Abstract
In order to investigate the gene expression pattern during adventitious root development, RNA of Pinus contorta hypocotyls, pulse-treated with the auxin indole-3-butyric acid and harvested at distinct developmental time points of root development, was hybridized to microarrays containing 2,178 cDNAs from Pinus taeda. Over the period of observation of root development, the transcript levels of 220 genes changed significantly. During the root initiation phase, genes involved in cell replication and cell wall weakening and a transcript encoding a PINHEAD/ZWILLE-like protein were up-regulated, while genes related to auxin transport, photosynthesis, and cell wall synthesis were down-regulated. In addition, there were changes in transcript abundance of genes related to water stress. During the root meristem formation phase the transcript abundances of genes involved in auxin transport, auxin responsive transcription, and cell wall synthesis, and of a gene encoding a B-box zinc finger-like protein, increased, while those encoding proteins involved in cell wall weakening decreased. Changes of transcript abundance of genes related to water stress during the root meristem formation and root formation phase indicate that the plant roots had become functional in water transport. Simultaneously, genes involved in auxin transport were up-regulated, while genes related to cell wall modification were down-regulated. Finally, during the root elongation phase down-regulation of transcripts encoding proteins involved in cell replication and stress occurred. Based on the observed changes in transcript abundances, we suggest hypotheses about the relative importance of various physiological processes during the auxin-induced development of roots in P. contorta.
Multicellular organisms require proper timing for control of their development. The transition between different stages of development implies changes in cell division rates and patterns of cell differentiation. Entering a new stage of development also requires a change in the balance of expression of many genes. While processes and genes regulating development in angiosperms, and especially in the model plant Arabidopsis, have been identified, hardly anything is known about development in gymnosperms. Few gymnosperm species have been subjected to intensive molecular genetic analysis. Gymnosperms have several disadvantages as experimental organisms. They have large genomes, about 200 to 400 times bigger than that of Arabidopsis (Somerville and Somerville, 1999). Furthermore, they have a large size and a long generation time. Molecular data suggest that extant seed plants (gymnosperms and angiosperms) share a last common ancestor about 285 million years ago (Savard et al., 1994). From an evolutionary point of view, it is important to learn more about the regulation of development in gymnosperms. Another reason to study gymnosperms, and especially conifers, is that they are of great commercial importance.
The regulation of root development, including lateral root formation, has been studied in Arabidopsis mutants affected in normal development, as well as by using laser ablation techniques (Bhalerao et al., 2002; Himanen et al., 2002; Scheres et al., 2002). By contrast, little is known about the regulation of root development in gymnosperms. Observations of the development of the radicle are technically cumbersome because the meristem is initiated in the embryo. In addition, no root-defective mutants have been described in gymnosperms. However, root meristem formation is experimentally accessible during the development of adventitious roots.
To investigate the temporal distribution of specifically regulated transcripts in adventitious root development, we exploited the simple and synchronized model system for adventitious root development of hypocotyl cuttings of Pinus contorta (Grönroos and von Arnold, 1987; Lindroth et al., 2001a, 2001b). Close to 100% of the hypocotyls develop roots after a pulse treatment with an optimal dose of indole-3-butyric acid (IBA). In a previous study, we isolated a PSTAIRE CDC2 cDNA, PcCDC2, and two S-adenosylmethionine synthase (SAMS) cDNAs, PcSAMS1 and PcSAMS2, from P. contorta (Lindroth et al., 2001a, 2001b). The expression pattern of PcCDC2 during auxin-induced adventitious root formation points toward a role of CDC2 in cell division competence. PcSAMS1 is preferentially expressed in roots and exhibits a specific expression pattern in the meristem at the onset of adventitious root development, whereas PcSAMS2 is expressed in both roots and shoots but is down-regulated during adventitious root formation.
To continue the analysis of root formation, a technique is needed to follow the changes of expression of many genes simultaneously, rather than a few selected ones. Microarray technology has become a useful tool for studying global gene expression during plant development. To date, few conifer cDNA libraries have been sequenced. Currently, Pinus taeda is the only conifer species for which extensive sequence information is available (http://pine.ccgb.umn.edu). Microarray analysis has previously been used for identifying genes involved in cell wall biosynthesis during xylogenesis in P. taeda (Whetten et al., 2001). Recently, we showed that arrays from P. taeda can be used for studying gene expression in Picea abies and Pinus sylvestris (van Zyl et al., 2002a, 2002b; Stasolla et al., 2003). At present, the possibility of sampling single cells or a small number of cells, and also obtaining synchronized populations of cells, is restricted (Hertzberg et al., 2001; van Zyl et al., 2002a). The opportunity to induce adventitious roots by the application of auxin allows the generation of large populations of developmentally well-controlled and synchronized meristems. The primary aim of this study has been to obtain an overall impression of gene expression during adventitious root development in a gymnosperm. This microarray study on adventitious roots characterizes the molecular basis of physiological processes during specific phases of root development. By necessity it is descriptive and will be followed by functional studies.
RESULTS
The Model System for Root Development
The process of adventitious root development of auxin-treated hypocotyl cuttings of P. contorta has been described earlier (Grönroos and von Arnold, 1987; Lindroth et al., 2001a, 2001b) and is shown in Figure 1. Close to 100% of the cuttings develop roots within 12 d after a 6-h pulse treatment with 1.23 mm IBA. The rooting is very efficient, with roots developing in several ranks along the whole length of the hypocotyl. Cuttings not treated with IBA produce neither meristems nor roots during the first month. Within 3 d after wounding and auxin treatment, the cortical and epidermal cells expand and the hypocotyl splits. Adventitious root primordia are visible after 6 d. On day 9 the meristems are well developed with procambium established basipetally. Twelve days after wounding and the auxin treatment, the emerging adventitious roots are fully developed with root cap, apex, and vascular connection. The root base is situated outside resin ducts or differentiating resin ducts located centrifugally to the primary xylem. Xylem differentiates on both sides of the resin duct. In 0.5-cm-long roots, mature short tracheids have been formed in the transition zone between the root and the hypocotyl. Metaxylem develops between the primary xylem strands of the root. Phloem develops outside the xylem.
Figure 1.
Auxin-induced adventitious root development in P. contorta. Four-week-old hypocotyl cuttings were treated with 1.23 mm IBA for 6 h. The consecutive development of adventitious roots was monitored at 3-d intervals, starting day 0 with untreated plants. A, Drawings illustrating the different stages of adventitious root development (adapted from Lindroth et al., 2001b, with permission from Kluwer Academic Publishers). B, Schematic drawings illustrating transverse sections of the hypocotyl during each stage in the development. The hypocotyl split after 3 d. The early stage of an adventitious root primordium (AP) is visible at day 6, and the adventitious root meristem (AM) is formed by day 9. Fully developed roots are formed by day 12, which then start to elongate (adapted from Lindroth et al., 2001a, with permission from Elsevier).
Samples for microarray analyses were taken at the same circadian time point at day 0 (before the auxin treatment), day 3 (during cell expansion phase), day 6 (when root primordia were formed), day 9 (when root meristems were formed), day 12 (when the roots were fully developed), and day 33 (when root elongation was in progress; Fig. 1).
Genes Differentially Expressed during Root Development
Alteration in gene expression pattern during root development was analyzed by comparing gene expression of pairs of samples from sequential developmental stages. This approach was taken based on previous results.
(1) Root development in hypocotyl cuttings is similar for 2- to 6-week-old seedlings (Grönroos and von Arnold, 1985; Lindroth et al., 2001a). From that we assume that any changes in gene expression in untreated seedlings are small and of low significance for the root development process that we are studying. Therefore, we have not included untreated controls at each time point.
(2) Close to 100% of the cuttings developed roots within 12 d after wounding and auxin treatment, while cuttings not treated with auxin produced neither roots nor meristems (Grönroos and von Arnold, 1987). The auxin treatment probably induced several changes in gene expression, especially during the first 6 d, probably not all of which were specific for root development. However, most of the successive changes in gene expression deduced from comparisons of pairs of sequentially harvested samples during later developmental phases are likely to be related to root development.
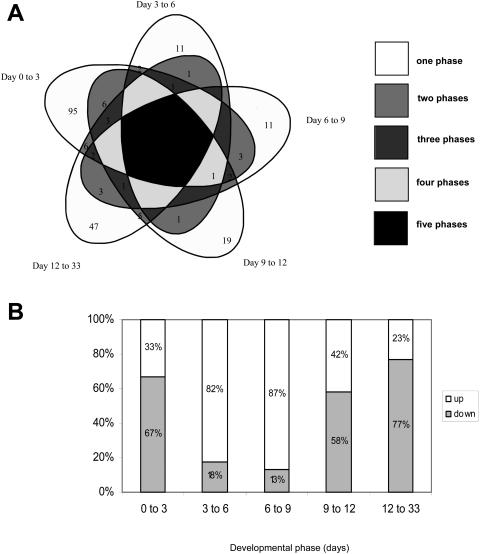
Out of 2,178 tested cDNAs, 220 were differentially expressed during the process of root development. The highest number of genes differentially expressed (121 genes) was observed between day 0 and day 3, while only 17 genes, and therefore the lowest number of significant fold changes in gene expression, occurred between day 3 and day 6 (Fig. 2A). Interestingly the majority (183 genes out of 220) of the genes differentially expressed showed changes during only one specific phase of development. Twenty-seven of the genes appeared to be differentially expressed during two phases, nine during three phases, and one during four phases. None of them was common to all five phases (Fig. 2A). Both up- and down-regulation took place during the whole process (Fig. 2B). However, up-regulation dominated from day 3 to day 6 and day 6 to day 9, while down-regulation dominated from day 0 to day 3, day 9 to day 12, and day 12 to day 33 (Fig. 2B).
Figure 2.
Genes differentially expressed during various phases of adventitious root development in P. contorta. A, Venn diagram showing the number of genes differentially expressed during specific developmental phases of adventitious root development. B, The 220 genes differentially expressed during specific developmental phases of root development were separated into two groups according to whether they were significantly up-regulated or down-regulated.
All 220 cDNAs were grouped into functional categories based on the categorization developed for Arabidopsis (http://pedant.gsf.de; Table I). In a few cases, when assignment was incomplete, genes were assigned to categories independently of the Arabidopsis system.
Table I.
Functional grouping of genes differentially expressed during adventitious root development in P. contorta
| Functional Category | Developmental Phase (Days)
|
||||
|---|---|---|---|---|---|
| 0 to 3 | 3 to 6 | 6 to 9 | 9 to 12 | 12 to 33 | |
| Cell cycle | 10% (12) | 24% (4) | 13% (3) | 11% (4) | 20% (14) |
| Cellular communication | 8% (10) | 24% (4) | 17% (4) | 11% (4) | 10% (7) |
| Cell fate | 10% (12) | 6% (1) | 30% (7) | 11% (4) | 10% (7) |
| Cellular transport | 1% (1) | 0% (0) | 9% (2) | 3% (1) | 6% (4) |
| Development | 6% (7) | 6% (1) | 4% (1) | 8% (3) | 7% (5) |
| Energy | 17% (20) | 12% (2) | 9% (2) | 13% (5) | 16% (11) |
| Stress | 11% (13) | 12% (2) | 13% (3) | 11% (4) | 10% (7) |
| Metabolism | 40% (48) | 41% (7) | 43% (10) | 32% (12) | 45% (31) |
| Transport facilitation | 8% (10) | 6% (1) | 13% (3) | 3% (1) | 4% (3) |
| Transcription | 16% (19) | 29% (5) | 35% (8) | 8% (3) | 23% (16) |
| Protein synthesis | 12% (15) | 12% (2) | 30% (7) | 24% (9) | 33% (23) |
| Protein fate | 15% (18) | 0% (0) | 22% (5) | 13% (5) | 16% (11) |
| Control of cell organization | 8% (10) | 18% (3) | 4% (1) | 16% (6) | 12% (8) |
| Cell rescue | 17% (20) | 24% (4) | 48% (11) | 32% (12) | 22% (15) |
A total of 220 genes differentially expressed during specific phases of root development were grouped into functional categories based on the categorization developed for Arabidopsis (http://pedant.gsf.de) and given as a percentage of all genes differentially expressed in that phase and as absolute numbers in parentheses. Each gene can belong to more than one category. The detailed list of the 220 genes is available at www.plantphysiol.org (Supplemental Table I). All data are available at the GEO database (http://www.ncbi.nlm.nih.gov/geo) and assigned with GEO accession numbers GSE1261, GSM19106, GSM19107, GSM19108, GSM19109, GSM19110, GSM19111, GSM19112, GSM19113, GSM19114, GSM19115, GSM19116, GSM19117, GSM19118, GSM19119,GSM19120, GSM19121, GSM19122, GSM19123, GSM19124, GSM19125, GSM19126, GSM419127, SM19128, GSM19129,GSM19130, GSM19131, GSM19132, GSM19133, and GSM19134.
A total of 184 out of 220 genes differentially expressed during root development were grouped according to the physiological process with which they are associated (Table II). Out of the 220 genes, 36 genes were not included because the sequences displayed no similarity to known proteins or were homologs to hypothetical proteins.
Table II.
Selected genes differentially expressed during root development in P. contorta
| Clone ID | Putative Function | E-Value | Developmental Phase (Days)
|
||||
|---|---|---|---|---|---|---|---|
| 0 to 3 | 3 to 6 | 6 to 9 | 9 to 12 | 12 to 33 | |||
| Genes Related to Protein Synthesis | |||||||
| ST29C09 | 60S ribosomal protein | 7E−65 | 1.8 | ||||
| ST25C01 | 60S ribosomal protein L6 | 4E−13 | 1.8 | −1.8 | |||
| NXSI117G02 | Ribosomal protein S2 | 6E−76 | 1.8 | ||||
| ST04G03 | Ribosomal protein L36 | 3E−35 | 1.8 | −4.6 | |||
| ST24H03 | 60S ribosomal protein L23A | 2E−52 | 1.9 | ||||
| ST37H05 | Ribosomal S29 protein | 2E−26 | 1.9 | ||||
| ST21A11 | Ribosomal protein P3A | 4E−16 | 2.0 | ||||
| ST25H11 | 40S ribosomal protein | 6E−51 | 2.2 | ||||
| ST30B07 | Ribosomal protein S4 | 2E−73 | 3.1 | ||||
| ST21F11 | Elongation factor 1-α 1 | 7E−62 | 2.1 | −4.6 | |||
| ST21A06 | Ribosomal protein L7 | 2E−64 | 1.9 | ||||
| ST13F05 | 26S ribosomal protein | 1E−180 | 8.6 | ||||
| NXNV096A02 | 40S ribosomal protein S15 | 1E−52 | −1.7 | ||||
| NXSI055E09 | 40S ribosomal protein S16 | 2E−61 | −2.2 | ||||
| NXCI009H11 | 60S ribosomal protein L2 | 9E−55 | −2.8 | ||||
| ST02E12 | Ribosomal protein L23 | 2E−72 | −2.3 | ||||
| NXSI114D12 | 60S ribosomal protein L22 | 8E−32 | −3.0 | ||||
| NXSI144H01 | 60S ribosomal protein L10 | 5E−86 | −2.0 | ||||
| ST23D04 | 60S ribosomal protein L17 | 2E−71 | −2.7 | ||||
| ST25B09 | 60S ribosomal protein L27A | 2E−64 | −5.4 | ||||
| NXNV183E12 | 60S ribosomal protein L27A | 1E−50 | −2.6 | ||||
| ST29H12 | 60S ribosomal protein L32 | 3E−20 | −2.6 | ||||
| ST32C07 | 60S ribosomal protein L32 | 4E−24 | −2.3 | ||||
| ST36G02 | Ribosomal protein L29 | 2E−12 | −3.9 | ||||
| ST18A08 | Ribosomal protein L36 | 5E−34 | −3.8 | ||||
| ST22A01 | Ribosomal protein L39 | 4E−33 | −3.6 | ||||
| ST01D03 | Ribosomal protein S18 | 5E−62 | −2.7 | ||||
| ST08B07 | Ribosomal protein S27 | 1E−35 | −3.8 | ||||
| NXCI070B10 | Translation initiation factor EIF-1A | 9E−33 | 2.8 | ||||
| ST34E10 | 18S ribosomal protein | 1E−164 | 22.3 | −3.6 | |||
| ST23B10 | 28S ribosomal protein | 1E−26 | 14.3 | −10.1 | |||
| NXCI096A09 | Translation initiation factor EIF-3b | 1E−40 | −4.2 | ||||
| NXCI094C11 | T-complex protein1, Θ subunit | 1E−17 | −3.2 | ||||
| NXSI065C08 | Translation initiation factor EIF-4A.7 | 4E−57 | −1.9 | ||||
| ST02C04 | ATP-dependent RNA helicase | 9E−5 | 3.3 | ||||
| Genes Related to Protein Assembly and Folding | |||||||
| NXCI045F10 | Calnexin precursor | 2E−95 | 1.7 | ||||
| NXSI045B09 | Protein disulfide isomerase precursor | 5E−30 | 2.0 | ||||
| ST09H11 | Chaperon | 1E−15 | −1.7 | ||||
| ST04D07 | Cyclophilin | 2E−75 | 2.5 | ||||
| ST39H08 | Peptidyl prolyl cis-trans isomerase | 2E−31 | 1.7 | −1.9 | |||
| Genes with Some Function Related to Protein Degradation | |||||||
| NXSI043G10 | Ubiquitin-like protein SMT3 | 2E−24 | −1.7 | ||||
| ST15D12 | Ubiquitin-like protein | 3E−28 | −2.3 | ||||
| NXSI103F08 | Ubiquitin-like protein SMT3 | 3E−35 | −1.9 | 1.8 | |||
| NXSI081D01 | Polyubiquitin | 6E−82 | 2.2 | −1.8 | −3.5 | ||
| NXSI102B05 | Ubiquitin extension protein | 4E−40 | 2.7 | 1.7 | −3.7 | ||
| ST24D04 | Ubiquitin extension protein | 4E−24 | −3.3 | ||||
| NXCI019E11 | Ubiquitin extension protein | 4E−37 | −2.5 | ||||
| NXNV117E03 | 20S proteasome subunit | 1E−77 | −2.0 | ||||
| ST03C12 | Proteasome endopeptidase complex | 1E−103 | −2.6 | ||||
| NXSI059H07 | Carboxyl-terminal proteinase | 2E−8 | −1.7 | ||||
| NXCI048E07 | Casein kinase | 3E−9 | −1.8 | ||||
| ST30B06 | Asparaginil-endopeptidase | 6E−45 | −2.3 | ||||
| Genes with Predicted Function in the Chloroplast | |||||||
| ST27A08 | PSII protein | 5E−16 | −2.6 | ||||
| ST20F08 | PSI reaction center subunit | 1E−71 | −2.7 | ||||
| ST31H04 | PSII reaction center protein | 9E−47 | −2.5 | ||||
| NXSI012D03 | PSII 10 kD polypeptide precursor | 2E−28 | −3.7 | ||||
| NXCI008C01 | PSII oxygen-evolving complex protein | 4E−30 | −3.0 | ||||
| NXSI007D08 | PSII reaction center protein | 1E−6 | −2.2 | −2.1 | 2.1 | ||
| NXCI085E04 | PSI subunit | 6E−33 | −6.2 | ||||
| ST12D01 | PSI subunit | 6E−20 | −3.1 | ||||
| ST36A10 | Chlorophyll a/b-binding protein | 2E−23 | −4.5 | ||||
| NXCI020A08 | Chlorophyll a/b-binding protein | 3E−43 | −3.9 | 5.1 | |||
| ST16C09 | 23-kD polypeptide of the oxygen evolving complex | 1E−24 | −3.8 | ||||
| ST06F07 | Plastid protein | 2E−8 | −4.4 | ||||
| ST39F03 | Ferredoxin precursor | 1E−27 | −2.0 | ||||
| NXSI113B09 | Heme oxygenase | 2E−33 | −1.6 | ||||
| ST04A02 | Oxoglutarate/malate translocator | 3E−20 | −3.6 | ||||
| NXSI092E10 | Thiazole biosynthetic enzyme precursor | 3E−82 | −3.0 | ||||
| NXSI131H02 | 1-Deoxy-d-xylulose-5-phoshate reductoisomerase | 8E−37 | −1.6 | ||||
| ST26D05 | ATP synthase C-chain | 1E−29 | −2.2 | ||||
| Genes Involved in Cell Replication | |||||||
| ST39C07 | Histone H3 | 2E−61 | 2.0 | 2.6 | −2.2 | ||
| ST08B05 | Tubulin alpha-2/alpha-4 chain | 5E−52 | 2.5 | −2.4 | |||
| ST13C07 | Histone H2B | 2E−32 | 2.8 | −2.5 | |||
| NXSI142F05 | Histone | 1E−8 | 1.7 | ||||
| NXSI113C10 | Cell division control protein cdc2 kinase | 2E−68 | 2.0 | ||||
| NXSI097D10 | β-tubulin | 2E−36 | −2.4 | ||||
| ST32B09 | Cell division cycle protein 48 | 1E−69 | 3.3 | ||||
| ST32G05 | Histone H2A | 3E−33 | −3.6 | ||||
| NXNV055B06 | Histone H2B | 6E−41 | −3.5 | ||||
| NXNV120E10 | Histone H2B | 2E−44 | −3.1 | ||||
| NXCI087F06 | Histone H3 | 3E−56 | −3.6 | ||||
| NXSI097G01 | Histone H2A | 7E−44 | −3.9 | ||||
| Genes with Some Function Related to Cell Wall Synthesis | |||||||
| NXCI106C10 | Sucrose synthase | 2E−3 | −1.9 | ||||
| NXSI055H08 | Caffeoyl-CoA-methyltransferase | 4E−34 | −2.7 | ||||
| ST14G06 | Lignin peroxidase | 8E−40 | −2.2 | ||||
| NXSI098C01 | Annexin | 7E−11 | −3.0 | ||||
| NXSI040D02 | Arabinogalactan protein | 4E−6 | −2.2 | ||||
| NXCI075E11 | Arabinogalactan protein | 3E−9 | 2.4 | ||||
| NXSI061G02 | Peroxidase | 1E−37 | 3.6 | ||||
| ST36F04 | Peroxidase | 2E−25 | 5.2 | ||||
| ST21A06 | UDP glucose 4-epimerase | 3E−43 | 1.9 | ||||
| ST23C10 | Catalase | 3E−53 | 1.8 | ||||
| NXSI104F05 | Porin Mip1 | 2E−68 | 1.9 | ||||
| ST34F04 | Cinnamoyl CoA reductase | 1E−32 | 1.8 | ||||
| Genes Related to Cell Wall Weakening and Modification | |||||||
| NXSI134F04 | Cellulase 1 precursor | 8E−35 | 2.1 | −3.3 | |||
| NXSI134E09 | Pectate lyase | 4E−51 | 1.8 | ||||
| ST34G01 | Pectinesterase | 2E−25 | 1.7 | ||||
| NXSI082H01 | Endoxyloglucan transferase | 6E−96 | 2.0 | ||||
| NXSI028A08 | Pectate lyase | 5E−34 | −1.6 | ||||
| NXCI094E12 | Pectate lyase | 5E−27 | −3.0 | ||||
| NXSI007F12 | Phytocyanin/early nodulin | 1E−19 | −1.9 | ||||
| NXNV095F04 | Coatomer protein delta-COP | 1E−21 | −1.9 | ||||
| NXCI047C05 | 2-Keto-3-deoxy-d-arabino-heptulosonate 7-phosphate synthase | 2E−62 | −2.2 | ||||
| NXNV153F09 | Basic blue protein phytocyanin | 4E−20 | −2.0 | 1.7 | |||
| Genes with Some Function Related to Stress | |||||||
| PC18B08 | Late embryogenesis abundant protein | 4E−7 | −3.3 | 11.5 | |||
| ST32C09 | Late embryogenesis abundant protein | 1E−33 | −2.9 | ||||
| NXSI008G11 | Late embryogenesis abundant protein | 2E−13 | 2.5 | −5.8 | |||
| NXNV096C08 | Intracellular pathogenesis-related protein | 1E−55 | 4.0 | 2.4 | −5.8 | ||
| ST03G08 | Non-specific lipid transfer protein precursor | 5E−15 | −2.3 | ||||
| NXCI132H04 | Water stress inducible protein | 3E−14 | −5.3 | ||||
| NXNV129E04 | Water stress inducible protein | 2E−31 | −6.8 | ||||
| ST34H09 | Water stress inducible protein | 2E−16 | −6.9 | ||||
| NXCI094C11 | Class VII chitinase precursor | 8E−49 | −3.2 | ||||
| ST37A06 | Aluminium induced protein | 3E−27 | −3.4 | ||||
| ST40F04 | Low Mr heat shock protein | 2E−36 | −2.2 | ||||
| ST14B10 | Class 1 heat shock protein | 2E−36 | −2.1 | ||||
| ST04C10 | Antimicrobial peptide 1 precursor | 3E−36 | −1.9 | ||||
| NXCI085B12 | Chaperon | 2E−34 | −1.9 | ||||
| ST09H11 | Chaperon | 1E−15 | −1.7 | ||||
| NXCI164H02 | Cys proteinase precursor | 3E−53 | −1.9 | ||||
| NXNV103E10 | Cys proteinase inhibitor | 4E−27 | −1.8 | ||||
| NXCI155G05 | Avr9. Cf-9 rapidly elicited gene | 9E−13 | −4.7 | ||||
| NXNV150F06 | Hypersensitive-induced response protein | 8E−53 | −2.1 | 2.4 | 1.8 | ||
| ST34A01 | 14-3-3-like protein | 7E−32 | 2.5 | −2.3 | |||
| ST04G06 | Antimicrobial peptide 1 precursor | 2E−37 | 3.6 | ||||
| NXSI128E05 | Copper chaperon | 4E−4 | −2.3 | ||||
| NXNV132H12 | Disease resistance protein | 2E−28 | −2.4 | ||||
| Genes Encoding Enzymes of the Flavonoid Pathway | |||||||
| NXCI098F10 | Chalcone-flavonone isomerase | 1E−34 | 5.3 | −2.5 | |||
| NXSI063D01 | Naringinin, 2-oxoglutarate 3-dioxygenase | 1E−36 | 1.9 | ||||
| ST28B11 | Naringinin, 2-oxoglutarate 3-dioxygenase | 1E−41 | −2.4 | 3.1 | 2.9 | −1.9 | |
| NXSI068H09 | Phenylcoumaran benzylic ether reductase | 1E−28 | −1.8 | ||||
| NXSI063D09 | Flavoprotein monooxygenase | 3E−35 | −2.7 | 19.3 | −4.3 | ||
| Genes Related to Hormone Transport, Metabolism, and Signaling | |||||||
| ST08H09 | Auxin-repressed protein | 1E−25 | −3.6 | ||||
| NXSI132F03 | Auxin-repressed protein | 2E−5 | −2.5 | ||||
| NXSI137E06 | Auxin-repressed protein | 8E−5 | −3.6 | 4.8 | |||
| ST28E05 | ABC transporter | 4E−96 | −2.4 | 2.8 | |||
| NXSI118D08 | AUX1-like protein | 8E−93 | −1.6 | ||||
| NXNV083G05 | Integral membrane transporter protein | 4E−4 | −2.9 | 13.1 | −5.2 | ||
| ST01G02 | Gasa5-like protein | 2E−32 | 2.4 | ||||
| ST34B04 | Gasa5-like protein | 2E−32 | 2.4 | ||||
| NXSI055B06 | Gasa5-like protein | 5E−22 | 3.1 | −3.3 | |||
| NXNV171G10 | Isopentenyl pyrophosphate dimethylallyl pyrophosphate isomerase | 8E−43 | 1.9 | ||||
| ST08F07 | S-adenosylmethionine synthetase | 2E−38 | −2.1 | ||||
| NXCI133B03 | S-adenosylmethionine synthetase | 3E−52 | −2.8 | ||||
| NXNV164F10 | S-adenosylmethionine synthetase | 1E−5 | −2.1 | ||||
| ST22G04 | S-adenosylmethionine synthetase | 1E−87 | −2.7 | ||||
| ST21H03 | S-adenosylmethionine synthetase | 9E−41 | 1.8 | ||||
| NXCI031E05 | S-adenosylmethionine synthetase | 3E−46 | 2.0 | ||||
| NXSI120A01 | Ethylene responsive element binding protein EREBP | 1E−17 | −1.9 | ||||
| Genes with Some Function Related to Signal Transduction | |||||||
| NXNV096G04 | PINHEAD/ZWILLE-like protein | 4E−35 | 2.6 | ||||
| NXSI101B01 | DNA binding protein | 6E−10 | 2.0 | ||||
| ST02D01 | Protein kinase PK1-like protein | 1E−131 | −2.0 | ||||
| NXSI065C12 | B-box zinc finger-like protein | 3E−28 | 2.6 | ||||
| NXSI120A01 | Ethylene responsive element binding protein EREBP | 1E−17 | −1.9 | ||||
| NXSI039E06 | GPMADS1-like protein | 1E−27 | −2.4 | ||||
| NXSI141G01 | Receptor protein kinase-like protein | 3E−68 | 3.1 | ||||
| NXSI021H06 | Homeobox protein HAT22-like | 1E−54 | 2.0 | ||||
| Other Genes Differentially Expressed during Root Development | |||||||
| NXSI066A02 | 2-Oxoglutarate dehydrogenase E2 subunit | 1E−43 | 2.2 | ||||
| ST02F01 | Aconitase | 2E−44 | −2.7 | ||||
| NXSI116A11 | ADP. ATP carrier protein | 1E−59 | −3.5 | ||||
| ST23C05 | ADP. ATP carrier protein | 3E-28 | −3.4 | ||||
| ST01E03 | Alcohol dehydrogenase | 2E−55 | 3.4 | ||||
| ST22F11 | Apospory-associated protein S18 | 4E−75 | −2.9 | ||||
| ST22E07 | Arginine decarboxylase | 9E−47 | 2.1 | ||||
| NXNV160F07 | Ascorbate peroxidase | 1E−76 | 3.1 | ||||
| NXSI002G12 | Ascorbate peroxidase | 2E−44 | −2.5 | ||||
| NXNV096C09 | Asparagine synthetase | 6E−32 | 1.8 | ||||
| NXSI128G04 | ATP synthetase β-chain | 2E−53 | 2.2 | ||||
| NXCI082D08 | Carbonate dehydratase | 8E−30 | −3.3 | ||||
| NXNV123H12 | Cytosolic ascorbate peroxidase | 8E−47 | 2.2 | 2.0 | |||
| NXSI025B12 | Dormancy associated protein | 6E−15 | 2.4 | ||||
| ST40D05 | Dormancy associated protein | 2E−22 | −4.4 | −2.2 | |||
| NXCI026G09 | Fructose-bisphosphate aldolase | 4E−41 | −2.1 | ||||
| NXCI071F03 | Glyceraldehyde-3-phosphate dehydrogenase | 2E−62 | 4.2 | ||||
| NXCI026F05 | Glycine dehydrogenase | 1E−54 | −2.6 | ||||
| ST12G02 | Internal transcribed spacer | 1E−180 | 13.1 | −5.2 | |||
| NXSI083G03 | Lipase | 2E−94 | |||||
| ST06H02 | Metallothionein-like protein | 2E−38 | |||||
| ST11C02 | Metallothionein-like protein | 5E−51 | −2.6 | −4.0 | |||
| ST14A10 | Metallothionein-like protein | 2E−39 | −2.3 | ||||
| ST21F12 | NADH dehydrogenase | 4E−49 | −2.1 | ||||
| NXSI067B12 | Nuclear RNA binding protein | 1E−16 | 2.0 | ||||
| NXSI139B08 | Nucleoside diphosphate kinase I | 8E−69 | |||||
| ST15H06 | Oxidoreductase | 1E−57 | −3.6 | ||||
| ST04A02 | Oxoglutarate/malate translocator | 3E−20 | −3.6 | ||||
| NXSI001B08 | Purple acid phosphatase precursor | 1E−36 | −2.6 | ||||
| ST15D07 | SAH7 protein | 6E−21 | 2.5 | ||||
| NXCI118F05 | SAR DNA binding protein | 2E−47 | 3.0 | −3.9 | |||
| NXSI023F11 | S-like ribonuclease | 1E−57 | −2.1 | ||||
| ST21A12 | Splicing factor rszp-22 | 6E−32 | 2.7 | ||||
A total of 184 out of 220 differentially expressed genes during different phases of root development were sorted into groups according to their relation to specific physiological processes. The putative function of the cDNAs was estimated according to the highest BLAST hits. Negative and positive ratios indicate down-regulation and up-regulation. For determination of significance, see “Material and Methods.” See Supplemental Table I for a complete list of all 220 genes.
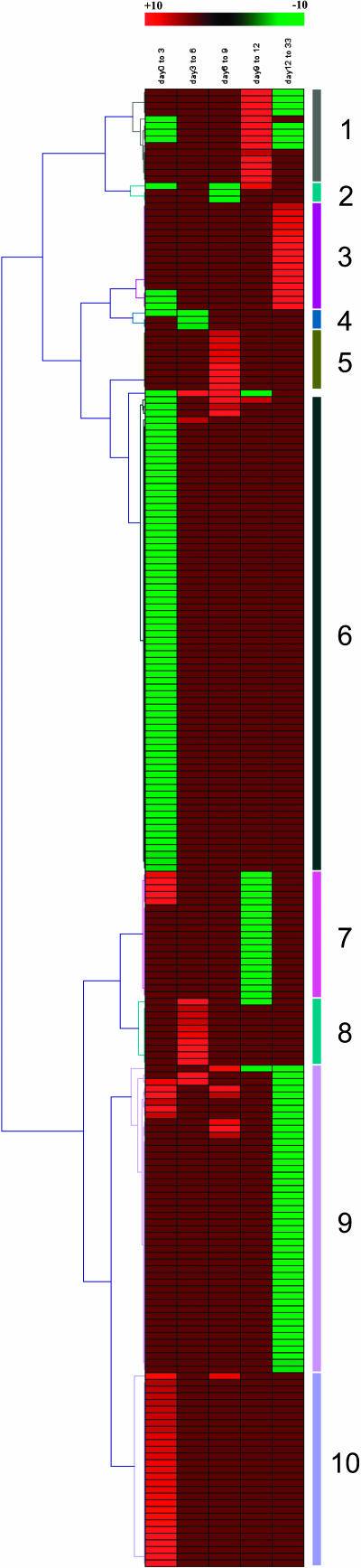
Cell Expansion Phase (Days 0 to 3)
During the first 3 d after wounding and auxin treatment, the cells expanded and the hypocotyl split as a result of excision and strong auxin treatment (Fig. 1). In total, 121 genes were up- or down-regulated (Fig. 2A). Seventy-one out of the 81 genes that were down-regulated are shown in cluster 6 (Fig. 3). Most of the genes differentially expressed belong to the functional categories metabolism (40%), energy, and cell rescue (each 17%; Table I). The most striking change in gene expression was related to down-regulation of transcripts encoding genes predicted to function in chloroplasts (Table II). In addition, transcripts encoding a PINHEAD/ZWILLE-like protein increased.
Figure 3.
Hierarchical clustering of 220 genes differentially expressed during adventitious root development in P. contorta. The fold changes of 220 genes differentially expressed during root development were supplied to the program Genesis (developed by Alexander Sturn, Institute for Biomedical Engineering, Graz University of Technology, 2000–2002). A genetree was created using the average linkage clustering method (Pearson correlation).
Root Primordia Formation Phase (Days 3 to 6)
During this period the root primordia were formed (Fig. 1). In total, 17 genes were differentially expressed (Fig. 2A) and 14 genes were up-regulated (Fig. 2B; Fig. 3, mainly cluster 8). Most genes that were differentially expressed belong to the functional categories metabolism (41%) and transcription (29%; Table I). Transcripts encoding three histones and one cdc2 kinase increased, while transcripts encoding an ethylene-responsive element-binding protein decreased (Table II).
Root Meristem Formation Phase (Days 6 to 9)
Root meristems were formed during this period (Fig. 1). In total, 23 genes were differentially expressed (Fig. 2A). Twenty genes were up-regulated (Fig. 2B; Fig. 3, mainly clusters 5 and 9). The majority of the genes differentially expressed belong to the functional categories cell rescue (48%) and metabolism (43%; Table I). Transcripts encoding an ABC transporter and a B-box zinc finger-like protein were up-regulated (Table II).
Root Formation Phase (Days 9 to 12)
Fully developed roots were formed during this phase (Fig. 1). In total, 38 genes were differentially expressed (Fig. 2A). Twenty-two genes were down-regulated (Fig. 2B; Fig. 3, mainly cluster 9). The majority of genes that were differentially expressed belong to cell rescue, metabolism (each 32%), and protein synthesis (24%; Table I). The transcript level of two genes encoding naringinin 2-oxoglutarate 3-dioxygenase decreased, while the transcript for a gene encoding a flavoprotein monoxygenase increased (Table II).
Root Elongation Phase (Days 12 to 33)
During this period the roots started to elongate (Fig. 1). In total, the expression of 69 genes was significantly changed during this phase (Fig. 2A). Fifty-three genes were down-regulated (Fig. 2B; Fig. 3, clusters 1 and 9). The majority of the genes belong to the functional categories metabolism (45%), protein synthesis (33%), and cell rescue (22%; Table I). Transcripts encoding two water stress-induced proteins and for a GPMADS1-like protein were down-regulated (Table II).
Real-Time PCR Data
To evaluate validity of analysis of gene expression during root development using cDNA arrays, we performed real-time PCR analysis for five genes. The results of expression data obtained by microarray analysis were in agreement (up- or down-regulation) with the ones obtained by real-time PCR (Table III).
Table III.
Quality control of microarray experiments
| Developmental Phase (Days) | Clone ID | Putative Function | Fold Change
|
|
|---|---|---|---|---|
| Microarray | Real-Time PCR | |||
| 0 to 3 | NXCI085E04 | Subunit of PSI | −6.2 | −15.8 ± 6.5 |
| 0 to 3 | NXNV096C08 | Intracellular pathogenesis-related protein | 4.0 | 663.0 ± 113.8 |
| 6 to 9 | NXNV096C08 | Intracellular pathogenesis-related protein | 2.4 | 3.3 ± 0.4 |
| 6 to 9 | NXSI065C12 | B-box zinc finger protein | 2.6 | 1.9 ± 0.03 |
| 12 to 33 | NXCI087F06 | Histone H3 | −3.6 | −4.8 ± 0.4 |
| 12 to 33 | NXCI031E05 | S-adenosylmethionine synthetase | 2.0 | 74.0 ± 3.4 |
Fold change differences of five cDNAs that appeared as significantly differentially expressed during specific phases of root development based on microarray analysis were confirmed by real-time PCR. Negative and positive ratios indicate down-regulation and up-regulation. All real-time PCR reactions were repeated three times, and the mean value and se are presented. The statistical significance of the microarray data is described under “Results” in the text.
DISCUSSION
Protein Synthesis and Degradation
During the first 3 d after auxin treatment, several transcripts encoding ribosomal proteins were up-regulated (Table II; Fig. 3, cluster 10), which indicates an increase of assembly of ribosomes and of protein synthesis. A slightly further increase of this process occurred when root primordia and meristems were formed (day 3 to day 9; Table II). By contrast, during the root formation and root elongation phases (day 9 to day 33) transcripts encoding several proteins involved in protein synthesis were down-regulated. The gene expression pattern of proteins related to protein assembly and folding was similar to those for protein synthesis (Table II). Genes involved in protein degradation were down-regulated during the first 3 d after auxin treatment, then up-regulated when the meristems were being formed (day 6 to day 9), and finally down-regulated again during root formation and root elongation phases (day 9 to day 33).
The general trend is an increased expression of genes involved in protein synthesis and a decrease in expression of genes related to protein degradation for the first 3 d after auxin treatment and the opposite trend when roots are formed and elongating.
Photosynthesis
For the first 3 d after auxin treatment, genes encoding proteins predicted to function in chloroplasts were down-regulated (Table II; Fig. 3, mainly cluster 6). This clearly shows that hypocotyl cells lose their potential to function as photosynthetic cells early during adventitious root formation.
Cell Replication
The auxin treatment stimulates cell division. Six days after auxin treatment, 3.5% of the cells in the pericycle of hypocotyl cuttings of P. contorta are in mitotis compared to 0.2% for non-auxin treated cuttings (Grönroos and von Arnold, 1987). In this study, we show that genes involved in cell replication were up-regulated during the first 6 d after the auxin treatment (Table II), which supports earlier investigations showing that the histone H2A and the PcCDC2 genes are strongly expressed during this period (Lindroth et al., 2001a) and that the expression of the S-phase-specific histone H3 gene increases within 6 to 8 h after induction of adventitious roots in Oryza sativa (Lorbiecke and Sauter, 1999). However, several genes involved in the cell replication were down-regulated during root formation and root elongation phases (day 9 to day 33; Table II).
Cell Wall Weakening/Cell Wall Synthesis
Plant morphogenesis requires mechanisms to control the balance between cell division, cell expansion, and cell adhesion. During the first 3 d after auxin treatment, the cell walls were undergoing modifications as shown by down-regulation of genes with the potential to be active in cell wall synthesis (Table II). At the same time genes involved in weakening cell walls and adhesion of cells were up-regulated (Table II). The opposite trend was observed during the root primordia, root meristem, and root formation phases (day 3 to day 12; Table II).
Stress Response
During the first 3 d after removal of the root and the auxin treatment, the transcript levels of two late embryogenesis-abundant proteins were reduced, and the transcript of a pathogenesis-related protein was more abundant. According to Bray et al. (2000) these changes can indicate that the plants were exposed to water stress. However, several other genes with some function related to stress were either up- or down-regulated (Table II). Increase in the transcripts encoding two late embryogenesis-abundant proteins while root meristems and roots were forming (day 6 to day 12) and reduction of the transcript abundance encoding a lipid transfer protein precursor (day 6 to day 9) point to a reduction in water stress beginning at day 6. Furthermore, down-regulation of the transcripts encoding two water stress-inducible proteins and a pathogenesis-related protein (day 12 to day 33; Table II) can indicate that the adventitious roots had then taken up their function.
A protein of the flavonoid pathway, chalcone synthase, and a pathogenesis-related protein contribute to a constitutive defense barrier in the root epidermis in pea (Mylona et al., 1994). A striking fact during the root primordia formation phase (day 3 to day 6) in P. contorta was the up-regulation of transcripts encoding enzymes of the flavonoid pathway. While root meristems are being formed (day 6 to day 9) transcript level of a naringinin,2-oxogluterate-3 dioxygenase, as well as transcript levels of an intracellular pathogenesis-related protein and a hypersensitive-induced response protein, were increased (Table II). We suggest that a defense barrier is built up inside the hypocotyl from day 3 to day 9.
Hormone Metabolism, Transport, and Signaling
During the first 3 d after auxin treatment, transcript levels of three auxin-repressed genes, an ABC transporter, and an AUX1-like gene were reduced (Table II). ABC transporters are involved in auxin transport (Luschnig, 2002). AUX1 is described as an auxin influx carrier that regulates root development in Arabidopsis by facilitation of auxin transport from leaf to root and unloading toward the primordia (Marchant et al., 2002).
In addition, transcripts of genes that are involved in flavonoid synthesis were up-regulated during the root primordia formation phase (day 3 to day 6; Table II). Flavonoids act as negative regulators of auxin transport in Arabidopsis (Brown et al., 2001). One possible explanation for our results is that active auxin transport is reduced from day 0 to day 6.
While root meristems are being formed (day 6 to day 9), the transcript level of a gene encoding an ABC transporter was up-regulated (Table II). This suggests that active transport of auxin starts during the root meristem formation phase (day 6 to day 9). Furthermore, an increased expression of genes involved in ubiquitin protein degradation machinery was detected during this phase (Table II). In Arabidopsis, Aux/IAA protein degradation is triggered by a ubiquitin-protein ligase. An increased degradation of Aux/IAA proteins leads to a higher concentration of active auxin response factors, which activate transcription by binding at the auxin response element DNA sequence, resulting in auxin-responsive genes showing higher levels of transcription (Dharmasiri and Estelle, 2002). Whether the ubiquitin protein degradation machinery is involved in auxin signaling during root development in P. contorta remains to be shown.
During the root formation phase (day 9 to day 12), the transcript levels of two genes involved in the flavonoid pathway that had been increased during earlier phases were now reduced, together with the transcript level of another gene involved in this pathway (phenylcoumaran benzylic ether reductase). Simultaneously, transcript levels encoding an integral membrane transporter protein and a flavoprotein monooxygenase that is responsible for degradation of flavoproteins were up-regulated. Auxin efflux carriers are composed of at least two polypeptides. One of them is thought to be an integral membrane transporter protein (Palme and Gälweiler, 1999). A continuous auxin transport to the root—probably with an enhanced rate—is likely during the phase of root formation (day 9 to day 12). In Arabidopsis, the process of lateral root formation consists of two major stages: cell cycle reactivation in the xylem pericycle and establishment of a new meristem (Himanen et al., 2002). Pericycle activation depends on a source of auxin inside the root, whereas the outgrowth of lateral roots is regulated by shoot-derived auxin (Bhalerao et al., 2002). Our results suggest that adventitious root formation in P. contorta is regulated by a similar mechanism, i.e. exogenous auxin supply stimulates pericycle activation, and actively transported endogenous auxin stimulates meristem establisment.
Gibberellin is believed to promote cell division and cell elongation (for review, see Harberd et al., 1998). During the first 3 d after auxin treatment, the transcript levels of three gasa-like genes (GA-up-regulated genes) were increased (Table II). Furthermore, transcript abundance of a protein involved in biosynthesis of isoprenoids (isopentenyl pyrophosphate dimethylallyl pyrophosphate isomerase) was up-regulated during the phase of root meristem formation (day 6 to day 9; Table II). During the root formation phase (day 9 to day 12), the transcript level of one gasa-like gene was down-regulated (Table II). Our results suggest that the GA signaling during root development coincides with the activity of the auxin-stimulated cell division.
Four transcripts encoding SAMS were down-regulated during the first 3 d after auxin treatment (Table II; Fig. 3, cluster 6). SAMS catalyze the formation of S-adenosylmethionine (SAM) from Met and ATP. SAM is involved in the methylation of several substances, including nucleic acids, proteins, carbohydrates, and membrane lipids (Ravanel et al., 1998), and it is thought to be an intermediate in ethylene biosynthesis. The importance of down-regulation of SAMS directly after the auxin treatment is unknown, but it might be related to a decrease in ethylene synthesis. This assumption is supported by the decrease of transcript abundance of a gene encoding an ethylene responsive element binding protein (EREBP)-like protein, when root primordia are being formed (day 3 to day 6; Table II).
Signal Transduction
Some genes that regulate cell fate and cell identity were differentially expressed during root development. An interesting fact is that all these genes were up- or down-regulated during a specific phase (Table II).
The level of a transcript encoding a PINHEAD/ZWILLE-like protein was increased during the first 3 d after auxin treatment. In Arabidopsis, mutations in the PINHEAD/ZWILLE gene block formation of shoot apical meristems (Lynn et al., 1999). Assuming that the PINHEAD/ZWILLE-like protein is also important for formation of root meristems, our data suggest that early stages of the root initials are laid down during the first 3 d before there is any sign of root primordia. In addition, the transcript level of a protein kinase-like protein was reduced during the first 3 d after auxin treatment. During the phase of root meristem formation (day 6 to day 9), the abundance of a transcript encoding a B-box zinc finger-like protein was increased (Table II).
During the root elongation phase (day 12 to day 33) transcripts levels of three genes with some function related to signal transduction changed significantly. The level of a transcript encoding a GPMADS1-like protein was reduced, while those encoding a homeobox gene H22-like protein and a receptor protein kinase-like protein were increased.
CONCLUSION
We have used cDNA arrays consisting of 2,178 selected sequences to analyze gene expression pattern during adventitious root development in the P. contorta model system (Grönroos and von Arnold, 1987). The transcript levels of 220 genes were significantly increased or reduced and the majority (183 out of 220) changed only during a specific phase of root development. By examining changes in the global gene expression, we have been able to determinate the timing of molecular events taking place during root development. Of course, changes in gene expression (mRNA levels) do not necessarily lead to changes in protein levels or to changes in developmental processes, but the special importance of transcription as a control point in development is well established for both plant and animal systems (Alberts et al., 2002). Based on the data obtained, we have generated the following hypotheses to be tested in future work.
During the first 3 d after removal of the seedling root and a strong auxin treatment, many processes change in the hypocotyl concomitant with an increase in protein synthesis and a decrease in protein degradation. The plants are exposed to water stress, fewer new cell walls are built, and existing cell walls are weakened. The photosynthetic machinery is down-regulated. The active auxin transport is reduced, including a decrease in transcript abundance of a protein kinase-like protein, which might be involved in regulation of auxin transport processes. The auxin treatment activates the cell replication machinery. Transcript abundance of a PINHEAD/ZWILLE-like gene believed to regulate cell fate is increased, indicating that the root development process has been initiated. During the next 3 d the root primordia are formed.
Root meristems differentiate from day 6 to day 9. This process coincides with an increase of a trancript encoding a B-box zinc finger-like protein. An activation of auxin transport and of auxin-responsive transcription takes place. Cell wall synthesis increases, cell wall weakening decreases, and a defense barrier is built up. The reduction of water stress during further development suggests that the adventitious roots are becoming increasingly functional.
The development of roots from meristems (day 9 to day 12) is accompanied by active auxin transport at a high rate. Cell wall reorganization decreases.
From day 12 the roots start to elongate. There is now an increase of transcripts encoding a HAT22-like protein and a receptor protein kinase-like protein and decrease of a transcript encoding a GPMADS1-like protein. The cell replication machinery is less active. Expression of stress-related genes decreases concomitant with the reduction of protein synthesis, degradation, and folding.
MATERIALS AND METHODS
Plant Material
Seeds of Pinus contorta Dougl. ex Loud. from a half-sib family were surface sterilized and germinated for 4 weeks as described before (Lindroth et al., 2001a) except that germination was performed in a growth chamber at 20°C and under 20 h light per day from fluorescent tubes (Philips TLD 58W/84; Eindhoven, The Netherlands) supplemented with incandescent light (90 μmol m−2 s−1, 400–700 nm). After 4 weeks the primary roots were cut off from the seedlings. The hypocotyl cuttings were treated in Hoagland nutrient solution at pH 5.8 (composed according to Eliasson, 1978) containing 1.23 mm IBA for 6 h. Thereafter, the hypocotyl cuttings were transferred to Hoagland nutrient solution lacking IBA. The hypocotyls (excluding the needles) were harvested at the following time points: 0 d (no IBA treatment), 3 d, 6 d, 9 d, 12 d, and 33 d after IBA treatment, corresponding to various stages of root development (Fig. 1), placed into liquid nitrogen, and stored at −80°C until further use. This experiment was performed three times, resulting in three samples for each developmental stage. For each of the four microarray datasets described below, the RNA samples corresponding to different developmental stages were derived from more than one biological replication (without mixing RNA samples from the same developmental stage). All samples were collected at the same time of day (13 h after the onset of light).
Microarray Procedure
A total of 2,178 Pinus taeda cDNAs were selected from expressed sequence tags obtained from five different cDNA libraries as described by Stasolla et al. (2003). Probe preparation and array printing were also performed in accordance with procedures published by Stasolla et al. (2003). Identity of the clones discussed in this study was confirmed for 99.5% of the cDNAs.
We have assigned functional designations for cDNAs included on the arrays based on homology to the inferred gene sequence of Arabidopsis using the predicted genes assigned by the Arabidopsis Genome Initiative (2000) to one of 12 major categories. The BLASTX e-value used as a cutoff was 10 E−5. A cutoff value of 10 E−10 gives similar results (Kirst et al., 2003).
Target Preparation
RNA was isolated according to the protocol of Chang et al. (1993). DNA was removed with DNase I (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations.
For first-strand synthesis, 20 μg of total RNA from distinct developmental stages, in a total volume of 40 μL, were reverse transcribed overnight using Superscript II RNase H− reverse transcriptase following the manufacturer's directions (Invitrogen). The resulting cDNA was precipitated by adding equal volumes of isopropanol and incubating overnight at −20°C. It was then spun down for 20 min (13,000 rpm, 4°C) and redissolved in 68 μL of DNAse and RNAse free water.
After denaturation the cDNA targets were labeled by incorporation of fluorescent nucleotide analogs (Cyanine 3-dUTP or Cyanine 5-dUTP; Perkin Elmer NEN, Foster City, CA). The targets were hybridized to microarrays using reciprocal labeling according to the experimental design. Labeling, hybridization, and stringency washes followed the protocol from North Carolina State University (van Zyl, U.S. Provisional Patent Application Nos. 60/372,872 and 60/390,142). The slides were scanned using a ScanArray 4000 Microarray Analysis System (GSI Lumonics, Ottawa, Canada), and raw nonnormalized intensity values were registered using Quantarray software (GSI Lumonics).
Experimental Design and Statistical Analysis
A fully balanced, incomplete loop experimental design was used in our experiment, as proposed by Kerr and Churchill (2001). Slides with hybridized microarrays were scanned sequentially for Cy3 and Cy5-labeled probes. Each slide contained the complete set of 2,178 cDNAs printed four times, i.e. giving four replicates. Only arrays with strong signals were considered for further data analysis. Four data sets were obtained: one with Cy3 and Cy5 (11 slides, 88 data points per gene), two with Cy5 alone (2 × 6 slides, 2 × 24 data points per gene), and one data set with Cy3 alone (6 slides, 24 data points per gene). In total, there were three biological replicates, four data sets, 29 slides, and 160 data points per gene. The three biological replicates were distributed over the four data sets such that for each time point, RNA from each biological replicate was represented without pooling the RNA samples.
Raw expression data were normalized for all sources of systematic variation using a modified method as proposed by Yang et al. (2002). This normalization method is based on a robust local regression and accounts for intensity and spatial dependence in dye biases. In brief, log2 transformed data were subjected to the following normalization model:
 |
(1) |
where Yx,y,s,g denotes the observed data in log-ratio, α(x,y,s) represents the effect of spatial (x,y) and signal intensity (s), (x,y) the coordinates of the spots on the slide (column and row, respectively), (αx,y,s) the effect of spatial and signal intensity, s the average log signal intensity, βg the gene effect, θ represents a scaling factor, and ɛ the stochastic random error. Gene significance was then estimated using a two-sample statistical test for comparison of treatments for each gene. Multiple-comparison correction was estimated based on a step-down false discovery rate method proposed by Benjamini and Liu (1999). The fold changes in gene expression between two subsequential stages of root development were estimated after normalization as the ratio of the mean signal intensities for each data set.
Based on the statistical analysis, a gene was considered significantly up- or down-regulated if it met all of four criteria: (1) P value ≤0.001; (2) fold change ≥1.6 for up-regulation or ≤−1.6 for down-regulation; (3) the trend (up- or down-regulation) was consistent in all data sets; and (4) there were significant fold changes in at least two of the four data sets. This resulted in a list of 220 genes that showed reproducible significant fold changes during at least one developmental phase. For the final analysis, fold changes of genes significantly differentially expressed were averaged. Fold changes were supplied to the program Genesis (developed by Alexander Sturn, University of Technology, Institute for Biomedical Engineering, Graz, 2000–2002) for hierachical clustering. A genetree was created using the average linkage clustering method (Pearson correlation). The genes were grouped into 10 clusters (Fig. 3). In a second approach, gene significance was estimated using the mixed model system developed by Wolfinger et al. (2001) and Jin et al. (2001). The results of both analyses were similar (data not shown).
Real-Time PCR
The transcript levels of five cDNAS (NXCI085E04, NXNV096C08, NXCI087F06, NXCI031E05, and NXSI065C12) that were significantly differentially expressed during different developmental phases in the microarray experiments were confirmed by real-time PCR. RNA was isolated according to the protocol of Chang et al. (1993). DNA was removed with DNase I (Sigma, St. Louis), and first-strand cDNA was reverse transcribed from 1.5 μg of RNA and 5,000 copies of kanamycin-RNA (Promega, Madison, WI) using Superscript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's recommendations. Gene-specific primers were designed by using the Primer Express 1.0 software (PE-Applied Biosystems, Foster City, CA). The relative transcript abundance was detected by the Applied Biosystems 7000 Sequencer using SYBR Green PCR Master Mix (PE-Applied Biosystems). The kanamycin and the 18S amplicons were used as internal controls for normalization.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers BE458164, CD016030, BE496352, BE496514, BE582230, BE582244, BE582286, CD016357, BE657064, CD016467, BE643881, BE761820, BE761917, BE761981, BE762153, BF010519, BF010530, BF010654, BE996976, BF010892, BF010912, BF010942, BE997080, BF049724, BF060488, BF060634, BF186115, BF186132, BF220950, BF221197, CD028195, CD027283, CD027448, CD027537, CD020656, AW736855, AW758800, AW758944, AW783947, AW783973, AW783974, AW784002, AW784136, AW784076, AW869967, AW869973, AW870090, AW870223, AW888125, AW985054, AW985250, AW985134, BE123653, BE123751, BE187211, BE209161, BE209361, BE241102, BE241158, BE241263, BF516621, BF516745, BF516963, BF516988, BF517070, BF517265, BF517448, BQ701198, BF517519, BF517621, BF609023, BF609096, BF609541, BF609340, BF609860, BF610137, BF610167, BF610201, BF610552, BQ701283, BQ701365, BQ701373, BQ701379, BQ701500, BQ701504, BF777162, BF777272, BF777380, BF778050, BF778402, BF778209, BF778753, BF778813, BG039084, BG039290, BG039318, BG039369, BG039795, BG039614, BG039757, BG039831, CD026141, BG040618, BG040627, BG040735, BG040865, BG041017, BQ701687, BQ70321, BQ702421, BQ702446, BQ702725, BQ702783, BQ702944, BQ702952, BQ703184, BG275465, BG275428, BG275332, BG275695, AW981744, AW010001, AW010012, AW010022, AW010040, AW010125, AW010132, AW042690, AW010150, AW010205, AW010245, AW010260, AW010288, AW010297, AW010306, AW010327, AW010330, AW010425, AW010443, AW010478, AW010516, AW010543, AW010545, AW010600, AW010624, AW010683, AW010707, AW010718, AW064810, AW010896, AW010925, AW010943, AW010793, AW010802, AW010843, AW010994, AW010999, AW011035, AW011066, AW011211, AW011289, AW011379, AW011429, AW011459, AW011462, AW011463, AW011488, AW011459, AW011525, AW011534, AW011583, AW011598, AW011602, AW042673, AW042679, AW042684, AW042690, AW042696, AW042651, AW042741, AW042769, AW042772, AW042777, AW042831, AW042868, AW042891, AW042917, AW043011, AW043038, AW043047, AW043096, AW043098, AW043150, AW043168, AW043169, AW043314, AW043330, AW043337, AW064862, AW043363, AW043384, AW064642, AW064656, AW064690, AW064693, AW064702, AW064717, AW064810, AW064853, AW064862, AW064886, AW064954, AW065071, AW065072, AW065096, AW065121, AW065158, and AW065178.
Supplementary Material
Acknowledgments
We thank the Forestry Research Institute (SKOGFORSK) for providing the seeds of P. contorta. A. Olson, C. Myburg, C. Shaw, and J. Osbourne are acknowledged for their work in the lab and valuable advice on data analysis.
This work was supported by grants from the Swedish Foundation for International Cooperation, Wenner-Gren Foundations, and the Swedish Research Council for Environment, Agricultural Science and Spatial Planning.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.032235.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular Biology of the Cell, Ed 4. Garland Science, New York
- Benjamini Y, Liu W (1999) A step-down multiple hypotheses testing procedure that controls the false discovery rate under independence. Journal of Statistical Planning and Inference 82: 163–170 [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E (2000) Responses to abiotic stresses. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 1158–1202
- Brown ED, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Dharmasiri S, Estelle M (2002) The role of regulated protein degradation in auxin response. Plant Mol Biol 49: 401–409 [PubMed] [Google Scholar]
- Eliasson L (1978) Effects of nutrients and light on growth and root formation in Pisum sativum cuttings. Physiol Plant 43: 13–18 [Google Scholar]
- Grönroos R, von Arnold S (1985) Initiation and development of wound tissue and roots on hypocotyls cuttings of Pinus sylvestris in vitro. Physiol Plant 64: 393–401 [Google Scholar]
- Grönroos R, von Arnold S (1987) Initiation of roots on hypocotyls cuttings of Pinus contorta in vitro. Physiol Plant 69: 227–236 [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE (1998) Gibberellin inhibitor of an inhibitor of…? Bioessays 20: 1001–1008 [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlén M, Teeri T, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G (2001) The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29: 389–395 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA (2001) Statistical design and the analysis of gene expression microarray data. Genet Res 77: 123–128 [DOI] [PubMed] [Google Scholar]
- Kirst M, Johnson AF, Baucom C, Ulrich E, Hubbard K, Staggs R, Paule C, Retzel E, Whetten R, Sederoff R (2003) Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A, Kvarnheden A, von Arnold S (2001. a) Isolation of a PSTAIRE CDC2 cDNA from Pinus contorta and its expression during adventitious root development. Plant Physiol Biochem 39: 107−114 [Google Scholar]
- Lindroth A, Saarikoski P, Flygh G, Clapham D, Grönroos R, Thelander M, Ronne H, von Arnold S (2001. b) Two S-adenosylmethionine synthetase-encoding genes differentially expressed during adventitious root development in Pinus contorta. Plant Mol Biol 46: 335–346 [DOI] [PubMed] [Google Scholar]
- Lorbiecke R, Sauter M (1999) Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol 119: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C (2002) Auxin transport: ABC proteins join the club. Trends Plant Sci 7: 329–332 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Moerman M, Yang WC, Gloudemans T, Van de Kerckhove J, van Kammen A, Bisseling T, Franssen HJ (1994) The root epidermis-specific pea gene RH2 is homologous to a pathogenesis-related gene. Plant Mol Biol 26: 39–50 [DOI] [PubMed] [Google Scholar]
- Palme K, Gälweiler G (1999) PIN-pointing the molecular basis of auxin transport. Curr Opin Plant Biol 2: 375–381 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job G, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard L, Li P, Strauss SH, Chase MW, Michaud M, Bousquet J (1994) Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc Natl Acad Sci USA 91: 5163–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L (2002) Root development. In CR Somerville and EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Somerville C, Somerville S (1999) Plant functional genomics. Science 285: 380–383 [DOI] [PubMed] [Google Scholar]
- Stasolla C, van Zyl L, Egertsdotter U, Craig D, Liu WB, Sederoff RR (2003) The effects of polyethylene glycol on gene expression of developing white spruce somatic embryos. Plant Physiol 131: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Van Zyl L, Bozhkov PV, Clapham DH, Sederoff RR, von Arnold S (2002. a) Up, down and up again is a signature global gene expression pattern at the beginning of gymnosperm embryogenesis. Gene 1: 83–91 [DOI] [PubMed] [Google Scholar]
- Van Zyl L, von Arnold S, Bozhkov PV, Chen Y, Egertsdotter U, MacKay J, Sederoff RR, Shen J, Zelena L, Clapham DH (2002. b) Heterologous array analysis in Pinacea: hybridization of Pinus taeda cDNA arrays with cDNA from needles and embryogenic cultures of P. taeda, P. sylvestris or Picea abies. Comp Funct Genomics 3: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten R, Sun YH, Zhang Y, Sederoff RR (2001) Functional genomics and cell wall biosynthesis in loblolly pine. Plant Mol Biol 47: 275–291 [PubMed] [Google Scholar]
- Wolfinger RD, Gibson E, Wolfinger L, Bennett H, Hamadeh P, Bushel C, Afshari C, Paules RS (2001) Asssessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Mgai J, Speed TP (2002) Normalization for cDNA microaray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.