Abstract
TERMINAL FLOWER is a key regulator of floral timing in Arabidopsis and other herbaceous species. A homolog of this gene, CsTFL, was isolated from the hybrid perennial tree crop Washington navel orange (Citrus sinensis L. Osbeck). The deduced amino acid sequence of CsTFL was 65% identical to the Arabidopsis TFL1 protein. Wild-type Arabidopsis plants ectopically expressing CsTFL showed late-flowering phenotypes similar to those described for overexpression of Arabidopsis TFL1. In addition, the 35S:CsTFL transgene complemented the tfl1-2 mutant. The severity of the overexpression phenotypes correlated with the amount of CsTFL transcript that accumulated. Unlike many model systems that have been studied, C. sinensis maintains two distinguishable CsTFL alleles. CsTFL transcripts from either allele were not detected in adult vegetative tissues using reverse transcription-PCR, but CsTFL RNAs were detected in all floral organs. In addition, real-time PCR determined that juvenility in citrus was positively correlated with CsTFL transcript accumulation and negatively correlated with the floral-regulatory genes, LEAFY and APETALA1, RNA levels.
Development of higher plants has two distinct phases, juvenile and adult. For all plants, the juvenile phase is characterized by an inability to initiate floral development in response to floral-inductive cues (Hackett, 1985; Poethig, 1990). The juvenile phase can last from 5 to 13 years in citrus depending on the variety and can extend 20 to 30 years for some other tree crops (Davies and Albrigo, 1994; Meilan, 1997). The long juvenile phase is a serious constraint for traditional or transgenic breeding practices. No details of the specific processes responsible for juvenility have been elucidated in perennial tree crops, despite the advantages juvenile phase reduction would have on genetic manipulation and improvement in agricultural systems.
Plants that reach the adult phase of vegetative development are reproductively competent, but most require an appropriate environmental signal to transition to reproductive development. The environmental stimuli necessary to induce flowering in perennial tree crops have been investigated by evaluating responses to phytohormone treatments (GA, auxin, etc.), growth retardant treatments (paclobutrazol, chloromequat), physical constraints (girdling, root restriction), cultural conditions (photoperiod, nutrition, temperature), and grafting (Luckwill, 1979; Pharis et al., 1980; Cobianchi, 1989; Mullins et al., 1989; Garcia-Luis et al., 1992; King et al., 1992; Eris and Barut, 1993; Meilan, 1997). However, documentation of the gene activities underlying vegetative and floral transition in perennials is relatively limited. Populus balsamifera, Malus × domestica, and Eucalyptus globulus are the only perennial tree species that have been extensively investigated (Kyozuka et al., 1997; Southerton et al., 1998; Sung et al., 1999; Kotoda et al., 2000; Rottmann et al., 2000).
In contrast, research over the past decade has resulted in the identification and characterization of numerous genes that disrupt vegetative phase transition or alter meristem identity in the herbaceous annual species, Arabidopsis (Bowman et al., 1993; Weigel and Nilsson, 1995; Blazquez et al., 1997; Telfer and Poethig, 1998; Liljegren et al., 1999; Ferrandiz et al., 2000; Pelaz et al., 2001). Alterations in the timing or location of expression of many of these gene products result in changes in vegetative phase length and/or flowering time (Weigel et al., 1992; Bradley et al., 1997). Among these genes, TERMINAL FLOWER 1 (TFL1) has been shown to be important for delaying flowering and regulating plant growth through the maintenance of indeterminancy of the shoot apex (Shannon and Meeks-Wagner, 1993; Ratcliffe et al., 1998). TFL1 is most similar to a class of mammalian phosphatidylethanolamine-binding proteins (PEBP) that can bind phospholipids (Bradley et al., 1996). A human PEBP (RKIP: Raf Kinase Inhibitor Protein) has been shown to specifically regulate the Raf-1/MEK/ERK signaling pathway (Yeung et al., 1999), suggesting a possible role for TFL1 in disruption of a floral-promotive signaling pathway in Arabidopsis. Overexpression of TFL1 causes a lengthening of the vegetative phase, increased secondary inflorescence production and a delay in flowering (Alvarez et al., 1992; Shannon and Meeks-Wagner, 1993; Ratcliffe et al., 1998). In contrast, strong tfl1 mutants have a reduced vegetative phase, flower early, produce fewer leaves, and prematurely terminate inflorescence development with a large compound flower (Shannon and Meeks-Wagner, 1993; Bradley et al., 1997). Phenotypes similar to those described for AtTFL1 have been reported for ectopic expression of the TFL1 homologs from rice (OsRCN1) and ryegrass (LpTFL) (Jensen et al., 2001; Nakagawa et al., 2002).
Whereas TFL1 maintains the Arabidopsis meristem in an indeterminate state, the production of determinate floral meristems is accomplished by the cooperative activities of the floral meristem identity genes LEAFY (LFY), APETALA1 (AP1), and CAULIFLOWER (CAL), among others (Mandel and Yanofsky, 1995; Pelaz et al., 2001). Loss-of-function mutations in either LFY or AP1 result in the replacement of flowers with shoots or shoot-like structures, whereas their ectopic expression under control of the 35S cauliflower mosaic virus (CaMV) promoter causes early flowering and shoot-to-flower conversion (Mandel and Yanofsky, 1995; Blazquez et al., 1997). Homologs of LFY and AP1 have been isolated from numerous diverse species and shown to cause similar phenotypes when overexpressed in Arabidopsis (Coen et al., 1990; Kelly et al., 1995; Berbel et al., 2001; Carmona et al., 2002; Pillitteri et al., 2004), indicating significant functional conservation of the floral-promotive signaling pathway among plants.
Recently, some of the details of the molecular interactions among floral regulatory genes have been elucidated (Liljegren et al., 1999; Samach et al., 2000; Pelaz et al., 2001). These studies determined that the opposing activities of TFL1 and LFY and AP1 are spatially separated within the meristem and are maintained through a mutual inhibition mechanism. TFL1 prevents floral development by blocking both the expression and activities of LFY and AP1 in the central dome of the shoot apex (Liljegren et al., 1999). Reciprocally, both LFY and AP1 have roles in the negative regulation of TFL1 along the flanks of the inflorescence to promote floral development (Ratcliffe et al., 1999). Loss-of-function mutations in TFL1 result in LFY and AP1 expression in the shoot apex and TFL1 is expressed in flanking meristem in LFY or AP1 mutants. In contrast, TFL1 expression is reduced when LFY or AP1 are constitutively expressed. Evidence suggests that the absolute amount of LFY or TFL1 transcript accumulation is less important than the ratio of LFY:TFL1 in determining meristem fate. A higher ratio results in shortening of the vegetative phase and production of floral meristems (Ratcliffe et al., 1999).
The time to flowering and the number and pattern of vegetative and reproductive shoot development along the stem can determine yield and other important agronomic traits of a given crop. To investigate some of the molecular mechanisms underlying juvenility and flower production in Citrus sinensis, the TFL1 homolog (CsTFL) from Washington navel orange was isolated. The functional similarity CsTFL relative to Arabidopsis (AtTFL1) was assessed using a 35S CaMV:CsTFL construct in both wild-type and tfl1-2 mutant plants. In addition, the accumulation of CsTFL, CsLFY, and CsAP1 transcripts were compared in juvenile (florally incompetent) and adult (florally competent) citrus trees prior to and after a floral-inductive treatment using real-time PCR.
RESULTS
Isolation of a TFL Homolog from C. sinensis
Using degenerate primers based on nucleotide alignments of the TFL sequences from Arabidopsis, Lycopersicon esculentum, Oryza sativa, and Brassica napus, a 1.1-kb genomic segment of the CsTFL gene was amplified from C. sinensis by PCR. This genomic fragment contained 254-bp of exon sequence, which shared 77% identity at the nucleotide level with the AtTFL1 cDNA. CsTFL-specific primers were used for genome walking to obtain a citrus genomic sequence spanning the entire TFL coding region (Fig. 1). Using CsTFL gene-specific primers within the 5′ and 3′ untranslated regions, a CsTFL cDNA was also isolated.
Figure 1.
Genomic organization of CsTFL and similarity of the deduced protein with other TFL homologs. A, The organization of the CsTFL gene, flanking regions (black boxes), coding regions (white boxes), and introns (thin lines) are illustrated. 5′-UTR and 3′-UTR lengths were not determined and are included in the flanking regions. Number 1 indicates the location of the translational start codon as determined by comparison with other TFL homologs. Coordinates for exons are given below the gene diagram. B, Comparison of the deduced amino acid sequence of CsTFL cDNA (AY344244) with those of O. sativa (OsTFL, AAD42895), L. esculentum self-pruning (LeSP, AAC26161), L. perenne (LpTFL, AAG31808), A. majus CENTRORADIALIS (AmCEN, CAC21563), Arabidopsis (AtTFL, BAB08610), and Arabidopsis FLOWERING LOCUS T (AtFT, BAA77838). ClustalW program was used to make the alignment. Identical residues are shaded in black; conserved residues are shaded in gray. Dashed lines indicate gaps introduced to achieve maximum alignment. Intron positions are indicated by black arrowheads below sequences. Asterisks above sequences indicate amino acids in which point mutations were described in Arabidopsis (Bradley et al., 1997; Ohshima et al., 1997) and tomato (Pnueli et al., 1998). The plus sign (+) indicates amino acids identified by Banfield and Brady (2000) to be ligand-binding sites in AmCEN by crystal structure. C, Maximum parsimony tree of different plant TFL-like homologs. The length of horizontal lines are proportional to the similarity between predicted protein sequences. Numbers above the branches indicate support from 200 bootstrap replicates. Tree includes TFL homologs from B and B. napus TFL1-1 and 1-3 (BnTFL1-1, BAA33415 and BnTFL1-3, BAA33417), Nicotiana tabacum CEN1, 2, and 4 (NtCEN1, Q9XH44; NtCEN2, Q9XH43; NtCEN4, Q9XH42), Brassica rapa TFL1-1 and TFL1-2 (BrTFL1-1, BAA33418 and BrTFL1-2, BAA33419), and C. unshiu FT (CuFT, AB027456).
Comparison of the CsTFL gene and cDNA sequences showed that CsTFL has 4 exons and 3 introns that have a conserved location among TFL homologs relative to the protein sequence (Fig. 1A). CsTFL encoded a 19-kD protein with 74% and 70% amino acid identity to Arabidopsis TFL1 and Antirrhinum majus CEN, respectively, but shared the highest identity (80%) with the O. sativa TFL homolog (Fig. 1B). Based on x-ray crystal structure, Banfield and Brady (2000) determined that there are 6 essential amino acids necessary for ligand binding in A. majus CEN (Fig. 1B). All of these residues are conserved in CsTFL with the exception of Ile-110. Ile-110 corresponded to Met-115 in the AmCEN protein (Fig. 1B). Arabidopsis, O. sativa, and Lolium perenne TFL homologs also have amino acid substitutions at this position relative to AmCEN.
Other proteins involved in the regulation of flowering share sequence identity with TFL. The FLOWERING LOCUS T (FT) protein shares 56% identity with TFL1, but functions antagonistically with TFL1 to promote flowering (Nilsson et al., 1998). CsTFL shares only 54% identity with AtFT and 61% identity with a FT homolog from a satsuma mandarin (Citrus unshiu) (Kobayashi et al., 1999). Maximum parsimony analyses of several plant TFL-like and FT-like proteins support the conclusion that the CsTFL is a TFL homolog (Fig. 1C).
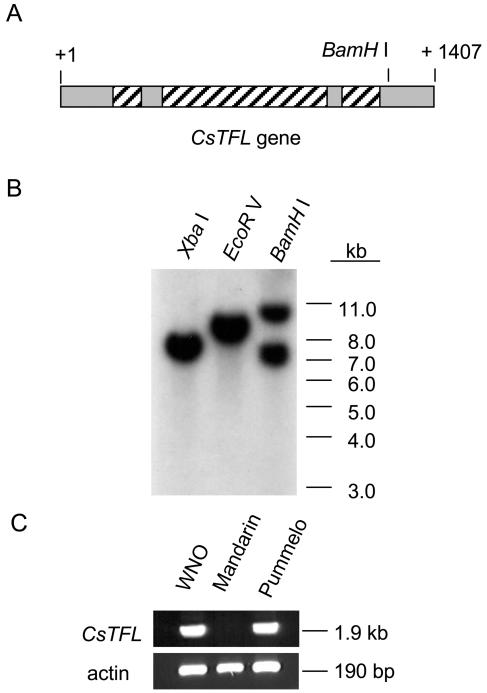
Genomic DNA hybridization using a CsTFL full-length cDNA probe under conditions of high stringency detected 1 or 2 DNA bands when C. sinensis DNA was digested with XbaI, EcoRI, or BamHI (Fig. 2, A and B). This pattern was consistent with CsTFL being a single-copy gene. AtTFL1 is single copy in the Arabidopsis genome (Bradley et al., 1997), whereas small gene families have been identified in ryegrass, rice, and Brassica spp. (Mimida et al., 1999; Jensen et al., 2001; Nakagawa et al., 2002). Previous studies have determined that C. sinensis maintains a relatively heterozygous genome due to its hybrid origin (Citrus maxima × Citrus reticulata; Federici et al., 1998; Pedrosa et al., 2000; Pillitteri et al., 2004). Restriction enzyme digests indicated that sweet orange had limited allelic variation in the flanking region at the TFL locus (Fig. 2B). This was in marked contrast to the heterozygosity detected at the CsLFY and CsAP1 loci (Pillitteri et al., 2003).
Figure 2.
Citrus genomic DNA blot analysis. A, Schematic diagram of the CsTFL gene with location of introns (striped boxes) and exons (gray boxes), and +1 corresponds to the translational start codon. A single BamHI site is located at nucleotide +1230. No XbaI or EcoRI sites are located within the CsTFL gene. B, DNA blot analyses. Washington navel orange DNA was digested to completion with XbaI, BamHI, or EcoRI. The blot was hybridized with a 32P-labeled CsTFL cDNA probe, spanning nucleotides −65 to +1471. Membrane washes were done under high stringency conditions as described in “Materials and Methods”. Size markers (kb) are shown to the right of the blot. C, PCR amplification of CsTFL from Washington navel (WNO), mandarin, or pummelo genomic DNA. PCR reactions were performed using primers designed to anneal to noncoding regions of CsTFL as described in “Materials and Methods”. PCR products were separated on a 0.8% agarose gel and stained with ethidium bromide for size determination. Product sizes are given to the right of the gel. Csβ-actin was used as a positive control for genomic PCR reactions.
To evaluate if the C. sinensis genome contains a single allele of TFL or if the parental alleles were more highly conserved than CsLFY and CsAP1, the CsTFL genomic region was further investigated using pairs of primers that spanned the CsTFL gene. CsTFL 5′- and 3′-flanking region primers designed to amplify a 1.9-kb genomic DNA fragment from Washington navel orange were used. These primers amplified the C. sinensis and C. maxima TFL genes, while a product from C. reticulata was not detected (Fig. 2C). These data indicated that the two C. sinensis parental alleles could be readily distinguished from each other using allele-specific primers, contrasting to the relative conservation of restriction site locations in the pummelo- and mandarin-derived alleles (Fig. 2B).
Expression of CsTFL in Mature Citrus Tissues
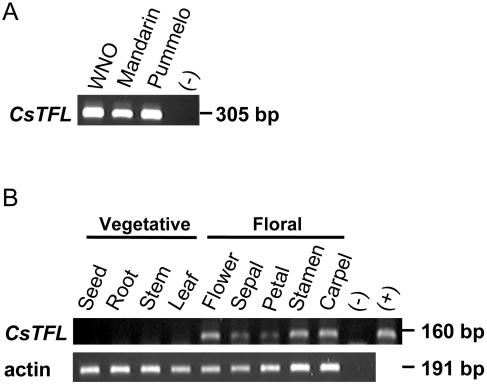
To determine if CsTFL transcripts accumulated in citrus vegetative and floral tissues, CsTFL RNAs were detected using reverse transcription (RT)-PCR. Because the two C. sinensis TFL alleles were diverged at the nucleotide level in the flanking regions (Fig. 2C), primers spanning the third intron were used for PCR. These primers amplified a single 305-bp product from navel orange, mandarin and pummelo genomic DNA (Fig. 3A). The CsTFL transcripts were not detected in any adult vegetative tissues tested, including root, stem, leaf, and seed (Fig. 3B), whereas CsTFL RNAs were present in stems of juvenile plants (see below). In addition, CsTFL was detectable in fully open flowers. Further examination of the individual floral organs showed that CsTFL was detectable in all four floral whorls (Fig. 3B). These data were in contrast to studies that have shown that the L. perenne and Brassica TFL-like genes are expressed in a variety of vegetative tissues in addition to floral organs (Jensen et al., 2001; Nakagawa et al., 2002).
Figure 3.
CsTFL mRNA levels in vegetative and floral tissues. A, Genomic DNA PCR using primers spanning nucleotides +1002 to +1022 of the CsTFL gene showing product amplification from Washington navel orange (WNO), Chandler pummelo, and Fairchild mandarin. No DNA was added to the negative control lane (−). B, Total RNA (4 μg) was used in the RT reactions. CsTFL and actin primer pairs and PCR conditions are described in “Materials and Methods”. All tissue was collected from Washington navel with the exception of seeds that were from navel variety CRC3306A. No RNA was added to the negative control lane (−). The CsTFL positive control (+) was pBSCsTFL-1 (10 ng).
CsTFL Delayed Flowering in Arabidopsis
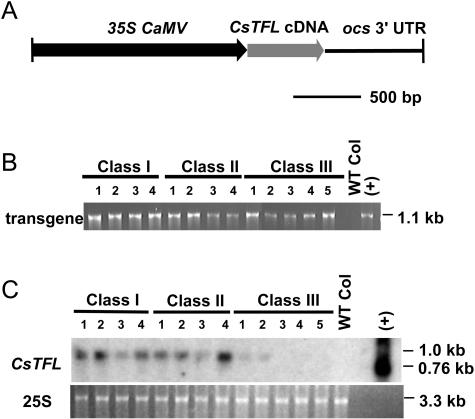
Consistent with AtTFL1 acting as a repressor of flowering, ectopic expression of AtTFL1 causes an extension of the vegetative phase and a delay in flowering in wild-type Arabidopsis. In addition, flowers are at least partially converted to shoots or shoot-like structures (Bradley et al., 1996; Bradley et al., 1997). To evaluate the similarity of CsTFL and AtTFL functions, a chimeric 35S:CsTFL:ocs gene was constructed (Fig. 4A) and introduced into both wild-type and tfl1-2 Arabidopsis plants. Ectopic expression of CsTFL cDNA produced phenotypes similar to those described for other TFL homologs (Ratcliffe et al., 1998; Nakagawa et al., 2002). All of the 32 independent BASTA-resistant T1 plants showed a delay in flowering compared to wild type. Based on flowering time, T1 plants were separated into 3 classes (Table I). On average 35S:CsTFL plants flowered 10 d later than wild-type Arabidopsis under long-day (LD) conditions. The delay in flowering was correlated with an increase in the number of rosette leaves produced prior to bolting.
Figure 4.
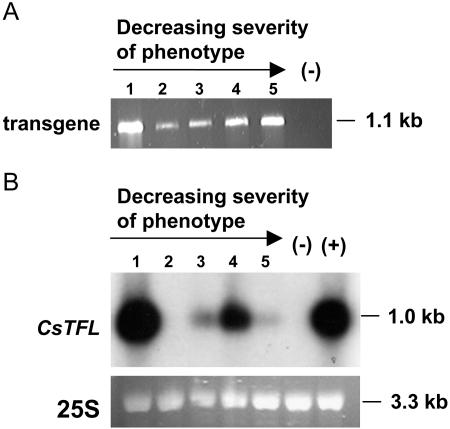
Analysis of Columbia plants expressing the 35S:CsTFL transgene. A, Diagram of the CsTFL transgene including 35S CaMV promoter (black arrow), CsTFL cDNA (−65 to +587; gray arrow), and octopine synthase 3′-untranslated region (ocs 3′-UTR; black line). B, Detection of the 35S:CsTFL transgene in transformed Columbia plants using a 35S forward primer, a CsTFL gene-specific reverse primer, and genomic DNA in PCR reactions. Negative control for transgene detection was wild-type Columbia genomic DNA (WT Col). The positive control (+) was the pPSCsTFL-1 transgene construct. C, RNA blot analysis of 35S:CsTFL T1 plants in wild-type Columbia background. Wild-type Columbia and representatives of three phenotypic classes of 35S:CsTFL T1 plants were analyzed. For RNA blot analyses, RNA was isolated from a mixture of inflorescence and rosette leaves. Total RNA (1 μg) was loaded per lane, blotted, and hybridized with a 32P-labeled CsTFL cDNA probe. The positive control (+) was 0.1 ng of in vitro-transcribed sense CsTFL RNA. The negative control was wild-type Columbia RNA. As a control for equal loading, a picture of the 25S ribosomal subunit from a gel stained with ethidium bromide is shown under the blots.
Table I.
Flowering characteristics of 35S:CsTFL plants
| Plant Genotype | Days to Floweringa | Number of Leavesb | Number of Plants |
|---|---|---|---|
| Landsberg erecta | 21.2 ± 0.40 | 6.2 ± 0.16 | 17 |
| Columbia | 24.0 ± 0.56 | 8.7 ± 0.28 | 20 |
| 35S:CsTFL | |||
| Class I | 44.1 ± 1.3* | 19.2 ± 0.82* | 5 |
| Class II | 36.2 ± 1.2* | 14.3 ± 0.32* | 8 |
| Class III | 29.6 ± 0.38* | 10.8 ± 0.21* | 19 |
| tfl1-2 | 21.0 ± 0.03 | 5.9 ± 0.01 | 36 |
| 35S:CsTFL tfl1-2 | 27.7 ± 0.19 | 8.8 ± 0.12 | 13 |
Plants were kept at 22°C on a 16-h day/8-h night light cycle.
Days from sowing to a 1-cm inflorescence (±sd).
Number of rosette leaves on plants with a 1-cm inflorescence (±sd).
, values are significantly different from wild-type Columbia plants using Student's t test (P > 0.01).
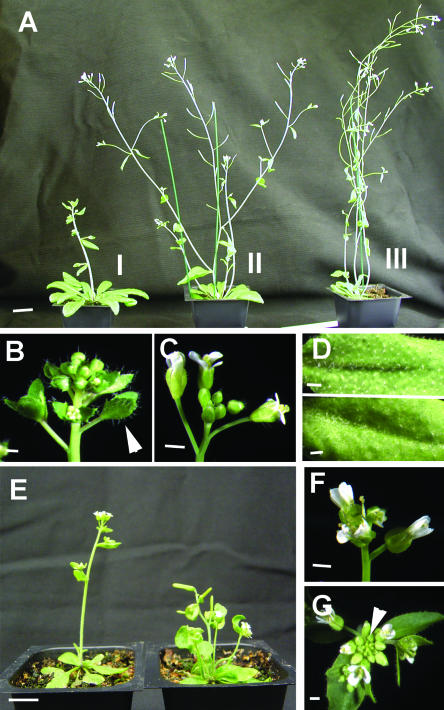
Class I plants had the most severe delay in flowering and increases in rosette leaf number (Table I, Fig. 5A). The number of nodes along the inflorescence was also increased (10.2 ± 0.60 versus 3.9 ± 0.18 for Class I and wild type, respectively). The increase in node number extended to the coflorescences produced along the primary inflorescence stem. The production of bractless inflorescences was common on Class I plants and shoot-like structures were produced in place of subapical flowers on all primary and axillary inflorescences (Fig. 5B). Class I 35S:CsTFL plants had a visible increase in trichome density on both the abaxial and adaxial side of the leaf compared to wild-type plants (Fig. 5D).
Figure 5.
Ectopic expression of CsTFL in wild-type and tfl1-2 mutant Arabidopsis. Transgenic wild-type Columbia (A–D) and tfl1-2 mutant (E–F) plants expressing 35S:CsTFL. A, The three observed phenotypic classes of 35S:CsTFL T1 plants 50 d after planting, showing delayed flowering and increased leaf production of Class I plants. B, 35S:CsTFL Class I primary inflorescence showing flower-to-shoot conversion at subapical floral positions (white arrowhead). C, Wild-type Columbia primary inflorescence. D, Comparison of 35S:CsTFL Class I (upper panel) and wild-type (lower panel) rosette leaf (abaxial side) showing increased trichome density of 35S:CsTFL plants. E, Comparison of a representative 35S:CsTFL tfl1-2 plant (left) and tfl1-2 plant (right) at 27 d after planting, showing extension of vegetative phase development in CsTFL overexpressing plant. F, tfl1-2 primary inflorescence with compound terminal flower. G, 35S:CsTFL tfl1-2 primary inflorescence showing secondary inflorescences and delayed terminal flower development. Terminal flower development had just initiated and is indicated by white arrowhead. Scale bars are 1 cm (A–B, E) and 1 mm (C–D, F–G).
Class II 35S:CsTFL plants showed an average 12-d delay in bolting compared to nontransformed wild-type plants. Floral conversion was always observed on the primary inflorescence. However, some secondary and axillary inflorescences on the same plant did not exhibit this phenotype. No differences in trichome density or inflorescence node number were observed for Class II plants. The late-flowering and flower-to-shoot conversion phenotypes were stably inherited by the successive generation of Class I and II plants.
Class III 35S:CsTFL plants showed an average 5-d delay in flowering. Both leaf number and days to flowering were statistically different from wild type (Table I). Flower-to-shoot conversion in Class III plants was observed, but sporadically on individual plants. The majority of flowers on these plants appeared normal, and they had no significant deviation from wild-type plants with regard to height, trichome density, or inflorescence node number.
Representative 35S:CsTFL plants from Class I, II, and III were tested for the presence of the CsTFL transgene and level of CsTFL transcripts. All transgenic plants examined were BASTA resistant and tested positive for the transgene as determined by genomic DNA PCR (Fig. 4B). The accumulation of CsTFL transcript in Class I, II, III, and Columbia wild-type plants was determined. Using RNA blot analysis under high stringency conditions, the CsTFL cDNA probe did not cross-hybridize with AtTFL1 RNA, since no transcript was detected in wild-type Columbia RNA samples (Fig. 4C). Class I and II plants, which had strong to moderate late flowering and flower-to-shoot conversion phenotypes, had higher levels of CsTFL transcripts than Class III plants. This suggested that CsTFL was sufficient to delay flowering and cause flower-to-shoot conversion in Arabidopsis in a manner similar to AtTFL1.
Ectopic expression of the CsTFL cDNA was also used to examine the ability of CsTFL to complement the phenotype of a strong TFL1 mutant, tfl1-2. tfl1-2 plants flowered earlier, produced a large compound terminal flower on all inflorescences, and produced increased numbers of secondary inflorescences (Alvarez et al., 1992; Page et al., 1999; Table I; Fig. 5E). Ectopic expression of CsTFL cDNA in the tfl1-2 mutant background significantly delayed terminal flower development as compared to the tfl1-2 mutant under LD conditions (Table I; Fig. 5, E–G). Several representatives of 35S:CsTFL tfl1-2 plants with a range of phenotypes were tested for the presence of the transgene and CsTFL transcript levels. All plants tested had the transgene present (Fig. 6A). CsTFL RNAs were detected in all plants exhibiting a delay in floral development with one exception (Fig. 6B). The 35S:CsTFL tfl1-2 plants that flowered latest and displayed the highest degree of flower-to-shoot conversion had CsTFL RNA levels similar to 35S:CsTFL Class I wild-type plants (Fig. 4C). In the studies presented here, there was not a strict correlation of high CsTFL RNA levels and the severity of the delay in flowering phenotype. Similar observations were made by Jensen et al. (2001) using the L. perenne TFL gene (LpTFL1). Ub:LpTFL1 plants with high LpTFL1 RNA levels had variable phenotypes. Five of these plants had a severe phenotype, where plants remained vegetative throughout their life cycle and never flowered. However, other transgenic plants with high LpTFL1 RNA levels flowered. The variation in 35S:CsTFL and Ub:LpTFL1 RNA levels and phenotypes may be due to the fact the it is not the absolute levels of TFL that dictates meristem fate; it is the ratio of LFY to TFL that is the best predictor of flowering (Ratcliffe et al., 1999).
Figure 6.
Analysis of tfl1-2 plants ectopically expressing the 35S:CsTFL transgene. A, Detection of the 35S:CsTFL transgene in representative 35S:CsTFL tfl-2 plants. Genomic DNA (100 ng) was used in a PCR reaction using the CaMV 35S primer and a gene-specific CsTFL primer. Negative control (−) was tfl1-2 genomic DNA. B, RNA blot analyses of 35S:CsTFL tfl1-2 plants. RNA was isolated from a mixture of inflorescence and rosette leaves. Total RNA (2 μg) was loaded in each lane, blotted, and hybridized with a 32P-labeled CsTFL cDNA probe (pBSCsTFL-1). The negative control (−) was tfl1-2 RNA. The positive control (+) was RNA from a 35S:CsTFL Class I plant shown in Figure 4C (lane 1). As a control for equal RNA loading, a picture of the 25S ribosomal subunit from the gel stained with ethidium bromide is shown under the blot. 35S:CsTFL tfl1-2 transgenic plants 1 to 5 were ordered by degree of shoot production with plant 1 having the largest number of shoots.
CsTFL, CsLFY, and CsAP1 RNA Levels in Juvenile and Adult Citrus Trees in Response to Floral-Inductive Treatments
The citrus homologs of the floral meristem identity genes LEAFY and APETALA1 have been described and shown to function in a similar manner to their Arabidopsis counterparts (Pillitteri et al., 2004). To begin to understand the endogenous roles for these genes and TFL1 in juvenility and floral induction in citrus, CsTFL, CsLFY, and CsAP1 RNA levels were compared in adult and juvenile tissue in response to floral-inductive conditions. Adult citrus trees flower profusely in response to 8 weeks of low temperature (15°C day/10°C night) and subsequent shift to warm temperatures (24°C day/19°C night). In contrast, juvenile citrus plants are not competent to flower and produce only vegetative shoots given the same conditions (Davies and Albrigo, 1994). Under floral-inductive conditions, 100% of the adult branches that were chosen according to the criteria given in “Materials and Methods” produced floral inflorescences. In contrast, no flowers were ever produced on juvenile plants under the floral-inductive conditions used in these experiments.
To quantitate the levels of CsLFY, CsAP1, and CsTFL RNAs that accumulate under floral-inductive conditions in adult and juvenile plants, real-time PCR was performed. Real-time RT-PCR measures the threshold cycle value (Ct), which is the PCR cycle at which a detectable increase in product amplification is observed. Therefore Ct is inversely related to the initial amount of template present in a sample. In this study, the Ct values for unknown samples were directly compared to standards amplified in parallel reactions to accurately measure RNA concentrations. A standard curve was produced for each target gene, CsTFL, CsLFY, and CsAP1 (“Material and Methods”). The slope coefficient values for CsTFL, CsLFY, and CsAP1 were −3.08, −2.90, and −2.92, respectively, indicating that PCR amplification efficiency for these products was less than 100%. For example, a reaction at 100% efficiency would double the amount of DNA in every cycle and have a slope coefficient of −3.32 (−1/log2). Variation in amplification efficiencies have been noted elsewhere (Dhar et al., 2001). The Ct values from 3 replicates of unknown samples were averaged and normalized against Csβ-actin to account for variation among RT-PCR reactions. Based on their Ct values, quantities of CsTFL, CsLFY, and CsAP1 transcripts from adult and juvenile tissue were extrapolated and are shown in Figure 7.
Figure 7.
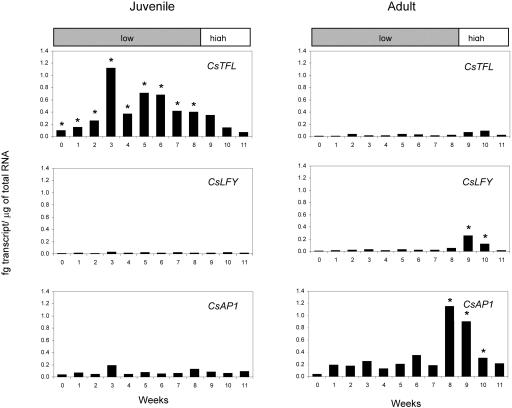
Extrapolated quantities of CsTFL, CsLFY, and CsAP1 transcripts in adult (right panels) and juvenile (left panels) citrus stems. Standard curves for CsLFY, CsTFL, and CsAP1 were constructed as described in “Materials and Methods” (data not shown). Quantities of CsTFL, CsLFY, and CsAP1 RNAs per 1 μg total RNA were extrapolated from these curves. Ct values were normalized using Csβ-actin as covariate. Each time point represents the average of three technical replicates and the least squared mean of 3 biological replicates as described in “Material and Methods”. Asterisks indicate a statistically significant difference (P < 0.05) using pairwise comparisons at each time point between adult and juvenile tissue for either CsTFL, CsLFY, or CsAP1. Gray and white bars indicate time points plants were subjected to low (15°C) or high (24°C) day-time temperatures, respectively.
CsTFL transcripts accumulated to higher levels in juvenile stem tissue compared to adult tissue (Fig. 7). The average concentration of CsTFL transcript across all time points was barely detectable for adult stems (0.02 fg/μg total RNA) compared to 0.40 fg/μg total RNA for juvenile stems. During the 8 weeks of low-temperature conditions, CsTFL RNAs were 7- to 32-fold more abundant in juvenile versus adult plants (Fig. 7). However, when plants were transferred from low to warm conditions (15°C to 24°C daytime temperature), CsTFL RNA levels decreased in juvenile plants. In contrast, a small increase (3-fold) in the CsTFL transcript level was observed in adult tissues under warm-temperature conditions.
The CsLFY and CsAP1 patterns of expression contrasted that seen for CsTFL. CsAP1 and CsLFY transcripts were present at low levels in juvenile tissue (Fig. 7). CsAP1 RNAs were more abundant in juvenile plants, ranging from 0.03 fg/μg to 0.19 fg/μg, than CsLFY RNA, which was detected at <0.05 fg/μg. No change in CsLFY or CsAP1 RNAs was detected when juvenile plants were shifted to warm temperature.
The responses of adult tissues to floral-inductive treatments were distinct from juvenile plants. CsLFY and CsAP1 transcripts accumulated to higher levels in adult tissues relative to juvenile tissues toward the end of the low-temperature induction period and after transfer to warm temperatures. Although the concentration of CsAP1 RNA was approximately 6 times that of CsLFY RNA in mature stems (average 0.34 fg/μg total RNA versus 0.052 fg/μg total RNA, respectively), both transcripts had a similar 6-fold increase in transcript accumulation in mature stems after week 7 of low-temperature treatment (Fig. 6). Two to 3 weeks after transfer to warm temperatures, both CsLFY and CsAP1 transcript accumulation declined. This was expected; this period corresponded to fruit set, where flowers were senescing and ovaries were expanding.
DISCUSSION
CsTFL Is Functionally Similar to the Arabidopsis TFL1 Gene
Citrus is a diploid hybrid derived from C. reticulata (mandarin) and C. maxima (pummelo) with a relatively stable heterozygous genome (Pedrosa et al., 2000). DNA blot hybridization analysis revealed that CsTFL is most likely a single-copy gene with more limited allelic variation compared to other loci such as CsLFY and CsAP1 (Pillitteri et al., 2004). Based on restriction enzyme digestion patterns, there was less heterozygosity around the 5′- and 3′-regions flanking the CsTFL locus than previously observed for CsLFY or CsAP1. The importance of heterozygosity for regulation of CsTFL, CsLFY, or CsAP1 is currently unknown. However, heterozygosity could be maintained by selection and have some unique impact on gene regulation within C. sinensis that is not present in either parent.
CsTFL encoded a 19-kD protein that shared high sequence identity with the AtTFL1 and AmCEN proteins that function to delay flowering and maintain the indeterminate fate of inflorescence meristems. Ten of the 11 residues that were identified to be important for protein function (Ohshima et al., 1997; Pnueli et al., 1998; Banfield and Brady, 2000) were conserved in CsTFL with the exception of Ile-110. Variation at this position was observed in other species as well, including Arabidopsis, ryegrass, and rice (Ohshima et al., 1997; Jensen et al., 2001; Nakagawa et al., 2002). Jensen et al. (2001) has speculated that the residue at position 110 could be partially responsible for the range in the severity of phenotypes in transgenic Arabidopsis plants ectopically expressing the TFL homologs. However, recently the rice TFL homologs, RCN1 and RCN2, have been isolated and determined to have a Ser at position 110. The phenotypes described for overexpression of either RCN1 or RCN2 in Arabidopsis were not as severe as those described for ryegrass, which also has a Ser at position 110 (Nakagawa et al., 2002). No data were presented describing the level of RCN1 or RCN2 transcript accumulation in transformed Arabidopsis plants. Therefore, although transcript accumulation between the LpTFL and rice RCN1/RCN2 cannot be directly compared, there does not appear to be a strict correlation with type of amino acid present at position 110 and severity of phenotype in plants ectopically expressing TFL homologs.
Ectopic expression of CsTFL caused a significant delay in flowering and increase in inflorescence production in both wild-type and tfl1-2 plants. These data indicated that CsTFL can function in a manner similar to the endogenous AtTFL1. In wild-type plants, the CsTFL transgene RNAs were more abundant in Class I and II transformants, which showed a more severe delay in flowering and more complete flower-to-shoot conversion relative to Class III plants. The observed phenotypes were correlated with CsTFL RNA accumulation. In addition, trichomes on the adaxial and abaxial side of the leaves of 35S:CsTFL Class I plants were visibly more dense than those on wild-type Columbia plants. This was not described for 35S:AtTFL1, but was observed when the ryegrass TFL homolog (LpTFL) driven by the Zea mays ubiquitin (Ub) promoter in Arabidopsis (Jensen et al., 2001). The loss of adaxial trichomes is a common morphological marker to identify juvenile to adult vegetative phase transition (Chien and Sussex, 1996; Telfer and Poethig, 1998). Therefore, the high density of trichomes on 35S:CsTFL and Ub:LpTFL plants may indicate a lengthening of the juvenile phase of development. This is consistent with the observation that loss of function tfl mutants have slightly shortened juvenile phases when grown under short-day conditions (Shannon and Meeks-Wagner, 1991).
CsTFL transcript did not accumulate in any of the citrus vegetative tissues examined in these experiments, including leaves, stems, and roots. These data were not consistent with studies using rice, apple, or ryegrass. In these plants, TFL RNAs were readily detected in vegetative tissues of adult plants (Jensen et al., 2001; Kotoda et al., 2001; Nakagawa et al., 2002). In citrus, CsTFL transcripts accumulated in all organs of fully developed flowers. The significance of CsTFL RNAs in flowers is unclear; however, the RNAs for the TFL homolog from apple were also detected in flowers (Kotoda et al., 2001). The possibility that some TFL homologs have been recruited for selected aspects of other developmental pathways has not been examined. However, among the small family of TFL1-like genes in Arabidopsis, Mimida et al. (2001) determined that there are some differences in the expression and function of TFL1 and another gene family member ARABIDOPSIS THALIANA CENTRORADIALIS (ATC). Loss-of-function of ATC does not cause tfl1-like phenotypes even though overexpression of ATC caused similar phenotypes to those noted for overexpression of TFL1. This suggested that ATC can functionally substitute for TFL1, but that these genes have divergent roles in vivo.
CsTFL Expression Is Correlated with Juvenility in Citrus
Arabidopsis grows monopodally, where the apical meristem remains indeterminate and produces both the vegetative and floral phases of development. In contrast, plants such as tomato and citrus have a sympodal growth habit. In sympodal development, the shoot apical meristem is terminated and future growth continues from the upper most lateral bud causing a typical zigzag stem pattern. Pnueli et al. (1998) determined that the functional ortholog of TFL1 from tomato, SELF-PRUNING (SP), prevents the activation of the floral developmental program in vegetative sympodal shoots and therefore influences the process that controls vegetative and reproductive shoots alternation. In situ hybridization determined that SP was expressed in all developing primordia, but the sp mutation showed little effect on development other than shoot determinancy. Pnueli et al. (1998) speculated that the expression of SP in leaf and flower primordia may reflect a systemic mechanism to inhibit floral promotive signaling. By similar speculation, the higher levels of CsTFL transcript observed in juvenile stems prior to and during cold-temperature induction is consistent with CsTFL having a direct and/or systemic role in preventing flowering in incompetent plants (juvenile). This scenario was also suggested by Jensen et al. (2001) in perennial ryegrass where the significantly higher levels of TFL (and other floral repressors) are maintained to prohibit precocious flowering before appropriate flowering time.
The observation that ectopic expression of CsTFL was sufficient to delay flowering and cause flower-to-shoot conversion in both wild-type and tfl1-2 Arabidopsis was also consistent for a role of CsTFL in maintaining juvenility. In addition, the quantitative RT-PCR studies presented here are consistent with a model where CsTFL, CsLFY, and CsAP1 function in a manner similar to the Arabidopsis TFL, LFY, and AP1. AtTFL1 inhibits the expression and activities of the floral identity genes, AtLFY and AtAP1; furthermore, the ratio of LFY and TFL products determines meristem fate (Ratcliffe et al., 1999). Three observations of juvenile and adult plants prior to and after floral inductive conditions are consistent with the opposing actions of CsLFY and CsTFL. First, CsTFL RNA levels were negatively correlated with flowering. CsTFL RNA levels were dramatically different in adult and juvenile plants exposed to low-temperature floral-inductive conditions. CsTFL RNAs were 13-fold more abundant in nonflowering juvenile plants when compared to adult trees. Second, CsLFY and CsAP1 RNAs were positively correlated with the propensity to flower. CsLFY and CsAP1 RNAs were 3- to 4-fold more abundant in adult plants compared to juvenile plants. Third, only adult plants had significant increases in CsLFY or CsAP1 transcript accumulation during the floral inductive conditions.
Citrus trees will flower when exposed to as little as 4 weeks of low-temperature induction (Lovatt et al., 1988). Therefore CsLFY and CsAP1 transcripts were expected to be detected earlier in adult trees than what was observed in these experiments. There are two possible explanations for this observation. First, 4-week cold treatments induce flowering in many but not all stems; only an 8-week cold treatment provides a uniform floral induction (Lovatt et al., 1988). Therefore, heterogeneity in tree responses could account for this variation. Second, due to the limited amount of material available for these studies, whole stems were used for RNA isolation. This may have diluted the target RNAs early during the low-temperature treatment when buds remained small. However, as inflorescence buds swelled and expanded (weeks 7–11), more cells expressing the target RNAs were present.
The analysis of many distantly related species has determined that TFL-like genes play a critical role in plant development primarily through regulating the timing of vegetative phase transition and the maintenance of indeterminate meristems. No details of the specific mechanisms responsible for phase change have been elucidated in perennial tree crops. These experiments determined that elevated levels of a functional TFL homolog was well correlated with juvenility in C. sinensis. It remains to be determined whether down-regulation of CsTFL expression through transgenic technologies could reduce this lengthy phase in citrus and other important agronomic crops.
MATERIALS AND METHODS
Plant Material and Tissue Collection
Leaves, roots, and stems used in RT-PCR were collected from 5-year-old potted Washington navel orange (Citrus sinensis L. Osbeck) scions on Carizzo citrange (C. sinensis × Poncirus trifoliata L. Raf.) rootstock. Flowers were collected at full bloom from 18-year-old trees located at the Agricultural Experimental Station at the University of California (UC; Riverside, CA). Floral organs were separated using forceps. First whorl sepal tissue included receptacle. Seeds were collected from fully mature fruit of navel variety CRC3306A. Young, pliable leaves were collected for DNA isolation. All tissue was frozen in liquid nitrogen immediately and stored at −80°C until further use.
For real-time PCR, five-year-old Washington navel orange trees (adult) and 4-month-old seedlings (variety CRC3306A, juvenile) were maintained at 15°C day/10°C night temperatures for 8 weeks (low temperature treatment), followed by 24°C day/19°C night temperatures (high temperature treatments) for an additional 3 weeks. A 16-h day/8-h night light cycle was used. Trees were watered regularly with nutrient solution. Stems with high probability of producing floral shoots were selected using the following criteria: shoot length ≤7 cm, leaf area ≤18 cm2, node number ≤8, and no thorns (Lord and Eckard, 1985). Uniform stems were collected at weekly intervals, cut immediately below the fourth node, and leaves were removed. Whole stems, including internodes, were collected for RNA isolation from both adult and juvenile plants.
Nucleic Acid Extraction from C. sinensis
C. sinensis genomic DNA used in PCR and genome walking was isolated by a modified cetyl-trimethyl-ammonium bromide (CTAB)-based method of Webb and Knapp (1990). DNA was resuspended in 100 μL of Tris-EDTA (10 mm Tris-HCl, pH 8.0, 1 mm EDTA). C. sinensis genomic DNA used for DNA blot analyses was isolated by CsCl banding according to Fischer and Goldberg (1982). The identity of the pummelo and mandarin parents of Washington navel origin are not known. Therefore representatives were chosen from the UC Riverside Citrus variety collection. Fairchild mandarin and Chandler pummelo DNAs were gifts from Virginia Alonzo (UC Riverside) and Drs. Mikeal Roose and Claire Federici (UC Riverside), respectively. RNA was isolated using a LiCl-based method described by Puthoff (1999). Total RNA was resuspended in water and stored at −80°C until further use.
Isolation of a CsTFL Genomic and cDNA Clones and Phylogenetic Analyses
A C. sinensis TFL homolog was isolated using degenerate forward (TFL F1; 5′-GTCT(A/T/C)(C/T)AATGG(A/C)CATGAG(C/T)TCT-3′) and reverse (TFL R3; 5′-CCT(A/G)TG(G/T)AT(C/T)CC(A/T)AT(A/G)(C/T)(G/T/C)GGC-3′) primers, which were designed based on alignments of the TFL nucleotide sequences from Arabidopsis (U77674), Antirrhinum majus (Bradley et al., 1996), Lycopersicon esculentum (Pnueli et al., 1998), Oryza sativa, and Brassica napus (Mimida et al., 1999). Genomic PCR was carried out using 100 ng of genomic DNA under the following conditions: 30 cycles of 15 s at 95°C, 30 s at 54°C, and 2 min at 72°C. A 1.1-kb genomic DNA CsTFL fragment was isolated from a 1.0% agarose gel and cloned into pGEM T-Easy vector (Promega, Madison, WI). All PCR products were sequenced using fmol DNA Cycle Sequencing System (Promega) or were sequenced at the UC Riverside Genomics Institute Core Facility.
Citrus genomic DNA (5 μg) was digested separately with DraI, EcoRV, PvuII, ScaI, and StuI and ligated to GenomeWalker adapters according to manufacturer's instructions (CLONTECH, Palo Alto, CA). A full description of primer sequences and product sizes are given in Pillitteri (2002). A minimum of 120 bp of overlapping sequence with 100% identity was used as the criteria for identification of overlapping GenomeWalker fragments. The assembled CsTFL gene sequence (accession no. AY344254) was used to design the forward (TFL finalF1; 5′-GGGGTACCGTTCTTACAATCTCTTTAGCG-3′) and reverse (TFL finalR1; 5′-GCTCTAGACATTATATTGCAGCAACAAGC-3′) primers. To facilitate subsequent cloning, an XbaI site was incorporated into the CsTFL reverse primer (underlined). These primers were used to isolate a CsTFL cDNA clone and genomic DNA sequence spanning the translational start and stop sites. Genomic PCRs used these primers and 100 ng of genomic DNA under the following conditions: 30 cycles of 15 s at 95°C, 30 s at 58°C, and 2 min at 72°C. The CsTFL first-strand cDNA synthesis used 4 μg of total RNA from whole flowers, 0.5 μm oligo(dT) (20-mer), and 4 units of ImProm-II reverse transcriptase (Promega) according to manufacturer's instructions and the PCR reaction conditions above. All products were ligated into the pGEM T-Easy vector (Promega). The PCR products were sequenced to ensure fidelity. The 1.9-kb CsTFL genomic sequence and 652-bp CsTFL cDNA sequences have GenBank accession numbers of AY344255 and AY344244, respectively. A maximum parsimony tree of different plant TFL and FT proteins was generated using PAUP 4.0b10 (Swofford, 1999). Branches were supported from 200 bootstrap replicates.
Citrus DNA Blots and PCRs to Evaluate CsTFL Parentage
To evaluate CsTFL gene copy number and the allele origins, genomic DNA blots with 10 μg of restriction-enzyme-digested citrus genomic DNA were hybridized with a 32P-labeled CsTFL cDNA probe (pBSCsTFL-1). Transfer, hybridization, and wash procedures were done according to Wahl et al. (1979). The CsTFL cDNA was PCR amplified, gel purified, and labeled using α[32P]dCTP and the Prime-a-Gene labeling kit (Promega). Membranes were exposed to Hyper-film-MP (Amersham, Buckinghamshire, UK) at −80°C for at least 2 d.
Genomic PCR was performed using DNA isolated from Washington navel orange, Fairchild mandarin orange and Chandler pummelo. The primer pair used for allele-specific CsTFL amplification was TFL finalF1 and TFL finalR1, which amplified nucleotides −65 to +1471. PCR was performed using the conditions described above.
RT-PCR
CsTFL RNAs were detected in various citrus tissues using RT-PCR. Forward and reverse primer pairs for CsTFL cDNA fragment amplification were 5′-GATTGTGACAGACATTCCAG-3′ (TFL sybrF1) and 5′-ATGATCTCTTGATGAAGGTG-3′ (TFL sybrR1), respectively. TFL sybrF1 and TFL sybrR1 primers corresponded to nucleotides +1002 to +1022 and +1288 to +1307 in the complete CsTFL gene sequence and detected TFL RNAs in mandarin, pummelo, and sweet orange. The positive control for the PCR reactions was pBSCsTFL-1. A citrus β-actin gene was used as a positive control for RT-PCR reactions as previously described (Pillitteri et al., 2004). Total RNA (2 μg) was used for first-strand synthesis using an oligo(dT) primer (20-mer) and 2 units of ImProm-II reverse transcriptase. PCR reaction conditions were 30 cycles of 30 s at 94°C, 30 s at 63°C (CsTFL) or 61°C (β-actin), and 2 min at 72°C.
Real-Time PCR
Total RNA (3 μg) from adult and juvenile stem tissue was treated with 3 units of RQ1 DNase (Promega) according to manufacturer's instructions and used in first-strand synthesis using an oligo(dT) primer (20-mer) and ImProm-II reverse transcriptase according to manufacturer's instructions. For real-time PCR, gene specific forward and reverse primers were used: the CsTFL primers were TFL sybrF1 and TFL sybrR1; the CsAP1 primers were 5′-ACCGCTCTCAAACACATCAG-3′ and 5′-GCAGCCTTCTCTCTCTCC-3′; the CsLFY primers were 5′-AGGTCCAGAACATCGCCAAG-3′ and 5′-TGAAAGCCCTCCTCAGTGC-3′, and the Csβ-actin primers were Csactin F1 and Csactin R1. The CsTFL, CsLFY, and CsAP1 primer pairs detected PCR products from both pummelo and mandarin, therefore were likely to detect RNAs from both alleles in C. sinensis. Product sizes were 160 bp (CsTFL), 137 bp (CsAP1), 185 bp (CsLFY), and 191 bp (Csβ-actin).
Real-time PCR products were amplified using 1 μL (Csβ-actin) or 4 μL (CsTFL, CsLFY, and CsAP1) of the RT reaction mixture, 10 μL 2 × SYBR Green Master Mix (Qiagen, Valencia, CA), 0.3 μm each of forward and reverse primer, and water to a 20 μL final volume. Reactions were run on an ABI PRISM 7700 Sequence Detector (PE-Applied Biosystems, Foster City, CA). Thermocycler conditions were 15 min at 95°C and 40 cycles of 15 s at 95°C, 30 s at 64°C (CsLFY) or 61°C (CsTFL, CsAP1, and Csβactin), and 1 min at 72°C. Confirmation of specific product amplification was done by Tm analysis using Dissociation Curve 1.0 program (PE-Applied Biosystems).
To establish a standard curve for quantification, sense-strand RNAs for CsTFL, CsLFY, and CsAP1 were synthesized in vitro using the MAXIscript T3 in vitro transcription kit (Ambion, Austin, TX) according to manufacturer's instructions. In vitro-transcribed RNA (1 ng) was reverse transcribed using ImProm-II reverse transcriptase and gene-specific RT primers. The gene-specific RT primers were 5′-TTTGGAGGTTATGTGGAG-3′ (CsTFL), 5′-TACCAAATGCCGAGACG-3′ (CsLFY), and 5′-AAGGCTACACGAACATAC-3′ (CsAP1). First-strand cDNAs were serially diluted ranging from 5 × 10−4 ng to 5 × 10−9 ng and used as template controls in real-time PCR experiments. All RT-PCR reactions using standards were done in parallel with unknown samples. Threshold cycle (Ct) value is the cycle number at which a significant increase in product amplification can be detected. The Ct value for each serial cDNA dilution was plotted against the log of the cDNA concentration to determine the concentrations of target-gene transcript in unknown samples.
Statistical Analysis of Real-Time PCR Data
At each weekly collection, 3 stems (biological replicates) were collected from both adult and juvenile citrus plants. The RNA isolated from each stem was used in 3 independent RT-real-time PCR reactions (technical replicate). Csβ-actin was amplified from one technical replicate from each biological replicate. Ct values from the 3 independent replications of RT-real-time PCR of unknown samples were averaged and statistical analyses were done across biological replicates. The main effects of age (adult or juvenile) and time (and their interaction) were included in an analysis of covariance using Csβ-actin as a covariate to control for sample variation in the total amount of RNA in different reactions. Least squared means were compared at each time point between the different age groups. These analyses were done using JMP statistical software version 4.0.3 (Statistical Analysis Software, SAS Institute, Cary, NC). The resulting Ct values were extrapolated to an averaged standard curve of the corresponding target gene to determine the quantities of transcripts present.
Construction of the Chimeric CsTFL Transgene
The complete coding region of the CsTFL cDNA was excised from pGEM T-easy (pGCsTFL-1) with EcoRI and ligated into EcoRI-digested pBluescript SK+ to create pBSCsTFL-1. The pBSK+ plasmid was digested with XbaI to excise the CsTFL cDNA using the XbaI site in the pBSK+ multicloning site and the XbaI site introduced by the reverse primer during amplification. The cDNA was ligated into the XbaI site in pCL0011 (C. Li and P. Springer, unpublished data) to create pPSCsTFL-1. The BASTA-resistant vector pCL0011 is described in Pillitteri et al. (2004). The pPSCsTFL-1 construct was transformed into Agrobacterium tumefaciens strain EHA105 using the freeze-thaw method of Gelvin and Schilperoot (1995).
Arabidopsis Seed Stocks, Transformation, and Evaluation of Transgenic Plant Phenotypes
Seed stocks were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (Columbus, OH). The tfl1-2 mutant (CS3091) was homozygous recessive in Landsberg erecta (Ler) background. All seeds were washed in 95% ethanol and rinsed 3 times with distilled water. Seeds were kept in water at 4°C for up to 5 d prior to planting in soil.
Arabidopsis plants were transformed with A. tumefaciens strain EHA105 using the floral dip method described by Clough and Bent (1998). Transformed seeds (T1) were planted in flats in soil and selected with BASTA (ammonium-dl-homoalanine-4-yl-(methyl)phosphinate) (AgroEvo, Monvale, NJ). For flowering-time experiments, untransformed control seeds were planted at the same time, but did not receive any BASTA applications. Plants were kept under long-day (LD) conditions (16-h day/8-h night) at 22°C. Days to flowering and rosette leaves were counted when plants had a 1-cm long inflorescence. Statistical analysis was done using Student's t test at P < 0.01. Tissue for DNA and RNA analysis was collected from BASTA-resistant T1 plants when siliques started to form.
Transgene Detection and RNA Blot Analyses
To detect the presence of the CsTFL transgene, genomic DNA was used in a PCR reaction using a CaMV 35S forward primer (5′-ACCTCCTCGGATTCCATTGCC-3′) and TFL FinalR1. PCR reactions were performed using 100 ng of genomic DNA under the following conditions: 27 cycles of 15 s at 95°C, 30 s at 63°C, and 2 min at 72°C. For RNA blot analyses, inflorescences and leaves were collected from representative transformed plants. Total RNA from all transgenic plants was isolated using the Qiagen RNAeasy Isolation kit (Qiagen). RNA blots and washes were performed according to Pautot et al. (1991) using 1 or 2 μg of total RNA per lane. In vitro transcribed RNAs were used as positive controls. In vitro CsTFL transcripts were produced from XhoI-digested pBSCsTFL-1 using the T3 MAXIscript in vitro transcription kit (Ambion) according to manufacturer's instructions.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY344244, AY344254, and AY344255.
Acknowledgments
We thank Virginia Alonzo and other Walling laboratory members for helpful discussions. We thank Drs. Michael Roose and Claire Federici (UC Riverside) for providing Chandler pummelo DNA and Virginia Alonzo for providing the Fairchild mandarin DNA. Statistical analysis of the real-time PCR data was done with the help of Drs. Leonard Nunney and Kurt Mckean (UC Riverside). We also thank Dr. Tracy Kahn (Citrus Variety Collection, UC Riverside) for locating and donating fruit from the seedy navel variety CRC3306A for use in these experiments. The sequencing of CsTFL was supported by UC Riverside Genomics Institute Core grants to C. Lovatt and L. Walling.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036178.
References
- Alvarez J, Guli CL, Yu XH, Smyth DR (1992) TERMINAL FLOWER: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2: 103–116 [Google Scholar]
- Banfield MJ, Brady RL (2000) The structure of Antirrhinum CENTRORADIALIS protein (CEN) suggests a role as a kinase regulator. J Mol Biol 297: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran J-P (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25: 441–451 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379: 791–797 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Cubas P, Martinez-Zapater JM (2002) VFL, the grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiol 130: 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cobianchi D (1989) Paclobutrazol and S3307 effects on cherry and pear trees. Acta Hortic 239: 292–296 [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) FLORICAULA: A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Davies FS, Albrigo LG (1994) Citrus. CAB International, Wallingford, UK
- Dhar AK, Roux MM, Klimpel KR (2001) Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR green chemistry. J Clin Microbiol 39: 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eris A, Barut E (1993) Decreasing severity of alternation using girdling and some plant regulators in olive. Acta Hortic 329: 131–133 [Google Scholar]
- Federici CT, Fang DQ, Scora RW, Roose ML (1998) Phylogenetic relationships within the genus citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theor Appl Genet 96: 812–822 [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Fischer RL, Goldberg RB (1982) Structure and flanking regions of soybean seed protein genes. Cell 29: 651–660 [DOI] [PubMed] [Google Scholar]
- Garcia-Luis A, Kanduser M, Santamarina P, Guardiola JL (1992) Low temperature influence on flowering in Citrus. The separation of inductive and bud dormancy releasing factors. Physiol Plant 86: 648–652 [Google Scholar]
- Gelvin SB, Schilperoot RA editors (1995) Plant Molecular Biology, Ed 2. Kluwer Academic, Norwell, MA
- Hackett WP (1985) Juvenility, maturation, and rejuvenation in woody plants. In J Janick, ed, Horticultural Reviews, Vol 7. AVI Publishing, Westport, CT, pp 109–147
- Jensen CS, Salchert K, Nielsen KK (2001) A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol 125: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner DR (1995) NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell 7: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Dawson IA, Speer SS (1992) Control of growth and flowering in two western Australian species of Pimelea. Aust J Bot 40: 377–388 [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kotoda N, Wada M, Kato H, Iwanami H, Masuda T, Soejima J (2001) The function analysis of MdMADS5 and MdTFL, apple homologues of APETALA1 and TERMINAL FLOWER1, in transgenic Arabidopsis. Hortscience 36: 441
- Kotoda N, Wada M, Komori S, Kidou S, Abe K, Masuda T, Soejima J (2000) Expression pattern of homologues of floral meristem identity genes LFY and AP1 during flower development in apple. J Am Soc Hortic Sci 125: 398–403 [Google Scholar]
- Kyozuka J, Harcourt R, Peacock WJ, Dennis ES (1997) Eucalyptus has functional equivalents of the Arabidopsis AP1 gene. Plant Mol Biol 35: 573–584 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord EM, Eckard KJ (1985) Shoot development in Citrus sinensis L. (Washington navel orange) I. Floral and inflorescence ontogeny. Bot Gaz 146: 320–326 [Google Scholar]
- Lovatt CJ, Zheng Y, Hake KD (1988) Demonstration of a change in nitrogen metabolism influencing flower initiation in citrus. Isr J Bot 37: 181–188 [Google Scholar]
- Luckwill LC (1979) The effects of daminozole and gibberellic acid on flower initiation, growth and fruiting in apple cv Golden Delicious. J Hortic Sci 54: 217–223 [Google Scholar]
- Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377: 522–524 [DOI] [PubMed] [Google Scholar]
- Meilan R (1997) Floral induction in woody angiosperms. New For 14: 179–202 [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336 [DOI] [PubMed] [Google Scholar]
- Mimida N, Sakamoto W, Murata M, Motoyoshi F (1999) TERMINAL FLOWER 1-like genes in Brassica species. Plant Sci 142: 155–162 [Google Scholar]
- Mullins MG, Plummer JA, Snowball AM (1989) Flower initiation: new approaches to the study of flowering in perennial plants. In CJ Wright, ed, Manipulation of Fruiting. Butterworth, London, pp 65–77
- Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750 [DOI] [PubMed] [Google Scholar]
- Nilsson O, Lee I, Blazquez MA, Weigel D (1998) Flowering-time genes modulate the response of LEAFY activity. Genetics 150: 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene TERMINAL FLOWER 1. Mol Gen Genet 254: 186–194 [DOI] [PubMed] [Google Scholar]
- Page T, Macknight R, Yang C-H, Dean C (1999) Genetic interactions of the Arabidopsis flowering time gene FCA, with genes regulating floral initiation. Plant J 17: 231–239 [DOI] [PubMed] [Google Scholar]
- Pautot V, Holzer FM, Walling LL (1991) Differential expression of tomato proteinase inhibitor I and II genes during bacterial pathogen invasion and wounding. Mol Plant-Microbe Interact 4: 284–292 [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Schweizer D, Guerra M (2000) Cytological heterozygosity and the hybrid origin of sweet orange. Citrus sinensis (L.) Osbeck. Theor Appl Genet 100: 361–367 [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Pharis RP, Ross SD, McMullen E (1980) Promotion of flowering in Pinaceae by gibberellin. Physiol Plant 50: 119–126 [Google Scholar]
- Pillitteri LJ (2002) Isolation and characterization of the floral regulatory genes TERMINAL FLOWER, LEAFY, and APETALA1 from ‘Washington’ navel orange (Citrus sinensis L. Osbeck). Dissertation. University of California, Riverside, CA
- Pillitteri LJ, Lovatt CJ, Walling LL (2004) Isolation and characterization of the floral-regulatory genes, LEAFY (LFY) and APETALA1 (AP1) from ‘Washington’ navel orange (Citrus sinensis L. Osbeck). J Am Soc Hort Sci (in press)
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Poethig RS (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Puthoff DP (1999) Plant-insect interactions: the tomato defense response following feeding by phloem-feeding whiteflies. PhD thesis. University of California, Riverside, CA.
- Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ (1998) A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Rottmann WH, Meilan R, Sheppard LA, Brunner AM, Skinner JS, Ma C, Cheng S, Jouanin L, Pilate G, Strauss SH (2000) Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J 22: 235–245 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1993) Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 5: 639–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southerton SG, Strauss SH, Olive MR, Harcourt RL, Decroocq V, Zhu X, Llewellyn DJ, Peacock WJ, Dennis ES (1998) Eucalyptus has a functional equivalent of the Arabidopsis floral meristem identity gene LEAFY. Plant Mol Biol 37: 897–910 [DOI] [PubMed] [Google Scholar]
- Sung S-K, Yu G-H, An G (1999) Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiol 120: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (1999) PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods), Vol 4. Sinauer Associates, Sunderland, MA
- Telfer A, Poethig RS (1998) HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125: 1889–1898 [DOI] [PubMed] [Google Scholar]
- Wahl, GM, Stern M, Stark, GR (1979) Efficient transfer of large DNA fragments from agarose gels to diazobenyloxymethyl-paper and rapid hybridization using dextran sulfate. Proc Natl Acad Sci USA 76: 3683–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DM, Knapp SJ (1990) DNA extraction from a previously recalcitrant plant genus. Plant Mol Biol Report 8: 180–185 [Google Scholar]
- Weigel D, Alverez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377: 495–500 [DOI] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, et al. (1999) Suppression of Raf-1 kinase activity and MAP kinase signaling by RKIP. Nature 401: 173–177 [DOI] [PubMed] [Google Scholar]