc (pSK300, TT3-26-1) and Ds gene trap (DsG, pSK200, TT6-101-1) transgenic lines described previously by Upadhyaya et al. (2002). Stable Ds insertion lines were identified by selection marker (hygromycin resistance), visual marker (green fluorescent protein), and gene-specific (iAc and Ds) PCRs as described previously (Upadhyaya et al., 2002). The iAc and DsG binary vector constructs, tissue culture, and Agrobacterium transformation procedures used have been described previously (Upadhyaya et al., 2000, 2002). Plants were grown under controlled glasshouse with 25°C ± 3°C day and 21°C night temperatures. Upon request and signing a normal biological material transfer agreement, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requester.
DNA Extraction and Ds FST Rescue
Genomic DNA was extracted from plants using the PureGene nucleic acid isolation kit (Gentra Systems, Minneapolis) according to the manufacturer's instructions. Ds flanking sequences were rescued using the built-in plasmid rescue system (Upadhyaya et al., 2000). Approximately 10 μg of genomic DNA were digested with SacI. The digested DNA was then phenol-chloroform extracted, precipitated, and self-ligated in 500 μL of ligation mix containing 5 Weiss units T4 DNA ligase at 16°C overnight. The ligated DNA was used for electroporation after ethanol precipitation. Plasmid clones were analyzed with an appropriate restriction enzyme digestion before being selected for sequencing using primer Ds3_6587+ in a reaction mix of the reagents from the ABI Prism BigDye termination cycle sequencing kit (Applied Biosystems, Foster City, CA). DNA sequence comparisons were performed using the NCBI GenBank BLAST programs (Altschul et al., 1990, 1997; http://www.ncbi.nlm.nih.gov). Alignment of MYB domains was performed using ClustalW (1.82; http://www.ebi.ac.uk/clustalw).
PCR Conditions
PCR primers (Table IV) were designed using the prime program of the Genetics Computer Group (Madison, WI) software suit (Devereux et al., 1984). Stable Ds insertion plants (Ac−Ds+) were identified by iAc (primers Ac_1931+ and Ac_2382−) and Ds (primers GUS_313− and GPAInt) specific PCR. The following PCR program was used: 30 cycles of 94°C for 2 min, 58°C for 30 s, and 72°C for 1 min, followed by a final 72°C for 5 min. To identify the presence or absence of Ds insertions corresponding to PACs P0675A05 and P0015E04, and BAC OSJNBa0035I03, a set of primers were designed for each insertion based on the PAC and BAC sequences available in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/). Three sets of PCRs were performed for each Ds insertion (see Table IV for the primers used): PCR1 amplified the region flanking the Ds insertion, and PCR2 and PCR3 amplified part of the 3′ and 5′ end of Ds and their adjoining genomic DNA, respectively. The primers used for the 3′ and 5′ end of the Ds insertion were Ds3_6587+ and Ds5_112−, respectively (Table IV; Fig. 4B). The PCR conditions were 30 cycles of 94°C for 2 min, 60°C for 30 s, and 70°C for 1 min, followed by a final 70°C for 5 min.
Southern Blot
For Southern-blot analysis, genomic DNA (approximately 10 μg) was extracted from F4 progeny of B4-1-1-1-8 and wild-type plants, digested with AvrII, fractionated on a 0.7% agarose gel, and blotted onto a Hybond-N+ membrane (Amersham Life Science, Buckinghamshire, UK) according to the manufacturer's instructions. A 744-bp genomic region (Fig. 4B) flanking the Ds insertion was amplified with primers OSJN_For and OSJN_Rev (Table IV) and used to prepare a radioactively labeled probe with the Megaprime DNA labeling system (Amersham Life Science) according to the manufacturer's instructions. The membrane was washed at 60°C with 0.1× SSC and 0.1% SDS and visualized by autoradiography using phosphor screens (Molecular Dynamics, Sunnyvale, CA).
RT-PCR and RACE
Total RNA was extracted from young panicles and leaves by using RNeasy mini kit (Qiagen, Valencia, CA) and treated with DNA-free (Ambion, Austin, TX). Partial 5′ cDNA and 3′ cDNA (two bands) of the AID1 gene were amplified by primer combination of AID1_fg27_For and AID1_Rev, and OSJN_For and AID1_tigr28_Rev (Fig. 5), respectively, using OneStep RT-PCR kit (Qiagen), according to the manufacturer's instructions. After purification using UltraClean (Mo Bio Laboratories, Solana Beach, CA), bands were cloned into pGEM-T Easy (Promega, Madison, WI) and sequenced using T7 and M13 reverse primers. One 3′ cDNA clone was found to contain premature stop codon after sequence analysis. 3′ RACE cDNA was isolated with primer AID1_fg27_For2 using SMART RACE cDNA amplification kit (BD Biosciences CLONTECH, Palo Alto, CA), according to the manufacturer's instructions. The rice Suc synthase gene RSs1 (Wang et al., 1992) was used as an internal control for RNA integrity and DNA contamination.
Investigation of Anther Dehiscence and Pollen Viability
Investigations on spikelet opening, filament elongation, anther dehiscence, and pollen viability were conducted on the day of anthesis. Spikelets were categorized into three types, based on the behavior of anther dehiscence and pollen viability: (1) no filament elongation—anthers did not elongate out of the spikelet because of no filament elongation and spikelet opening, and showed pollen sterility; (2) anther indehiscence—anthers elongated out of spikelet, but did not dehisce or showed delayed dehiscence, and pollen grains were viable; and (3) anther dehisced—anthers dehisced normally and showed normal pollen viability.
Pollen viability was examined by staining with 1% iodine-potassium iodide solution. For wild-type and normal dehisced anthers of the aid1 mutant (type 3), pollen grains were directly collected on a slide by shaking dehiscing anthers. For indehiscence anthers of the aid1 mutant (types 1 and 2), pollen grains were manually squeezed onto a slide. Observations were immediately made by microscopic investigation following the addition of a drop of 1% iodine-potassium iodide solution. Ten spikelets were checked for each type of spikelet.
Microscopic Observation
For scanning electron micrograph observation, anthers were collected from a spikelet at its peak stage of anthesis and processed essentially as described by Keijzer et al. (1996) and observed with JSM-6400 scanning electron microscopy. For anther cross-section observation, spikelets of different developmental stages were collected, based on the distance between the auricle of the flag leaf and the second leaf, and fixed in formaldehyde-acetic acid, and then dehydrated by a methanol-butanol series and embedded in paraffin. Ten-micrometer-thick sections were cut and stained with toluidine blue-O and observed with Leica DMR upright compound microscopy (Wetzlar, Germany). Ten spikelets (60 anthers) were observed for each developmental stage.
JA Rescue Experiment
Plants of wild type and the aid1 mutant were tagged immediately prior to booting. Tissue paper was wrapped outside the leaf sheath of the flag leaf. Three panicles of each plant were given one of the following treatments: (1) no treatment (control); (2) water; and (3) 500 μm MeJA (Sigma-Aldrich, St. Louis). The three treatments were applied twice daily (morning and dusk) by applying 2 mL of water or MeJA solution to the tissue paper using a syringe. The treatment lasted for 15 d. Each treatment included five plants (replications). Fertile and sterile spikelets were counted separately for each tagged panicle to determine the percentage of spikelet sterility.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY429017, AY429020, AY429019, and AY429018.
Acknowledgments
The authors thank Celia Miller, Shu-Ting Pan, Shamsul Hoque, and Kathryn Smith for their invaluable technical assistance. We also thank Drs. John Watson, Tony Pryor, Ming-Bo Wang, and Andrew Eamens for their critical reading and discussion. We are grateful to our collaborators, Chellian Santhoshkumar and Kottaram K. Narayanan, for providing pSK200 and pSK300 constructs.
This work was supported in part by GrainGene, Australia.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041459.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham CR (1978) Progress report on inactivated Ms* alleles. Maize Genet Coop Newsl 52: 71–72 [Google Scholar]
- Chen CM, Wang CT, Ho CH (2001) A plant gene encoding a Myb-like protein that binds telomeric GGTTAG repeats in vitro. J Biol Chem 276: 16511–16519 [DOI] [PubMed] [Google Scholar]
- Costa e Silva O, Klein L, Schmelzer E, Trezzini GF, Hahlbrock K (1993) BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J 4: 125–135 [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12: 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Sawhney VK (2001) Ultrastructural characterization of male sterile33 (ms33) mutant in Arabidopsis affected in pollen desiccation and maturation. Can J Bot 79: 118–129 [Google Scholar]
- Feldbrügge M, Sprenger M, Hahlbrock K, Weisshaar B (1997) PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J 11: 1079–1093 [DOI] [PubMed] [Google Scholar]
- Gale M, Morre G, Devos K (2001) Rice: a central genome for the genetics of all cereals. In GS Khush, DS Brar, B Hardy, eds, Rice Genetics IV. Proceedings of the Fourth International Rice Genetics Symposium, International Rice Research Institute, Los Banos, Philippines, pp 79–88
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E, Royo J, Guo Y, Thompson R, Hueros G (2002) Establishment of cereal endosperm expression domains: identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 14: 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett EA, Eslick RF (1971) Genetic male-sterile genes useful in hybrid barley production. In DA Reid, ed, Barley Genetics II. Proceedings of the Second International Barley Genetics Symposium, Washington State University Press, Pullman, WA, pp 298–307
- Hwang MG, Chung IK, Kang BG, Cho MH (2001) Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett 503: 35–40 [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE 1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer CJ (1987) The processes of anther dehiscence and pollen dispersal. I. The opening mechanism of longitudinally dehiscing anthers. New Phytol 105: 487–498 [DOI] [PubMed] [Google Scholar]
- Keijzer CJ, Leferink-ten Klooster HB, Reinders MC (1996) The mechanism of the grass flower: anther dehiscence and pollen shedding in maize. Ann Bot (Lond) 78: 15–21 [Google Scholar]
- Kinoshita T (1997) Gene symbols and information on male sterility. Rice Genet Newsl 14: 13–22 [Google Scholar]
- Korzun V, Malyshev S, Voylokov A, Borner A (1997) RFLP-based mapping of three mutant loci in rye (Secale cereale L.) and their relation to homologous loci within the Gramineae. Theor Appl Genet 95: 468–473 [Google Scholar]
- Ku S, Yoon H, Suh HS, Chung YY (2003) Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217: 559–565 [DOI] [PubMed] [Google Scholar]
- Lu CA, Ho TH, Ho SL, Yu SM (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert T, Werr W (1994) A novel DNA-binding domain in the Shrunken initiator-binding protein (IBP1). Plant Mol Biol 25: 493–506 [DOI] [PubMed] [Google Scholar]
- Marian CO, Bordoli SJ, Goltz M, Santarella RA, Jackson LP, Danilevskaya O, Beckstette M, Meeley R, Bass HW (2003) The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro. Plant Physiol 133: 1336–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Omasa K, Horie T (1999) Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann Bot (Lond) 84: 501–506 [Google Scholar]
- Nagato Y, Yoshimura A (1998) Report of the committee on gene symbolization, nomenclature and linkage groups. Rice Genet Newsl 15: 13–74 [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Raghavan V (1988) Anther and pollen development in rice. Am J Bot 75: 183–196 [Google Scholar]
- Rose A, Meier I, Wienand U (1999) The tomato I-box binding factor LeMYBI is a member of a novel class of myb-like proteins. Plant J 20: 641–652 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE 1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34: 519–528 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Upadhyaya NM, Surin B, Ram K, Gaudron J, Schünmann PHD, Taylor W, Waterhouse PM, Wang M-B (2000) Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust J Plant Physiol 27: 201–210 [Google Scholar]
- Upadhyaya NM, Zhou X-R, Zhu Q-H, Ramm K, Wu L, Eamens A, Sivakkumar R, Kato T, Yun D-W, Santhoshkumar C, et al (2002) An iAc/Ds gene and enhancer trapping system for insertional mutagenesis in rice. Funct Plant Biol 29: 547–559 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Zhang H, Menke FL, Deneka M, Memelink J (2000) A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol Biol 44: 675–685 [DOI] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wang M-B, Boulter D, Gatehouse JA (1992) A complete sequence of the rice sucrose synthase-1 (RSs1) gene. Plant Mol Biol 19: 881–885 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK (2000) Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem 275: 24208–24214 [DOI] [PubMed] [Google Scholar]