Abstract
Differentiation of xylem cells in dicotyledonous plants involves expansion of the radial primary cell walls and intrusive tip growth of cambial derivative cells prior to the deposition of a thick secondary wall essential for xylem function. Expansins are cell wall-residing proteins that have an ability to plasticize the cellulose-hemicellulose network of primary walls. We found expansin activity in proteins extracted from the cambial region of mature stems in a model tree species hybrid aspen (Populus tremula × Populus tremuloides Michx). We identified three α-expansin genes (PttEXP1, PttEXP2, and PttEXP8) and one β-expansin gene (PttEXPB1) in a cambial region expressed sequence tag library, among which PttEXP1 was most abundantly represented. Northern-blot analyses in aspen vegetative organs and tissues showed that PttEXP1 was specifically expressed in mature stems exhibiting secondary growth, where it was present in the cambium and in the radial expansion zone. By contrast, PttEXP2 was mostly expressed in developing leaves. In situ reverse transcription-PCR provided evidence for accumulation of mRNA of PttEXP1 along with ribosomal rRNA at the tips of intrusively growing xylem fibers, suggesting that PttEXP1 protein has a role in intrusive tip growth. An examination of tension wood and leaf cDNA libraries identified another expansin, PttEXP5, very similar to PttEXP1, as the major expansin in developing tension wood, while PttEXP3 was the major expansin expressed in developing leaves. Comparative analysis of expansins expressed in woody stems in aspen, Arabidopsis, and pine showed that the most abundantly expressed expansins share sequence similarities, belonging to the subfamily A of α-expansins and having two conserved motifs at the beginning and end of the mature protein, RIPVG and KNFRV, respectively. This conservation suggests that these genes may share a specialized, not yet identified function.
Woody tissues appeared early in land plant evolution and enabled plants to maintain a large photosynthetic biomass high above the ground by providing mechanical support and the means for long-distance transport. During wood differentiation, newly divided cambium cells undergo cell enlargement before they develop a secondary cell wall (for review, see Larson, 1994; Mellerowicz et al., 2001). The direction and extent of cell enlargement differs among the various cell types in the xylem and ultimately facilitates the specialized function of each cell type. Conductive vessel elements and tracheids and supporting xylem fiber cells develop from already much-elongated fusiform initials. They increase in diameter by extension of their radial walls, which, in the case of vessel elements, is accompanied by splitting of adjacent cell files to allow expansion in a tangential direction. In addition, developing fiber cells elongate by intrusive tip growth, which results in up to a severalfold increase in cell length (Wenham and Cusick, 1975; Larson, 1994). Poplar fibers elongate intrusively in the radial-expansion zone, reaching 150% of their initial cell length at the average when fully differentiated (for review, see Mellerowicz et al., 2001). Ray initials of the vascular cambium provide precursors for xylem ray-parenchyma cells. These cells elongate radially by diffuse growth. Thus, development of different cell types in the wood involves cell-specific global and local changes in properties of their primary cell walls.
Expansins are proteins that, without requiring other active proteins, can induce extension of cell walls in a pH-dependent manner and, therefore, are considered to be primary regulators of plant cell enlargement (McQueen-Mason et al., 1992; McQueen-Mason and Cosgrove, 1994; Whitney et al., 2000; for recent review, see Cosgrove, 2000a, 2000b; Darley et al., 2001). Expansin genes comprise a superfamily, which is divided into three major families, α-, β-, and γ-expansins, on the basis of sequence divergence (Li et al., 2002, 2003). However, cell wall-loosening activity has only been reported for α- and β-expansins. Each of these gene families contains several members that are differentially expressed in various cell types and/or during different developmental stages, thus having different roles during plant development (Reinhardt et al., 1998; Brummell et al., 1999a; Lee et al., 2001; Wu et al., 2001; Li et al., 2002; http://www.bio.psu.edu/expansins). There is, however, a possibility that each gene encodes a protein with distinct properties. For example, Rose et al. (2000) observed differences in antibody epitopes between elongation-specific and fruit ripening-specific expansins. The basis for this epitope difference has not been determined, and no correlation between the structure of the coding sequence of expansin genes and their specific expression patterns has been reported so far.
Expansins were found to be expressed during the primary xylem formation in zinnia (Zinnea elegans; Im et al., 2000), but nothing is known about expansins involved in secondary xylem (wood) development. In this article, we demonstrate a high level of expansin activity in the stem tissues of a model tree species hybrid aspen (Populus tremula × Populus tremuloides Michx) forming secondary xylem. We identified expansin cDNAs from developing wood tissues and carried out transcript localization studies of a highly expressed expansin (PttEXP1) in these tissues. Interestingly, the aspen wood-expressed expansin genes are structurally similar and related to wood-expressed expansin genes of pine and Arabidopsis. This suggests that expansins of the wood-forming tissues share a specific, but not yet fully elucidated, function.
RESULTS
Expansin Activity in Wood-Forming Tissues of Aspen
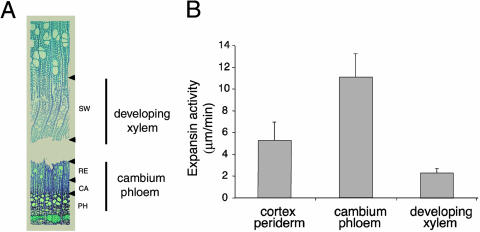
Expansin activities of mature stem tissues were measured using cellulose-hemicellulose composites (Fig. 1). Mature stem tissues were separated into three fractions: cortex/periderm, cambium/phloem, and developing xylem. The cambium/phloem fraction contained mostly cambial zone cells, recently formed phloem, and radial-expansion zone xylem cells, while the developing xylem fraction contained xylem cells depositing a secondary cell wall, as determined by microscopic examination (Fig. 1A). Cell wall expansion in a radial direction and intrusive tip growth of xylem fibers are known to occur in the cambial and radial-expansion zones (Mellerowicz et al., 2001) that were included in the cambium/phloem fraction. Proteins extracted from this fraction possessed the highest expansin activity (Fig. 1B).
Figure 1.
Expansin activity of proteins isolated from secondary tissues of aspen stem measured by creep of cellulose-xyloglucan composites. A, Micrograph showing wood-forming tissues isolated by bark peeling and scraping of tissues from exposed bark and wood sides. The bark separated in the late radial-expansion zone. Developing xylem fraction was collected from the exposed wood side and contained mostly secondary wall-depositing xylem (SW), while the cambium/phloem fraction was collected from the exposed bark side and comprised cambial zone (CA), radial expansion zone (RE), and some phloem (PH). B, Expansin activity of proteins extracted from different secondary tissues and used at the concentration of 1 mg mL−1. Results are averages and ses from 12 measurements.
Cloning of Expansin Genes from the Cambial Region
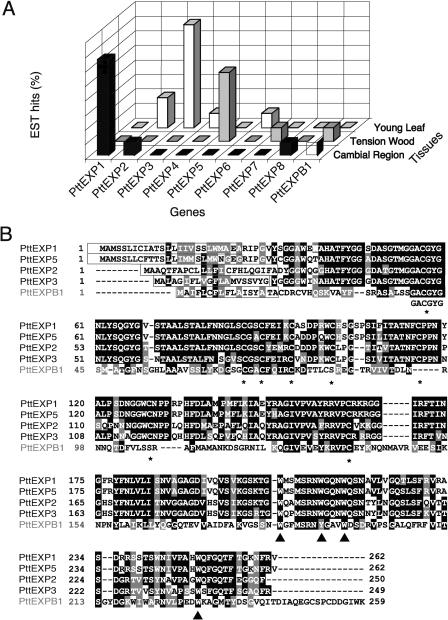
To isolate genes encoding expansins responsible for the observed activity, we searched the aspen expressed sequence tag (EST) library of cDNAs from the cambial region (Sterky et al., 1998). Three different α-expansin cDNAs were found among the 5,692 ESTs. One of them, PttEXP1, appeared eight times in the library, while two others, PttEXP2 and PttEXP8, were represented by only one EST each. In addition, one β-expansin, PttEXPB1, belonging to a β2 clade (Li et al., 2002), also known as expansin-like clade (http://www.bio.psu.edu/expansins), was represented by one EST. Searches in the EST libraries from developing tension wood and young leaves (http://poppel.fysbot.umu.se; Bhalerao et al., 2003) resulted in the identification of 18 additional EST sequences. In all, we found eight putative α-expansin genes, referred to as PttEXP1 to 8, and one putative β-expansin gene, referred to as PttEXPB1. Figure 2A summarizes the distribution and frequency of each expansin gene in the three libraries. Each library contained a different set of expansin genes and a different dominant expansin.
Figure 2.
Expansin genes from aspen. A, Survey of expansin genes in aspen EST databases. The y axis represents the percentage of EST hits in each library. GenBank accession numbers of EST sequences and clones: PttEXP1 (AI161578, AI161567, AI161713, AI164678, AI166095, AI165200, AI165287, and clone B016P63U; E.J. Mellerowicz and B. Sundberg, unpublished data), PttEXP2 (BI070667, BI068323, AI163583), PttEXP3 (BI071119, BI071981, BI071873, BI073018, BI070515, BI072608, BI070744), PttEXP4 (BI072263), PttEXP5 (BI131728, BI127499, BI130398, BI131229, BI131887), PttEXP6 (BI072028), PttEXP7 (BI127490), PttEXP8 (AI161876), PttEXPB1 (BI127601, AI162918). B, Sequence alignment of PttEXP1, PttEXP2, PttEXP3, PttEXP5, and PttEXPB1. The boxed sequences represent N-terminal signal peptide sequences. The conserved Cys and Trp residues are indicated by asterisks and arrowheads, respectively.
We sequenced the clones to obtain the full-length coding sequence of PttEXP1, PttEXP2, and PttEXP3, while it was possible to predict full-length sequences of PttEXP5 and PttEXPB1 by assembling corresponding EST sequences (Supplemental Fig. 1, available at www.plantphysiol.org). The amino acid sequence alignment of the predicted proteins encoded by these genes (Fig. 2B) shows that the sequences contain key features of α- and β-expansins, such as N-terminal signal peptides, four conserved Trp residues that might be involved in the interaction with cellulose, eight conserved Cys residues, and other conserved motifs such as GACGYG near the N terminus (Cosgrove, 2000b; Li et al., 2002). Amino acid sequences of PttEXP1 and PttEXP5 were very similar to each other and exhibited 91% amino acid sequence identity (96% identity after the removal of signal peptides), while PttEXP2 was more divergent with 64% identity to PttEXP1 (70% identity without the signal peptide).
Differential Expression of PttEXP1 and PttEXP2
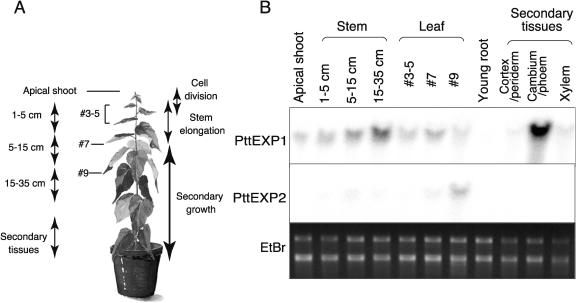
Expression of PttEXP1 and PttEXP2 in various aspen organs and tissues was studied by northern-blot analysis. Gene-specific DNA probes were designed from 3′ untranslated regions, and their specificity was confirmed by dot-blot hybridization against plasmid DNAs containing each cDNA sequence (data not shown). Total RNA was extracted from organs and tissues shown in Figure 3A. A weak signal for the PttEXP1 transcript was detected in the RNA obtained from the apical shoots (Fig. 3B). This signal increased progressively in more mature stems, and the highest level of signal was detected from stem tissues exhibiting active secondary growth, among which cambium/phloem tissues exhibited the strongest signals. PttEXP1 transcripts were also present in developing leaves, although the signal intensity was much weaker than that of mature stems. Little or no PttEXP1 mRNA signals were detected in young roots. Northern-blot hybridization using the PttEXP2 probe yielded much weaker signals than those of PttEXP1 in all the tissues examined. The strongest signal for this transcript was observed in RNA from the young leaves that were still expanding where signal intensity was similar (or slightly higher) to that of PttEXP1.
Figure 3.
Northern-blot analysis of PttEXP1 and PttEXP2. A, Diagram showing location of the tissues from which RNA was extracted. Stem, leaf, and root tissues were collected from 3-month-old aspen plants. Distance from apex was used to compare different developmental stages of stems and leaves. Apical shoots were obtained from the tissues within 1 cm of the apex and contained the apical meristem, immature leaves, and immature stems. Stems 1 to 5 cm from the shoot apex were going through active elongation and exhibited little, if any, secondary growth. After this point, stems ceased to elongate and secondary growth became more prevalent. Young roots contained root tips and elongation zones. Secondary tissues were obtained from mature stem and separated into periderm/cortex, cambium/phloem, and developing xylem. B, Northern blotting using gene-specific probes. Twenty micrograms of RNA were loaded to each lane. Ethidium bromide staining of RNA sample showing equal loadings is shown at bottom.
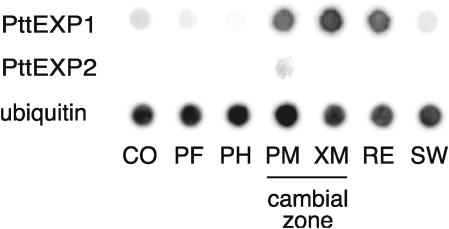
To increase the precision of expression analysis in mature stem tissues, tangential sections containing two to four cell layers (Uggla et al., 1996) from seven locations across the wood-forming tissues were collected (Fig. 4). Transcripts obtained from each location were amplified by reverse transcription (RT)-PCR, blotted on a membrane, and hybridized against PttEXP1 and PttEXP2 probes. Strong PttEXP1 hybridization signals were detected in cDNAs obtained from the cambial zone and expanding xylem cells, with the strongest signal found in xylem mother cells. Weak hybridization signals were also detected in cDNAs obtained from the cortex, phloem fiber, and secondary cell wall formation zone. By contrast, the PttEXP2 transcripts were detected only in cambial zone cells containing phloem mother cells.
Figure 4.
High-resolution transcript profiling of PttEXP1 and PttEXP2. Amplified cDNA libraries were obtained from tangential sections across the cambial region, blotted onto a nylon membrane, and probed with gene-specific 32P-labeled DNA probes. CO, Cortex; PF, phloem fibers; PM, cambial zone containing phloem mother cells; XM, cambial zone containing xylem mother cells; RE, radial expansion zone; SW, secondary wall formation zone. A polyubiquitin probe was used as a positive control and gave strong hybridization signals against all the stages of secondary tissue development.
In Situ Localization of PttEXP1 mRNA
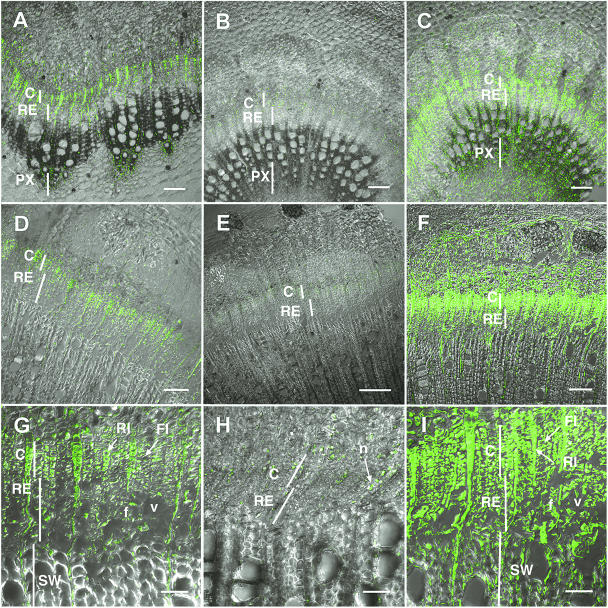
Cellular and subcellular transcript localization of PttEXP1 was determined using in situ RT-PCR in the wood-forming tissues of young and mature stem internodes (Fig. 5). The PttEXP1 transcripts were detected mostly in the cambial and radial-expansion zones in both stem ages, and some signal was found in paratracheal parenchyma of primary xylem in young stems (Fig. 5, A, D, and G). In the cambial zone, signals were detected in both ray and fusiform initials (Fig. 5G). Signals in ray initials and ray cells were uniformly distributed in the cytoplasm, whereas signals in fusiform initials and developing fiber and vessel cells were detected only in the peripheral region of the cytoplasm (Fig. 5G), reflecting differences in vacuolization between cells of axial and radial systems. These cytoplasmic signals were absent in negative controls in which DNA polymerase was omitted in the reactions (Fig. 5, B, E, and H). The negative controls detected nonspecific signals in nucleus and nucleolus (arrow in Fig. 5H). As a positive control for labeling reactions, RT-PCR reactions were performed using primers specific for mitochondrial 26S rRNA. This resulted in signals from almost all the cells that retained cytoplasmic contents (Fig. 5, C, F, and I), including wood cells in the secondary wall-forming phase (Fig. 5, compare I to G), phloem (Fig. 5, compare D to F), and pith (Fig. 5, compare A to C), in which PttEXP1 signals were absent. The signal intensity for the constitutive probe also varied according to the degree of vacuolization. This demonstrates the importance of including a constitutively expressed reference when comparing gene expression in cells differing in cytoplasm content as discussed in Gray-Mitsumune et al. (2004).
Figure 5.
In situ RT-PCR localization of PttEXP1 transcripts in the wood-forming tissues in the cross-sections of the 10th (A–C) and 30th (D–I) internode from the apex. Images are projections along the z axis of optical sections obtained by confocal laser scanning microscopy. A, D, and G, Tissue distribution of PttEXP1 mRNA. B, E, and H, Negative control in which DNA polymerase was omitted from RT-PCR reactions. C, F, and I, Tissue distribution of 26S rRNA. C, Cambial zone; f, fiber; FI, fusiform initial; n, nucleolus; PX, primary xylem; RE, radial expansion zone; RI, ray initial; v, vessel element. A to D and F scale bars = 100 μm; E scale bar = 200 μm; G and I scale bars = 50 μm.
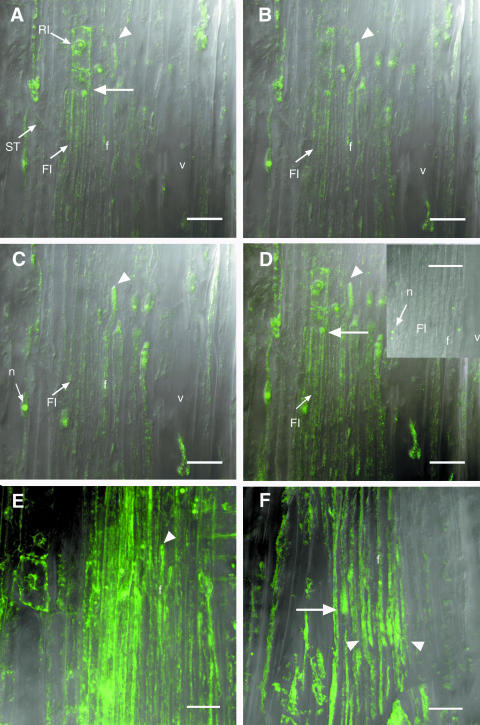
Figure 6, A to C, are serial optical sections in the longitudinal radial orientation allowing a three-dimensional reconstruction of developing wood cells. These sections reveal strong PttEXP1 mRNA signals near the tips of fusiform initials (arrow in Fig. 6A) and even stronger signals in the intrusively growing tips of developing xylem fiber cells (arrowheads in Fig. 6, A–C). The apparent distribution of the signal at the fiber tips was not due to local accumulation appearing at random in some sections because a composite figure, including all optical sections through these cells, projected into one plane shows the same intracellular label pattern (Fig. 6D). Negative controls with DNA polymerase omitted from the reaction (inset, Fig. 6D) detected only nonspecific signals in the nucleolus. In situ RT-PCR signals for mitochondrial 26S rRNA were not only more uniformly distributed in fusiform initials but also were frequently accumulated at the tips of intrusively growing fibers (Fig. 6E). By contrast, the 18S rRNA signal specific for cytoplasmic ribosomes was more prominent at the tips of fusiform initials and intrusively growing fibers (arrow and arrowhead in Fig. 5F, respectively). These observations suggest targeting of PttEXP1 transcript and ribosomes to the intrusively growing tips of differentiating xylem fibers.
Figure 6.
In situ RT-PCR localization of PttEXP1 transcripts in the wood-forming tissues in the longitudinal radial sections. A to C, A series of successive optical sections obtained by confocal laser scanning microscopy and showing longitudinal distribution of PttEXP1 transcripts. A, Arrow points to tips of fusiform initials (FI). A to C, Arrowheads show the tip of an intrusively growing fiber (f) with strong signals. D to F, Projections of all optical sections through the cells shown in A to C onto one plane demonstrating tip-localized signals of PttEXP1 (arrowhead in D), more uniformly distributed signals of mitochondrial 26S rRNA (E), except for intrusively growing fiber tips (arrowhead), and tip-localized signals from cytoplasmic 18S rRNA (F) in fusiform initials (arrow) and intrusively growing fibers (arrowhead). Inset in D represents typical negative control with DNA polymerase excluded from the PCR reaction. In the negative control, only nucleoli (n) show nonspecific signals. RI, ray initial; ST, sieve tube. Scale bars = 20 μm.
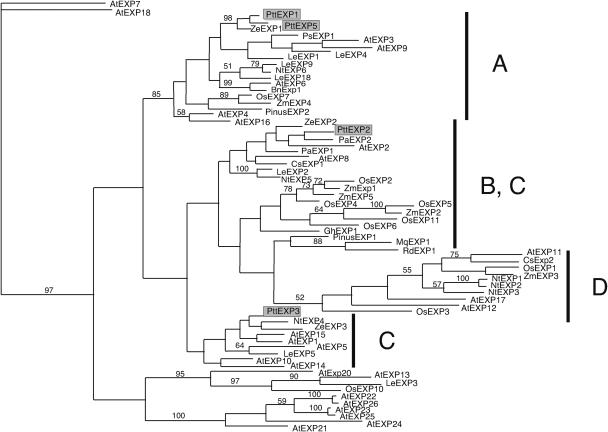
Phylogenetic Analysis of α-Expansins
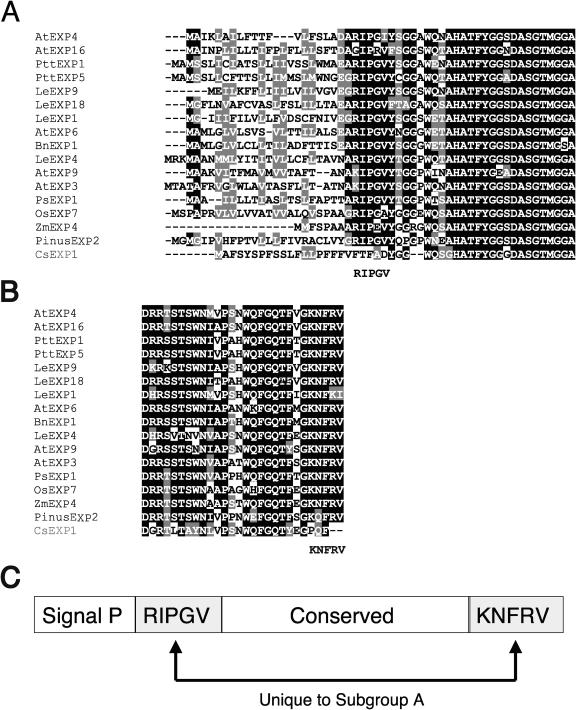
The α-expansin gene family splits into four or more major phylogenetic branches (Fig. 7; Link and Cosgrove, 1998). The two expansin genes abundant in wood-forming tissues, PttEXP1 and PttEXP5, aligned with subgroup A expansin genes previously described by Link and Cosgrove (1998). PttEXP2 and PttEXP3 were more divergent and grouped with subgroup B and C expansin genes, respectively. However, it should be noted that these subgroups were not supported by significant bootstrap levels and they were not separated into distinct branches (Fig. 7). By contrast, subgroup A expansin genes exhibited striking sequence conservation among themselves and formed a phylogenetically distinct branch. This subgroup contained genes from plant species of diverse origins, such as monocotyledonous species rice (OsEXP7) and corn (ZmEXP4) and gymnosperm species pine (PinusEXP2), suggesting that the divergence of the subgroup had occurred before the divergence of these vascular plants. The PinusEXP2 was found in abundance in EST libraries from wood-forming tissues of loblolly pine (Supplemental Fig. 2), with 75% of all expansin clones belonging to this gene, and, therefore, appears to be orthologous to the PttEXP1/PttEXP5 group. In addition to the sequences conserved among all α-expansin genes, subgroup A genes exhibited unique sequence conservation among themselves. Particularly striking is the amino acid sequence immediately after predicted signal peptide cleavage site RIPGV, already noted by Link and Cosgrove (1998), and the C-terminal sequence KNFRV (Fig. 8, A–C). These two motifs can serve as signature sequences of subgroup A.
Figure 7.
Phylogenetic analysis of α-expansin genes. The phylogenetic tree was constructed using the PAUP software. Bootstrap numbers above 50% were indicated. Subgroups A to D as defined by Link and Cosgrove (1998). GenBank accession numbers (protein or nucleotide) of expansin genes from Arabidopsis: AtEXP1 (AAG60095), AtEXP2 (BAB09972), AtEXP3 (AAC23634), AtEXP4 (AAB97125), AtEXP5 (BAA95756), AtEXP6 (AAC33223), AtEXP7 (AAF79645), AtEXP8 (AAB87577), AtEXP9 (CAB85531), AtEXP10 (AAF61712), AtEXP11 (AAF79895), AtEXP12 (AAF35403), AtEXP13 (AAF26104), AtEXP14 (BAB11259), AtEXP15 (AAC32927), AtAtEXP17 (AAC72858), AtEXP18 (AAF75810), AtEXP19 (AP001309), AtEXP20 (CAB37561), AtEXP21 (BAB09381), AtEXP22 (BAB09382), AtEXP23 (BAB09383), AtEXP24 (BAB09386), AtEXP25 (BAB09385), AtEXP26 (BAB09384); Brassica napus: BnEXP1 (AJ000885); Cucumis sativus: CsEXP1 (U30382), CsEXP2 (U30460); Gossypium hirsutum: GhEXP1 (AF043284); Lycopersicon esculentum: LeEXP1 (U82123), LeEXP2 (AF096776), LeEXP3 (AF059487), LeEXP4 (AF059488), LeEXP5 (AF059489), LeEXP6 (AF059490), LeEXP7 (AF059491), LeEXP9 (AJ243340), LeEXP18 (AJ004997); Marsilea quadrifolia: MqEXP1 (AF202119); Nicotiana tabacum: NtEXP1 (AF049350), NtEXP2 (AF049351), NtEXP3 (AF049352), NtEXP4 (AF049353), NtEXP5 (AF049354), NtEXP6 (AF049355); Oryza sativa: OsEXP1 (Y07782), OsEXP2 (U30477), OsEXP3 (U30479), OsEXP4 (U85246), OsEXP5 (AF247162), OsEXP6 (AF247163), OsEXP7 (AF247164); Prunus armeniaca: PaEXP1 (U93167), PaEXP2 (AF38815); Pinus taeda: PinusEXP1 (U65891), PinusEXP2 (assembled from the following EST sequences: AA556812, BQ107011, AW290022, AW011524, BG275593, BF186094, BF609532, BF517250, AW290718, BE187483, BF779049, BE187464, AW226065, BE762031, and BF049828); Regnellidium diphyllum: RdEXP1 (AF202120); Zea mays: ZmEXP1 (AF332169), ZmEXP2 (AF332170), ZmEXP3 (AF332171), ZmEXP4 (AF332172), ZmEXP5 (AF332173).
Figure 8.
Subgroup A α-expansins exhibit unique sequence conservation at N and C termini. A, Alignment of N-terminal amino acid sequences of subgroup A α-expansins. Sequence of CsEXP1 of subgroup B is shown at the bottom as a reference sequence. B, Alignment of C-terminal amino acid sequences. C, Diagram showing conserved motifs of subgroup A α-expansins.
Survey of Expansin Genes in Wood-Forming Tissues of Arabidopsis
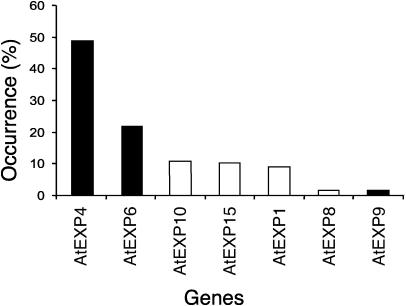
Structural conservation of expansin genes expressed abundantly in developing wood from two evolutionally distinct species, aspen and pine, suggested that the process of wood formation may specifically involve expansins from subgroup A. To further explore this possibility, we examined expression of α-expansin genes in wood-forming tissues of Arabidopsis. Arabidopsis hypocotyls develop extensive secondary growth when grown under specific growth conditions (Chaffey et al., 2002). Using degenerate primers to the conserved motifs in Arabidopsis α-expansins, we cloned cDNA sequences expressed in Arabidopsis hypocotyls exhibiting active secondary growth. We detected transcripts from seven α-expansin genes: AtEXP1, AtEXP4, AtEXP6, AtEXP8, AtEXP9, AtEXP10, and AtEXP15, among which AtEXP4, AtEXP6, and AtEXP9 belonged to subgroup A (Fig. 9). AtEXP4 and AtEXP6 represented 70% of expansin cDNAs found in wood-forming hypocotyls, supporting the idea that subgroup A is of particular significance for secondary growth.
Figure 9.
Survey of α-expansin genes expressed in Arabidopsis hypocotyls exhibiting active secondary growth. The α-expansin mRNA signals were amplified using RT-PCR and the frequency of each expansin gene fragment in the amplified cDNA pool was scored as described in “Materials and Methods.” The y axis represents the percentage of each gene in total number of α-expansin genes identified. Black bars represent subgroup A α-expansin genes.
DISCUSSION
Expansin Activity in Wood-Forming Tissue Indicates a Role for Expansin in Xylogenesis
Wood cell differentiation in a dicotyledonous plant requires differently controlled cell wall extension, where different cell wall regions undergo various degrees and various directions of stretching (Mellerowicz et al., 2001). We demonstrate that wood-forming tissues exhibit expansin activity, as tested in the creep assay on cellulose-XG composites (Whitney et al., 2000). Such an activity has been previously demonstrated for other tissues and organs, such as developing leaves, roots, young stems, and ripening fruits (McQueen-Mason et al., 1992; Wu et al., 1996; Caderas et al., 2000; Rose et al., 2000; Harrison et al., 2001; Pien et al., 2001). Our results, along with the previously published observations of expansin gene expression in developing primary vascular tissues (Cho and Kende, 1997; Im et al., 2000; Reidy et al., 2001), support the involvement of expansins in the regulation of wall modification during differentiation of vascular elements. The question arises as to whether the same expansin genes are involved during primary and secondary vascular tissue development and whether these genes are functionally distinct.
Characterization of Aspen Expansin Genes of Wood-Forming Tissues
Nine expansin genes were detected in aspen EST libraries from the cambial region, developing tension wood, and expanding leaf tissue. Each tissue contained a different dominant expansin. PttEXP1 was the most abundant expansin in the cambial region, PttEXP5 was the most important in the developing tension wood, whereas PttEXP3 was a dominant expansin in the young leaf. EST frequency of PttEXP1 and PttEXP2 in the cambial region was shown by northern-blot analysis to reflect their expression pattern, suggesting that the abundance of ESTs PttEXP3 and PttEXP5 may also reflect their high expression in leaf and tension wood libraries, respectively. Why did the normal wood- and tension wood-forming tissues have different dominant expansin genes? Tension wood is produced when stems respond to gravitational stimuli due to tilting or bending, which results in formation of morphologically and chemically distinct wood cells, the so-called gelatinous fibers (for review, see Jourez, 1997a, 1997b). In addition, the cellular composition of the two wood types is different, with tension wood having fewer vessel elements and more fiber cells. PttEXP5 may be specifically involved in gelatinous fiber formation. It is not clear if PttEXP1 and PttEXP5 proteins are functionally different because they exhibit high sequence similarity to each other. However, they might have evolved from a recent gene duplication event, where the differential expression has led to the acquisition of a novel function (or expression profile).
Expansin Genes Highly Expressed in Differentiating Xylem Belong to Subgroup A of α-Expansin Genes
Two expansin genes highly expressed in wood-forming tissues of aspen, PttEXP1 and PttEXP5, belong to subgroup A of the α-expansin gene family. This subgroup also includes the pine expansin PinusEXP2, which accounted for the majority of expansin ESTs found in wood-forming tissues in the loblolly pine. In addition, subgroup A expansin genes AtEXP4 and AtEXP6 accounted for 70% of expansin transcripts that could be detected with degenerate primers in wood-forming hypocotyls of Arabidopsis. The abundance of AtEXP4 and AtEXP6 transcripts in Arabidopsis stems with secondary growth was also observed in microarray experiments using Affymetrix chip technology (S. Turner, data published online at http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl). Subgroup A also includes genes that are expressed in primary vascular tissues, such as zinnia expansin ZeEXP1 (Im et al., 2000) and tomato LeEXP18 (Reinhardt, et al., 1998). Aspen PttEXP1 expression has also been detected in young shoots and leaves that have not yet developed secondary growth but contain primary vascular tissues. These observations suggest that the functions of subgroup A α-expansins are overlapping in development of primary and secondary vascular tissues.
Although subgroup A α-expansin genes appear to be of particular importance for xylogenesis, they are not restricted to this process since they also include genes encoding expansins involved in fruit ripening, such as tomato LeExp1 (Brummell et al., 1999a, 1999b; Rose et al., 2000). The function of expansin encoded by LeExp1 was proved to entail wall degradation as demonstrated by overexpression in transgenic plants (Brummell et al., 1999b). Wall degradation and rearrangement processes might be similar between fruit ripening and xylogenesis. Several genes encoding enzymes that have the ability to break down cell wall carbohydrates, including 1,3-β-glucosidase, pectin methylesterase, and pectate lyase, were up-regulated in the radial-expansion zone of developing wood, similar to PttEXP1 (Hertzberg et al., 2001), suggesting participation in a common process. Moreover, the formation of perforations in vessel elements, which is a wall-degrading process, is similar between the primary and secondary xylem differentiation, and it is possible that similar expansins are involved in these developmental processes.
Subgroup A α-expansins exhibit unique sequence conservation at both the N and C end of mature proteins. BLAST analysis of GenBank sequences revealed that similar AA motifs were found in many nonrelated proteins, but a common presence of both RIPVG and KNFRV motifs was restricted to expansin proteins (data not shown). These sequence motifs may endow unique properties and functions to these proteins. Alternatively, these motifs could be important for posttranslational processing, such as targeting to a specific compartment of cell walls, or for interaction with other proteins and/or carbohydrates.
While subgroup A α-expansins are most abundant in xylem-forming cells, it is important to note that xylogenesis also involves other types of α-expansins. For example, two other zinnia expansin genes, ZeEXP2 and ZeEXP3, (Im et al., 2000), Festuca pratensis FpEXP2 (Reidy et al., 2001), and three rice expansin genes OsEXP1 to 3 (Cho and Kende, 1997) are all expressed in vascular tissues but do not belong to subgroup A.
Cellular and Subcellular Distribution of PttEXP1 mRNA in Wood-Forming Tissues Indicate a Role in Radial Xylem Cell Expansion and in Intrusive Tip Growth
Preferential expression of PttEXP1 in the vascular cambium and its derivative tissues was demonstrated by the high abundance of PttEXP1 ESTs in the cambial region library. This was confirmed by northern-blot analysis. High-resolution transcript profiling together with in situ RT-PCR analysis specified the high abundance of PttEXP1 transcripts in the wood-forming tissues to the xylem mother cells and expanding xylem cells. This expression pattern confirms the cDNA microarray analysis reported by Hertzberg et al. (2001). In situ analysis demonstrated that PttEXP1 is expressed in all types of meristematic and expanding cells. High similarity of PttEXP1 to the primary xylem-expressed zinnia ZeEXP1 (Im et al., 2000) suggests that these genes are orthologous and the expression data from zinnia and poplar suggest that they may function in both primary and secondary xylem development. However, the expression of ZeEXP1 contrasted with that of PttEXP1 because it was restricted to paratracheal parenchyma cells. Moreover, there were differences in subcellular distribution of the signals in these expansin genes. ZeEXP1 transcripts were concentrated at the apical ends of paratracheal parenchyma cells (Im et al., 2000). PttEXP1 mRNA signals were also concentrated in cell tips, but in contrast to ZeEXP1 they were more diffuse, were detected at both ends of fusiform cells (data not shown), and were also present throughout the cell length. Optical sectioning by confocal microscopy demonstrated particularly strong PttEXP1 mRNA signals near the tips of intrusively growing fibers. The accumulation of ribosomes (detected using an 18S rRNA probe), along with PttEXP1 mRNA at the growing tips, suggests preferential translation of PttEXP1 near the tips.
The occurrence of mRNA targeting is well established in animal and plant cells (for review, see Okita and Choi, 2002); for example, tip-growing root hairs polarly target profilin mRNA to the tip (Baluska et al., 2000). Expansin protein has also been localized in the growing root hair tip, but its transcript localization has not been studied (Baluska et al., 2000). The polar expansin mRNA accumulation in zinnia xylem parenchyma cells has been interpreted as a means to facilitate expansin protein synthesis during intrusive tip growth of xylem cells (Im et al., 2000). However, paratracheal parenchyma cells do not elongate by intrusive growth (Esau, 1965), and no other cell type within the primary xylem of zinnia stems is known to elongate by intrusive growth. Possibly, the accumulation of expansin mRNA at the extremities of the parenchyma cells may be a part of a not yet described mechanism that facilitates wall stretching of primary xylem cells during internodal elongation or modifies cell walls during xylogenesis. By contrast, the developing fibers in the secondary xylem of aspen undergo substantial intrusive tip growth (Mellerowicz et al., 2001) and PttEXP1 is likely to be involved in this process. In addition, the presence of the transcripts throughout the cell length also supports the participation of PttEXP1 protein in radial fiber cell expansion.
MATERIALS AND METHODS
Plant Materials
Hybrid aspen (Populus tremula × Populus tremuloides Michx, clone T89) trees were grown in a greenhouse under natural light conditions supplemented with metal halogen lamps. They were grown under an 18-h-light/6-h-dark photoperiod at a temperature of 22°C (day) and 15°C (night). They were watered daily and fertilized once a week with a SuperbraS nutrient solution (Supra Hydro AB, Landskrona, Sweden).
For RNA extraction, young stem, leaf, and root tissues were collected from 3-month-old trees. For collection of secondary tissues, the bark was peeled from the woody core to expose developing xylem. Tissue scrapings from the xylem side (xylem scrapings) contained xylem undergoing secondary wall thickening, and tissue scrapings from the inner side of the bark (phloem scrapings) contained radially expanding xylem, cambium, and phloem tissues. Tissues were immediately frozen in liquid nitrogen and stored at −80°C.
Expansin Assay Using Cellulose-Hemicellulose Composite
Xylem and phloem scrapings similar to those used for the RNA preparations described above were used for protein extraction. Cell wall proteins were extracted and assayed for expansin activity using a protocol based on that described by McQueen-Mason et al. (1992). Portions of frozen tissues were homogenized in a Waring blender with 4 mL g−1 (fresh weight) of 25 mm HEPES, 1% polyvinylpyrolidone (40,000 MW), 1% Triton X-100, 3 mm sodium metabisulfite, 2 mm EDTA, and 2 mm dithiothreitol, pH 6.8. The homogenate was collected on a 50-micron nylon mesh and excess liquid removed by squeezing. Wall fragments were washed three times in the same volume of the homogenizing buffer without Triton X-100. Wall fragments were retained on the nylon mesh and then resuspended in 2 mL g−1 (original fresh weight) of 1 m NaCl, 25 mm HEPES, 3 mm sodium metabisulfite, 2 mm EDTA, and 2 mm dithiothreitol, pH 6.8, and left to extract for 1 h at room temperature. The salt extract was filtered through the nylon mesh, its volume measured, and proteins precipitated by the gradual addition of 0.39 g mL−1 of solid ammonium sulfate. After 10 min on ice, protein precipitates were recovered by centrifugation at 10,000g for 10 min at 4°C. The supernatant was removed and the pellets stored at −20°C until they were assayed for expansin activity. Immediately prior to expansin assays, precipitates were resuspended in 1 mL of 50 mm sodium acetate, pH 4.5, and desalted through a 5-mL column of Sephadex G25 (Amersham Bioscience, Buckinghamshire, UK) in the same solution, and brought to a final volume of 1 mL g−1 original fresh weight. Expansin assays were carried out in a purpose-built extensometer using a cellulose-xyloglucan composite as described by Whitney et al. (2000). Briefly, 2-mm-wide strips of composite were placed between the clamps of the extensometer, bathed in 50 mm sodium acetate, pH 4.5, and extended under by the application of a 5-g weight to the lower clamp. After 15 min extension, the bathing solution was replaced with 100 μL of the solution of desalted cell wall proteins at a concentration of 1 mg mL−1. Extension rates were measured for the 10 min before and 10 min after protein addition. Expansin activity was calculated as the rate of extension after protein addition minus the rate prior to protein addition.
RNA Extraction and Northern Hybridization
Plant tissues were ground into fine powder in liquid nitrogen. Two hundred milligrams of powdered tissues were suspended in 0.9 mL of the RNA extraction buffer described by Chang et al. (1993; 2% CTAB, 2% polyvinylpyrolidone, 100 mm Tris-HCl, 25 mm EDTA, 2 m NaCl, 0.5 g L−1 spermidine, and 2% β-mercaptoethanol, pH 8.0) and incubated at room temperature for 5 min. The solution was mixed with an equal volume of chloroform:isoamylalcohol (24:1) and vortexed for complete suspension. The aqueous phase was separated from the organic phase by centrifugation at 17,000g for 5 min. Nucleic acids in the aqueous phase were precipitated by adding 1.2 volume of isopropanol, followed by incubation at −20°C for 30 min, and were collected by centrifugation at 17,000g for 20 min at 4°C. The precipitate was dissolved in RNeasy RNA extraction buffer (Qiagen, Valencia, CA) and RNA was purified using RNeasy columns according to the manufacturer's recommendation.
Twenty micrograms of RNA were separated by electrophoresis using a formaldehyde agarose gel and blotted to a Hybond-N nylon membrane (Amersham Biosciences AB, Uppsala). Gene-specific probes of PttEXP1 and PttEXP2 were synthesized using PCR fragments amplified from 3′ end noncoding regions, and corresponded to nucleotide sequences of 891 to 1,190 and 722 to 1,033, respectively. The probe for the polyubiquitin gene was synthesized using plasmid DNA containing aspen polyubiquitin cDNA sequence (accession no AI162905.1, clone A081P57u). The fragments were labeled with α-32P-dATP using Strip-EZ DNA labeling kit (Ambion, Austin, TX). Hybridizations were performed in ULTRAHYB (Ambion) buffer at 42°C overnight, followed by washing in 2× SSC twice, 0.5× SSC twice, and 0.1× SSC twice. The temperature of the final wash was raised to 65°C. Hybridization signals on the membrane were analyzed using a GS-525 molecular image storage phosphor imaging system (Bio-Rad Spectroscopy Group, Cambridge, MA).
High-Resolution Transcript Profiling
Tangential sections of mature stem were obtained according to the method of Uggla et al. (1996); 30-μm-thick tangential sections of the stem were obtained with a cryomicrotome (HM505E microtome, Microm Laborgeräte, Walldorf, Germany). The sample sections were pooled into the following groups representing different developmental zones: CO, cortex, between cork and phloem; PF, phloem fibers; PH, functional phloem; PM, part of cambial zone containing phloem mother cells; XM, part of cambial zone containing xylem mother cells; RE, radially expanding xylem cells; and SW, zone of visible secondary wall formation. The distinction between different zones was based on size and shape of the cells as well as presence and thickness of secondary walls observed under the light microscope using Nomarski optics.
From each pool of sections, poly(A)+ mRNA was extracted directly using paramagnetic oligo(dT)25 Dynabeads (Dynal AS, Oslo, Norway) according to the manufacturer's recommendation. The bead-bound mRNA was transcribed into the first-strand cDNAs using Superscript II reverse transcriptase (GibcoBRL, Invitrogen, Carlsbad, CA) and the CUT-3T primer (TTGCATTGACGTCGACTATCCAGGT15). The bead mixture containing cDNA/mRNA hybrid was processed as described by Hertzberg and Olsson (1998). Free oligo(dT) remaining on the beads was removed using T4 polymerase (GibcoBRL), and RNA was removed from the cDNA/RNA hybrid using RNAse H (GibcoBRL). Oligo(dA) was added to the bead-bound cDNA using terminal deoxytransferase (GibcoBRL). The obtained cDNA was PCR amplified using eLONGase (GibcoBRL) and the primer CUT-3T (as above) for 10 cycles of 30 s at 94°C, 45 s at 40°C, and 5 min at 68°C. A second PCR reaction was performed using the primer CUT-3P (TTGCATTGACGTCGACTATCCAGG) for an additional 27 to 31 cycles. Approximately 500 ng of PCR product were dotted onto a Hybond-N nylon membrane (Amersham Biosciences AB) and hybridized using 32P-labeled DNA probes as described above.
In Situ RT-PCR
Detailed procedures of in situ RT-PCR have been described elsewhere (Gray-Mitsumune, et al., 2004). Stem segments were cut into pieces of approximately 10 mm in longitudinal dimension and 2 mm in tangential and radial dimensions, and placed immediately in 3.7% p-formaldehyde solution prepared in RNase-free phosphate-buffered saline (PBS; 10 mm Na phosphate, 130 mm NaCl, pH 7.2) for 3 h. The stem segments were then removed from the fixative, frozen in liquid nitrogen, and stored at −80°C. Frozen segments were sectioned using an HM505E cryomicrotome (Microm Laborgeräte, Walldorf, Germany). Transverse or radial sections of 70 μm in thickness were cut and immediately transferred to a PCR tube containing 0.5 units μL−1 RNase inhibitor (RNAguard; Amersham-Pharmacia Biotech, Uppsala) and washed three times with fresh RNase inhibitor solution.
For the first-strand cDNA synthesis, RNase inhibitor solution was removed from the PCR tubes containing tissue sections and replaced with RT buffer mixture (1× RT buffer [GibcoBRL], 10 mm dithiothreitol, 1 mm each of dNTP, and 5 ng μL−1 random oligohexamer). RNase inhibitor (RNAguard) and M-MLV reverse transcriptase (GibcoBRL) were added to give a final concentration of 0.5 units μL−1 and 10 units μL−1, respectively. The tubes were incubated at 37°C for 60 min, followed by 90°C for 5 min, and then brought down to 4°C. The RT mixture was replaced with PCR mixture (1× Advantage 2 PCR buffer [CLONTECH Laboratories, Palo Alto, CA], 0.2 mm each of dNTP, 0.4 μm each of gene-specific primers, and 1× Advantage 2 DNA Polymerase Mix [CLONTECH Laboratories]). Sequences of gene-specific primers are: PttEXP1, GGGGTGCTTGGGAAAATGCT (forward), CGCCTCAGCCTCAGTCTCCT (reverse); mitochondrial 26S rRNA, TCCCATGGTTCGATCCTTCC (forward), GCAGGGCGATCGTGTTTTTC (reverse). In addition, universal plant 18S rRNA primers (Ambion, Cambridgeshire, UK; Intermedica, Stockholm, Sweden) specific for cytoplasmic ribosomal 18S RNA were used. The PCR reaction was performed according to the following thermal regime: one cycle of 5 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, 1 min at 68°C; and one cycle of 10 min at 68°C. The PCR mixture was then replaced with labeling PCR mixture (1× Advantage 2 PCR buffer and 1× Advantage 2 DNA Polymerase Mix, 0.2 mm each of dCTP, dGTP, and dATP, 60 μm dTTP, 20 μm ChromaTide Alexa Fluor 488-5-dUTP [Molecular Probes, Eugene, OR], and 0.4 μm each of gene-specific primers). The labeling PCR reaction was continued for 20 cycles and one cycle of 5 min at 68°C. After the labeling reaction, sections were washed once in Tris-EDTA buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.0) and three times in PBS and then stored in PBS overnight at 4°C.
Tissue sections were carefully transferred to a glass slide, mounted in 50% glycerol, and observed in a confocal laser scanning microscope (LSM-510; Zeiss, Jena, Germany), using an excitation wavelength of 488 nm and detection wavelengths between 505 and 530 nm to detect Alexa signals. The transmitted light channel was used to allow detection of anatomical detail.
Database Search and Sequence Analysis
EST clones that exhibit homologies to α-expansin genes were identified from the EST database of Populus (PopulusDB, http://poppel.fysbot.umu.se; Bhalerao et al., 2003) by BLAST search. Full-length cDNA sequences of PttEXP1, PttEXP2, and PttEXP3 were further obtained by sequencing clones A067P09U, A044P14U, and C091P77, respectively. For each clone, plasmid DNA containing cDNA insert was purified according to standard procedures and sequenced by cycle sequencing using Big-Dye terminator enzyme mix (Perkin-Elmer Applied Biosystems, Foster City, CA). The fluorescent PCR products were size separated on an ABI 3700 sequencer (Perkin-Elmer Applied Biosystems). The GenBank accession numbers for fully sequenced cDNAs of PttEXP1, PttEXP2, and PttEXP3 are AY435099, AY435100, and AY435101, respectively. The cDNA sequences of PttEXP5, PttEXPB1, and PineEXP2 were obtained by assembling corresponding EST sequences using the CAP3 contig assembly program at the Centre de Resources INFOBIOGEN Web server (http://www.infobiogen.fr/services/analyseq/cgi-bin/cap_in.pl).
Pine expansin EST sequences were first identified via TBLASTN search in GenBank, using PttEXP1 and PttEXP2 amino acid sequences as query sequences. EST clones that belong to the same gene were identified by BLASTN search. The EST sequences exhibiting a sequence identity of 98% or more were clustered and given arbitrary names. The number of EST hits per gene was scored to calculate the proportion of each gene transcript among all α-expansin EST hits.
Amino acid sequences deduced from cDNA sequences were aligned using the ClustalW program at the BCM search launcher (Baylor College of Medicine, Houston, TX) Web server (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) using their default setting for protein multiple alignment (weight matrix, BLOSUM; gap opening penalty, 10.0; gap extension penalty, 0.05; residue-specific gap penalty, ON; hydrophilic gaps, ON). N-terminal signal peptide cleavage sites were predicted using the SignalP program at the Center for Biological Sequence Analysis (Technical University of Denmark, Lyngby, Denmark) Web server (http://www.cbs.dtu.dk/services/SignalP-2.0). A phylogenetic tree of α-expansin genes was constructed by parsimony analysis using the PAUP 4.0 software (Swofford, 1999). A bootstrap method with heuristic search was used with the following options: 100 random trees, simple stepwise addition, reference sequence AtEXP7, tree bisection reconnection and branch swamping with the MULPARS, steepest descent OFF. Trees were unrooted.
Survey of Expansin Genes Expressed in Arabidopsis Hypocotyls Exhibiting Active Secondary Growth
For induction of secondary growth, Arabidopsis ecotype Columbia plants were grown for 2 months under a long-day condition, while preventing senescence by removing flowers. Presence of secondary xylem was confirmed by observing cross-sections of hypocotyls stained with saturated phloroglucinol solution in 18% HCl. Total RNA was extracted from the hypocotyl segments as described above. A set of degenerate primers for RT-PCR, TGAYGCHTCHGGHACHATGGG (forward) and KTGCCARTTYTGYCCCCARTT (reverse), where Y = C or T, H = A, C, or T, K = G or T, and R = A or G, was designed to target two conserved amino acid sequences of α-expansins, DASGTMG and NWGQNWQ, respectively. Obtained PCR fragments were cloned into pGEM-T Easy vector system (Promega, Madison, WI) and introduced into Escherichia coli. Expansin gene fragments were then PCR amplified from single E. coli colonies and used for sequence diagnosis. The gene fragments were assigned to known Arabidopsis expansin genes by performing RFLP analysis using restriction enzymes HpaII and MseI. Out of 82 PCR fragments analyzed, 80 were identified as known Arabidopsis expansin genes.
Distribution of Materials
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Sequence data from this article have been deposited with the EMBL/GenBank libraries under accession numbers AY435099, AY435100, and AY43510.
Acknowledgments
We thank Kjell Olofsson for technical assistance and Gun Lövdahl for the artwork.
This work was supported by The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, the Foundation for Strategic Research, Enzyme Discovery in Hybrid Aspen for Fiber Engineering (EDEN; European project no. QLK5–CT–2001–00443), Kempe Foundation (postdoctoral fellowship to M.G.-M.), and Wood Ultrastructure Research Centre (WURC).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039321.
References
- Baluska F, Salaj J, Mathur J, Braun M, Jasper F, Samaj J, Chua N-H, Barlow PW, Volkman D (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin enriched bulges. Dev Biol 227: 618–632 [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Jonsson Birve S, Karlsson I, Gardeström P, Gustafsson P, Lundeberg J, et al (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999. b) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11: 2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Dunsmuir P (1999. a) Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol 39: 161–169 [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JKC, McQueen-Mason S, Kuhlemeier C (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123: 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey N, Cholewa E, Regan S, Sundberg B (2002) Secondary xylem development in Arabidopsis: a model for wood formation. Physiol Plant 114: 594–600 [DOI] [PubMed] [Google Scholar]
- Chang SJ, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Cho H-T, Kende H (1997) Tissue localization of expansins in deepwater rice. Plant J 15: 805–812 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000. a) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000. b) New genes and new biological roles for expansins. Curr Opin Plant Biol 3: 73–75 [DOI] [PubMed] [Google Scholar]
- Darley CP, Forrester AM, McQueen-Mason SJ (2001) The molecular basis of plant cell extension. Plant Mol Biol 47: 179–195 [PubMed] [Google Scholar]
- Esau K (1965) Plant Anatomy, Ed 2. John Wiley & Sons, New York
- Gray-Mitsumune M, Abe H, Takahashi J, Sunberg B, Mellerowicz EJ (2004) Liquid-phase fluorescence in situ RT-PCR analysis for gene expression analysis in woody stems. Plant Biol 6: 47–54 [DOI] [PubMed] [Google Scholar]
- Harrison EP, McQueen-Mason SJ, Manning K (2001) Expression of six expansin genes in relation to extension activity in developing strawberry fruit. J Exp Bot 52: 1437–1446 [DOI] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlén M, Teeri TT, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Olsson O (1998) Molecular characterization of novel plant homeobox gene expressed in the maturing xylem zone of Populus tremula × tremuloides. Plant J 16: 285–295 [DOI] [PubMed] [Google Scholar]
- Im K-H, Cosgrove DJ, Jones AM (2000) Subcellular localization of expansin mRNA in xylem cells. Plant Physiol 123: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourez B (1997. a) Le bois de tension. 1. Définition et distribution dans l'arbre. Biotechnol Agron Soc Environ 1: 100–112 [Google Scholar]
- Jourez B (1997. b) Le bois de tension. 2. Évaluation quantitative, formation et rôle dans l'arbre. Biotechnol Agron Soc Environ 1: 167–177 [Google Scholar]
- Larson PR (1994) The Vascular Cambium. Springer-Verlag, Berlin
- Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4: 527–532 [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Link BM, Cosgrove CJ (1998) Acid-growth response and α-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol 118: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unraveling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Okita TW, Choi SB (2002) mRNA localization in plants: targeting to the cell's cortical region and beyond. Curr Opin Plant Biol 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy B, McQueen-Mason S, Nosberger J, Fleming A (2001) Differential expression of α- and β-expansin genes in the elongating leaf of Festuca pratensis. Plant Mol Biol 46: 491–504 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C (1998) Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell 10: 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB (2000) Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol 123: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, et al (1998) Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA 95: 13330–13335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL (1999) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA
- Uggla C, Moritz T, Sandberg G, Sundberg B (1996) Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA 93: 9282–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenham MW, Cusick F (1975) The growth of secondary fibers. New Phytol 74: 247–261 [Google Scholar]
- Whitney SEC, Gidley MJ, McQueen-Mason SJ (2000) Probing expansin action using cellulose/hemicellulose composite. Plant J 22: 327–334 [DOI] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Thorne ET, Sharp RE, Cosgrove DJ (2001) Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol 126: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]