Abstract
The cellular polyamines putrescine, spermidine, and spermine are ubiquitous in nature and have been implicated in a wide range of growth and developmental processes. There is little information, however, on mutant plants or animals defective in the synthesis of polyamines. The Arabidopsis genome has two genes encoding spermidine synthase, SPDS1 and SPDS2. In this paper, we describe T-DNA insertion mutants of both of these genes. While each mutant allele shows normal growth, spds1-1 spds2-1 double-mutant seeds are abnormally shrunken and they have embryos that are arrested morphologically at the heart-torpedo transition stage. These seeds contain significantly reduced levels of spermidine and high levels of its precursor, putrescine. The embryo lethal phenotype of spds1-1 spds2-1 is complemented by the wild-type SPDS1 gene. In addition, we observed a nearly identical seed phenotype among an F2 seed population from the cross between the spds2-1 allele and SPDS1 RNA interference transgenic lines. These data provide the first genetic evidence indicating a critical role of the spermidine synthase in plant embryo development.
Polyamines are ubiquitous nitrogen compounds that are generally recognized as being necessary for orderly patterns of growth and development in most organisms (Tabor and Tabor, 1984; Cohen, 1998). Due to their positive charge, polyamines interact with various macromolecules and membranes. They show specific and differential binding to phospholipids and affect membrane rigidity (Schuber, 1989). Polyamines promote polymerization of cytoskeletal components, influence DNA conformation and stability, and activate the ribosomal ternary complex during protein synthesis (Igarashi and Kashiwagi, 2000). In higher plants, extracellular polyamines may interact with negatively charged cell wall components (Mariani et al., 1989). Growth-promoting activity of polyamines in plants was first demonstrated for callus production in explants of Jerusalem artichoke tubers (Bagni, 1966). Since then many studies on plants have implicated polyamines in a variety of cellular processes, including response to stress, control of cell division, root formation, flowering, and retardation of senescence (Evans and Malmberg, 1989; Galston and Sawhney, 1990; Kumar et al., 1997; Walden et al., 1997; Bouchereau et al., 1999). Involvement of polyamines in the stabilization of thylakoid membranes has also been reported for some plant species (Besford et al., 1993). However, the mechanisms underlying regulation of intracellular levels of polyamines and their localization have not been elucidated.
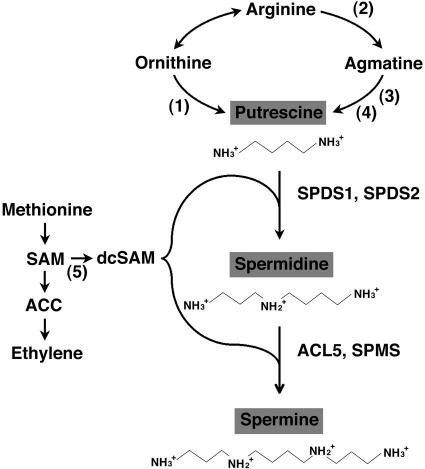
While the biosynthesis of polyamines in most organisms is initiated by decarboxylation of Orn to form putrescine via Orn decarboxylase (ODC), plants and some microorganisms can generate putrescine from Arg, via Arg decarboxylase (ADC) and agmatine ureohydrolase or agmatine iminohydrolase (Fig. 1). The respective functions of the two pathways of putrescine biosynthesis via ODC and ADC are still not clear. The genome of Arabidopsis contains no gene sequence for ODC (Hanfrey et al., 2001). Putrescine is converted to the triamine spermidine, and the tetramine spermine by the repetitive addition of an aminopropyl moiety from decarboxylated S-adenosylmethionine (SAM). These steps are mediated through the action of two closely related but specific enzymes, spermidine synthase and spermine synthase. Decarboxylated SAM is formed from SAM through the action of SAM decarboxylase (SAMDC), considered to be the rate-limiting step in polyamine biosynthesis. Because SAM is also the precursor of aminocyclopropane carboxylic acid, an important source of ethylene which is known to be the senescence hormone in higher plants, the balance between these two antagonistic compounds, ethylene and polyamines, has been thought to play a crucial role in plant development (Evans and Malmberg, 1989). Polyamines are further conjugated with a variety of compounds by the formation of an amide linkage. Conjugates with hydroxycinnamic acids accumulate in young leaves after floral induction and flower buds in various plants (Hamasaki and Galston, 1990). On the other hand, regulation of the level of intracellular polyamines involves degradation of putrescine by copper-containing diamine oxidase and of spermidine and spermine by FAD-containing polyamine oxidase. These enzymes predominantly occur in the apoplast and may also function in peroxidative lignification of the cell wall, cell wall stiffening, and cellular defense (Sebela et al., 2001).
Figure 1.
Pathway of biosynthesis of the major polyamines (putrescine, spermidine, and spermine) in plants. Enzymes shown in numbers are (1) ODC (EC 4.1.1.17), (2) ADC (EC 4.1.1.19), (3) agmatine iminohydrolase (EC 3.5.3.12), (4) N-carbamoylputrescine amidohydrolase (EC 3.5.1.53), and (5) SAMDC (EC 4.1.1.50). Spermidine is synthesized from putrescine by spermidine synthase (EC 2.5.1.16). Spermidine is further metabolized to spermine by spermine synthase (EC 2.5.1.22). The enzymes in Arabidopsis, SPDS1, SPDS2, ACL5, and SPMS, are indicated. ACC, 1-aminocyclopropane-1-carboxylic acid; dcSAM, decarboxylated S-adenosylmethionine.
Genetic analyses have revealed that the yeast spe3 mutant, which has no spermidine synthase activity, has an absolute requirement for spermidine or spermine for growth, while deletion of the SPE4 gene encoding spermine synthase has no effect on growth (Hamasaki-Katagiri et al., 1997, 1998). In Escherichia coli, spermidine-deficient mutants are still able to grow at near-normal rates without exogenous polyamines (Xie et al., 1993). In mouse fibroblast cell cultures, disruption of a spermine synthase gene that causes a lack of spermine and a higher accumulation of spermidine had no significant effect on the growth of cells (Mackintosh and Pegg, 2000). In higher plants, disruption of the ACAULIS5 (ACL5) gene, which encodes spermine synthase in Arabidopsis, has been shown to result in a severely dwarfed phenotype (Hanzawa et al., 2000). Furthermore, our recent study has demonstrated that spermine is not essential for survival of Arabidopsis, at least under normal growth conditions (Imai et al., 2004). However, no direct genetic evidence indicating the significance of spermidine in plant cells has been obtained. In this paper, we show that the genes encoding spermidine synthase are essentially required for embryonic development of Arabidopsis.
RESULTS
Identification of T-DNA Insertion Mutants in SPDS1 and SPDS2
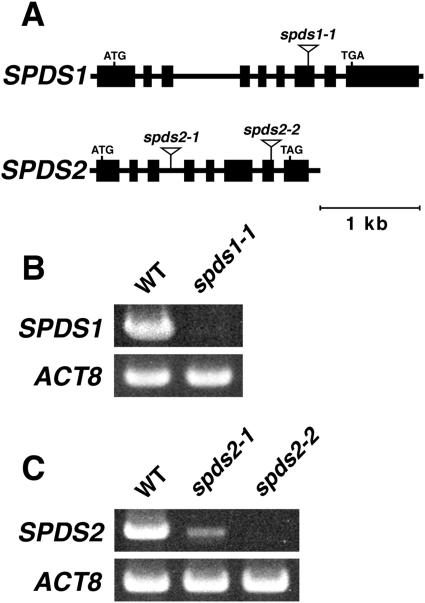
The Arabidopsis genome contains two genes encoding spermidine synthase, SPDS1 and SPDS2. These gene products have been shown to possess spermidine synthase activity (Hanzawa et al., 2002; Panicot et al., 2002b). We screened a library of about 74,000 independent T-DNA tag lines of the Kazusa DNA Research Institute for insertion mutants of SPDS1 and SPDS2 by a PCR-based screening strategy. We isolated insertion alleles designated spds1-1 and spds2-1 (Fig. 2A). The spds1-1 allele contains a T-DNA insertion in the seventh exon of the SPDS1 gene, 1,916 bp downstream of the translation start codon, and the spds2-1 allele contains a T-DNA insertion in the third intron, 660 bp downstream of the translation start codon of SPDS2. We also obtained another T-DNA insertion allele of the SPDS2 locus from the SALK Institute T-DNA tag lines (SALK_139824) and named it spds2-2. In the spds2-2 allele, the T-DNA was inserted into the seventh exon of the SPDS2 gene, 1,673 bp downstream of the translation start codon (Fig. 2A). Reverse transcription (RT)-PCR analysis of total RNA from these mutant seedlings was performed using gene-specific primers designed outside of the 5′ and 3′ ends of each T-DNA insertion. As shown in Figure 2B, no SPDS1 transcript was detected in the spds1-1 mutant. On the other hand, the SPDS2 transcript of correctly spliced size was detected in the spds2-1 mutant, but the relative level of the SPDS2 transcript was much lower in spds2-1 than in the wild type (Fig. 2C). This is probably because of the T-DNA insertion into an intron of SPDS2 in the spds2-1 allele. In spds2-2, no SPDS2 transcript was detected (Fig. 2C). These three mutant plants displayed no phenotypic alteration with respect to plant size and morphology under normal growth conditions.
Figure 2.
T-DNA insertion mutants of SPDS1 and SPDS2. A, Schematic diagrams of the spds1-1, spds2-1, and spds2-2 alleles indicating locations of the T-DNA insertion. The positions and lengths of exons and introns are indicated by black rectangles and lines, respectively. B, RT-PCR analysis of SPDS1 expression in wild-type (WT) and spds1-1 plants. C, RT-PCR analysis of SPDS2 expression in wild-type, spds2-1, and spds2-2 plants. Total RNA was prepared from 10-d-old seedlings. The level of Actin8 (ACT8) was used as an internal control.
Phenotype of the spds1 spds2 Double Mutant
To determine whether the normal growth of each single mutant is attributable to the functional redundancy between SPDS1 and SPDS2, we made spds1-1 spds2-1 double mutants. Since SPDS1 and SPDS2 belong to the same linkage group (chromosome I), we first selected spds1-1/spds1-1 spds2-1/+ and spds1-1/+ spds2-1/spds2-1 plants in an F2 generation of the reciprocal cross between spds1-1 and spds2-1 homozygous mutants by PCR-based genotyping. We then examined the siliques of these self-pollinated F2 plants and found that there are aborted seeds randomly distributed throughout the silique (Fig. 3A). These seeds, which become dark brown and shrink as they mature, apparently fail to germinate. The segregation ratio of these abnormal seeds was approximately a quarter of the total F3 seeds harvested (Table I). We confirmed that the rest of the seeds show normal germination and growth. These normal seedlings were found to contain at least one copy of the wild-type SPDS1 or SPDS2 gene depending on the parental F2 genotype (Table I). No seedlings homozygous for both spds1-1 and spds2-1 alleles were obtained, suggesting that the double mutant is embryonically lethal. Similar results were obtained from the reciprocal cross between spds1-1 and spds2-2 homozygous mutants (data not shown).
Figure 3.
Phenotype of the spds1-1 spds2-1 double mutant. A, Silique of a self-fertilized spds1-1/spds1-1 spds2-1/+ plant that was harvested 9 d after pollination. Arrows indicate spds1-1 spds2-1 double-mutant seeds that appeared brown and shrunken. Bar represents 0.5 μm. B and C, Nomarski images of a wild-type embryo (B) and an spds1-1 spds2-1 double-mutant embryo (C). Seeds were harvested from 3-d-old siliques after pollination. D and E, Light microscopy images of a wild-type embryo (D) and an spds1-1 spds2-1 double-mutant embryo (E) isolated from the mature dry seed. Bars represent 50 μm in B to E. F, Diagnostic PCR of the T-DNAs inserted in spds1-1 spds2-1. Genomic DNA was extracted from normal and arrested embryos. S1F and S1R, gene-specific primers for SPDS1; S2F and S2R, gene-specific primers for SPDS2; LB, primer specific to the T-DNA LB. Lanes 1, 3, 5, and 7, DNA from normal embryos; lanes 2, 4, 6, and 8, DNA from arrested embryos; lane M, DNA markers of indicated length.
Table I.
Segregation analysis of the progeny of self-fertilized spds1-1/spds1-1 spds2-1/+, spds1-1/+ spds2-1/spds2-1, and spds1-1/spds1-1 spds2-1/spds2-1 containing one copy of the wild-type SPDS1 transgene
| Parental Genotype | Observed Seeds
|
Normal Seeds
|
Shrunken Seeds
|
χ2 |
|---|---|---|---|---|
| No. | No. | No. | ||
| spds1-1/spds1-1 spds2-1/+ | 395 | 297 | 98 | 0.0076 |
| spds1-1/+ spds2-1/spds2-1 | 454 | 340 | 114 | 0.0029 |
| spds1-1/spds1-1 spds2-1/spds2-1 with a wild-type SPDS1 transgene | 405 | 302 | 103 | 0.040 |
χ2 calculation is based on expected ratio of three wild-type and one shrunken seed phenotypes. The χ2 values indicate no significant deviation (P > 0.05) from the expected ratio.
To characterize the nature of the seed lethality, we examined the embryos in the developing F2 siliques under a microscope. Approximately one-quarter of the immature seeds contained embryos defective in development. Figure 3, B and C, show that while a normal embryo reaches the bent-cotyledon stage at 3 d after pollination, a defective embryo remains at the heart stage. The defective embryos were eventually arrested at the heart-torpedo transition stage in mature dry seeds (Fig. 3, D and E). PCR-based genotyping analysis revealed that these arrested embryos are spds1-1 spds2-1 double mutants (Fig. 3F).
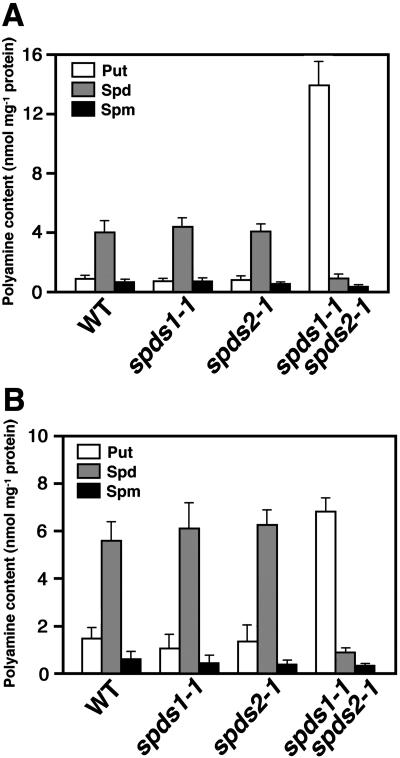
We further investigated the levels of putrescine, spermidine, and spermine in these embryos. Polyamines were extracted from dry seeds, dansylated, and analyzed by HPLC. The bound polyamine levels (perchloric acid [PCA]-insoluble fraction) in Arabidopsis seeds were just at the limits of our ability to detect them, and we therefore focused on the free and conjugated polyamines (PCA-soluble fraction; Fig. 4, A and B). In both free and conjugated forms, no significant difference was found between the wild type and each single mutant with respect to the levels of the three major polyamines. In contrast, the free spermidine and conjugated spermidine levels in the spds1-1 spds2-1 double-mutant seeds were decreased to 20.1% and 17.1% of those in the wild-type seeds, respectively. Spermine levels in spds1-1 spds2-1 seeds were also reduced. On the other hand, the free and conjugated putrescine levels in the double-mutant seeds were 17.0- and 4.8-fold greater than those in the wild-type seeds, respectively.
Figure 4.
Polyamine content in wild-type and mutant seeds. A, Free polyamines; B, polyamines conjugated in the PCA-soluble fraction. Polyamines were extracted from dry seeds of each plant line and quantified by HPLC. Bars indicate ± se (n = 3).
We performed a genetic complementation experiment with the wild-type SPDS1 gene. A genomic fragment encompassing the entire coding region of SPDS1 and its flanking 5′ sequence was used for transformation of spds1-1/spds1-1 spds2-1/+ plants. We obtained spds1-1 spds2-1 double mutant plants that contained a wild-type SPDS1 transgene and exhibited normal development in the progeny of the transformants. The progeny of self-fertilized spds1-1 spds2-1 plants with one copy of the SPDS1 transgene showed a segregation ratio that was a good fit to the expected ratio of normal seeds:shrunken seeds (3:1; Table I).
RNA Interference Analysis of SPDS Genes
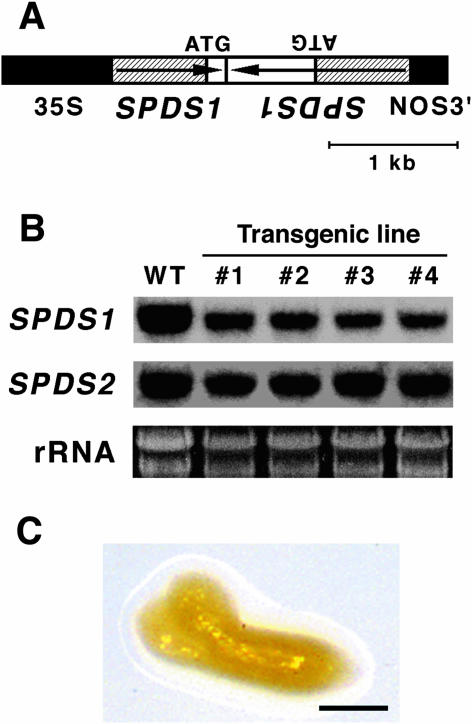
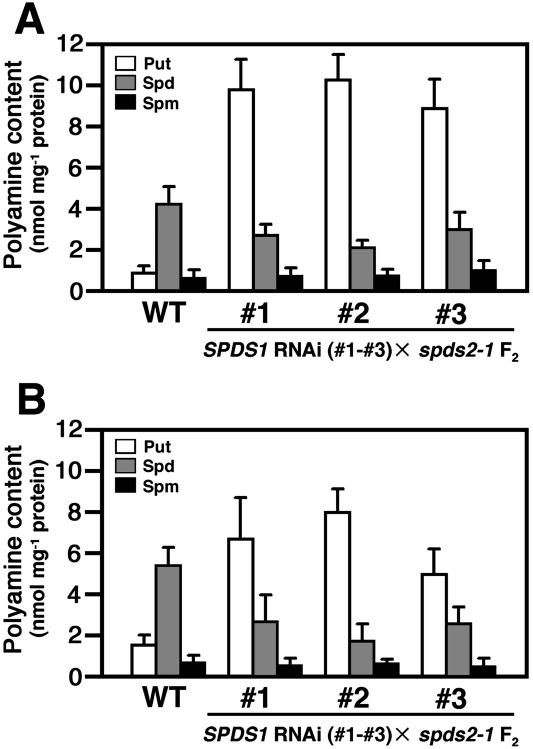
To further confirm that the embryonic arrest is associated with a defect in spermidine synthesis, we examined the effect of suppression of SPDS1 expression by RNA interference (RNAi) analysis (Chuang and Meyerowitz, 2000). A T-DNA construct in which the inverted repeat of the promoter region and a part of the N-terminal coding region of SPDS1 are placed under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 5A) was introduced into wild-type plants using Agrobacterium-mediated transformation. RNA gel-blot analysis revealed that all of the transgenic lines carrying the RNAi construct showed a reduction in the steady-state level of the SPDS1 endogeneous transcript, but no effect on the SPDS2 transcript level was seen (Fig. 5B). These transgenic plants were phenotypically normal. The effect of the RNAi was further investigated by crossing spds2-1 homozygous plants with a transgenic line, #1, carrying one copy of the SPDS1 RNAi construct. The F1 plants that were heterozygous for spds2-1 and carried the RNAi construct were then grown to the flowering stage. The self-pollination resulted in a segregation of shrunken seeds, which appeared to be nearly identical to those observed in spds1-1 spds2-1 double-mutant seeds in the F2 generation. Embryos dissected from these seeds appeared to be arrested at the torpedo-cotyledon stage (Fig. 5C). Although it was expected that 3/16 (18.75%) of the F2 seeds would be shrunken and fail to germinate, only 111/1,596 (7.0%) of the F2 seeds showed the aberrant seed phenotype (Table II). We also found that a low percentage of F2 plants (164/1,596 = 10.3% of the total F2 seeds examined) were homozygous for spds2-1 and carried the RNAi construct but showed normal growth. Similar results were obtained from crosses utilizing two additional transgenic lines (Table II and data not shown). When we measured the free and conjugated polyamine profiles of these shrunken seeds, markedly reduced levels of spermidine and increased levels of putrescine were detected (Fig. 6, A and B), supporting the conclusion that the embryonic arrest is associated with a defect in spermidine synthesis.
Figure 5.
Effect of SPDS1 RNAi in spds2-1. A, Scheme of the SPDS1 RNAi construct. The transcription cassette contains a 0.7-kb promoter region and a part of the N-terminal coding region of SPDS1, the 35S promoter of CaMV, and the nopaline synthase terminator (NOS3′). The hatched boxes represent the promoter region of SPDS1. Arrows indicate the orientation of the SPDS1 gene. B, RNA gel-blot analysis of SPDS1 and SDPS2 transcripts in 10-d-old seedlings of wild-type (WT) and SPDS1 RNAi lines (nos. 1–4). Each lane contained 10 μg of total RNA. rRNA is shown as a loading control. C, Phenotype of the arrested embryo segregated in F2 from the cross between spds2-1 and an SPDS1 RNAi line (no. 1). Bar represents 50 μm.
Table II.
Segregation analysis of the F2 progeny from the cross between spds2-1 and SPDS1 RNAi transgenic lines
| Cross | Observed Seeds
|
Normal Seeds
|
Shrunken Seeds
|
|---|---|---|---|
| No. | No. | No. (%) | |
| spds2-1 × no. 1 | 1,596 | 1,485 | 111 (7.0) |
| spds2-1 × no. 2 | 1,455 | 1,366 | 89 (6.1) |
| spds2-1 × no. 3 | 1,576 | 1,492 | 84 (5.3) |
Figure 6.
Polyamine content in the arrested embryos segregated in F2 from the cross between spds2-1 and SPDS1 RNAi lines (nos. 1–3). A, Free polyamines; B, polyamines conjugated in the PCA-soluble fraction. Polyamines were extracted from dry seeds of each plant line and quantified by HPLC. Bars indicate ± se (n = 3).
DISCUSSION
Physiological data obtained in various systems support the role of polyamines as regulators of cell proliferation and differentiation. Large amounts of spermidine and spermine accumulate during the transition from the G1 to the S phase of the cell cycle in both plants and animals (Fuller et al., 1977). The fact that inhibition of polyamine biosynthesis results in cell cycle arrest at the G1 phase suggests the involvement of polyamines in DNA synthesis (Kramer et al., 2001; Chattopadhyay et al., 2002). In tobacco (Nicotiana tabacum) ovary tissues, dramatic synchronous increases in polyamine titers and in rates of polyamine biosynthesis, macromolecular synthesis, and cell division occur for a few days after fertilization (Slocum and Galston, 1985). Taken together with these findings, our observation that spds1-1 spds2-1 and spds1-1 spds2-2 embryos did not develop beyond the torpedo stage indicates that spermidine synthase has an essential role in cell proliferation during embryogenesis.
Since a T-DNA in spds1-1 is located within the SPDS1 coding sequence for the large C-terminal catalytic core domain (Korolev et al., 2002) and since no SPDS1 transcript was detected in the spds1-1 seedlings at the level of RT-PCR, spds1-1 is most likely a null mutant. The spds2-1 allele has a T-DNA in the third intron of SPDS2. RT-PCR analysis revealed that the SPDS2 transcript is properly processed in spds2-1 with low efficiency. Thus, the low levels of spermidine accumulated in the double-mutant seeds (Fig. 4, A and B) might be produced from a limited amount of the full-length SPDS2 transcript in the spds2-1 allele. It is also possible that they reflect maternal carryover. In either case, these levels of spermidine may not be sufficient for the embryo development.
On the other hand, a markedly increased level of putrescine in the double mutant embryo may be due to the blockage of its conversion to spermidine. This raises the possibility that embryonic arrest in the double mutant is associated with toxicity of overaccumulated putrescine. Previous studies have shown that overexpression of oat ADC in tobacco leads to abnormal phenotypes such as short internodes, thin stems and leaves, leaf necrosis, and reduced root growth, the severity of which is correlated with putrescine content (Masgrau et al., 1997; Panicot et al., 2002a). In contrast, Noury et al. (2000) generated transgenic rice plants that show normal growth but up to a 10-fold increase in the amount of putrescine in the seeds compared to that in wild-type seeds by overexpressing the oat ADC cDNA. In yeast spe3 mutants that are defective in spermidine biosynthesis and in which putrescine overaccumulates, cell growth was rescued by the addition of spermidine or spermine to the culture medium (Hamasaki-Katagiri et al., 1997). We attempted to rescue the double mutant phenotype by allowing the embryos dissected from dry seeds to grow in a culture medium supplemented with spermidine or spermine, but the experiments were unsuccessful. This is presumably because of the experimental difficulty in applying spermidine or spermine to the dissected embryo. Alternatively, such exogenous polyamines might not be adopted in a proper intracellular location in the embryo. We observed that wild-type mature embryos showed abortion by the addition of 1 mm of spermidine or spermine to Murashige and Skoog liquid medium, while they could grow normally to seedlings in the presence of up to 5 mm of putrescine (data not shown). These data suggest that Arabidopsis embryos are insensitive to relatively high concentration of exogenous putrescine. However, further studies using transgenic Arabidopsis plants will be needed to determine whether the embryo lethality in the double mutant is caused by spermidine depletion, putrescine overaccumulation, or both.
The wild-type phenotype of spds1-1, spds2-1, and spds2-2 single-mutant plants can be interpreted by functional redundancy between SPDS1 and SPDS2. Both SPDS1 and SPDS2 transcripts are present in all organs and are present at higher levels in the root tissue than in other organs (Hanzawa et al., 2002). Urano et al. (2003) reported that, unlike SPDS1, SPDS2 shows barely detectable levels of expression in the upper stems and mature siliques. Moreover, treatment of wild-type seedlings with cytokinin results in up-regulation of SPDS2 expression but has no effect on SPDS1 expression (Hanzawa et al., 2002). Thus, it is still possible that each gene plays an individual role in different aspects of cellular processes or under different growth conditions. In pea plants, two genes encoding spermidine synthases are up-regulated in different time courses during the period of early fruit development (Alabadí and Carbonell, 1999). The Arabidopsis genome has two genes encoding spermine synthase, ACAULIS5 (ACL5) and SPMS (Hanzawa et al., 2000, 2002; Panicot et al., 2002b). While loss-of-function mutants of ACL5 show a severe defect in stem elongation (Hanzawa et al., 2000), those of SPMS show normal growth. Interestingly, acl5 spms double mutants contain no spermine but display no additional phenotype to the stem phenotype of acl5, suggesting that the spermine produced only through the ACL5 function is involved in stem elongation (Imai et al., 2004). The stem growth of acl5 plants is not rescued by exogenous spermine (Hanzawa et al., 2000). These findings suggest that each polyamine molecule needs to be targeted to a specific intracellular location and/or a specific conjugated form.
We used RNAi to examine whether the morphological defect of double-mutant embryos can also be realized by the suppression of SPDS1 gene expression in spds2-1. The construct was made to produce a double-stranded RNA containing the SPDS1 promoter sequence for triggering transcriptional gene silencing by de novo methylation of the target promoter (Mette et al., 2000; Paszkowski and Whitham, 2001). Although the gene-specific reduction of SPDS1 transcript levels was confirmed in each transgenic line, the expression was not completely suppressed, presumably because the degree of the silencing effect varied between cells. We detected shrunken seeds in the F2 progeny of the cross between spds2-1 and these RNAi lines. The low ratio of segregation (111/1596 = 7.0%; Table II) may be due to the incomplete suppression of SPDS1 expression. In fact, approximately 10.3% (164/1596) of the F2 population appeared to be spds2-1 homozygous plants that carried the RNAi construct and showed normal growth, suggesting that the total number (275/1596) corresponds to the expected ratio (3/16 = 18.75%).
In conclusion, the isolation of SPDS1 and SPDS2 T-DNA insertion mutants and the construction of their double mutants enabled us to prove the importance of these genes in plant survival. A previous study demonstrated that the stem elongation of acl5 mutants could be restored in a heat shock-dependent manner by introducing heat shock-inducible ACL5 cDNA (Hanzawa et al., 2000). For a more complete understanding of polyamine functions in plant development, experiments using an inducible RNAi system are currently under way. It will also be necessary to address the basis of polyamine homeostasis and metabolism, including transport, conjugation, and oxidation, which remains unclear in plants and animals.
MATERIALS AND METHODS
Plant Growth Conditions
The Columbia (Col-0) ecotype of Arabidopsis (L.) Heynh was used as the wild type. Plants were grown under continuous fluorescent light of 120 μmol photons m−2 s−1 at 22°C on rock-wool bricks supplemented with vermiculite or on 0.8% (w/v) agar plates containing Murashige and Skoog salts (pH 5.8) and 3% Suc after surface sterilization of seeds.
Identification of T-DNA Insertion Mutants
The spds1-1 and spds2-1 mutant lines were obtained by screening a DNA pool of the Arabidopsis (ecotype Col-0) T-DNA insertion lines deposited at the Kazusa DNA Research Institute. PCR reactions included gene-specific primers, S1F (5′-CATTTCTCGGAGATATTCACCAG-3′) and S1R (5′-CCTCCAGATTAGTTTTCTTTCCC-3′) for SPDS1 and S2F (5′-CTAATCTCTTACTCACTGTCTCTCT-3′) and S2R (5′-CTAGTTGGCTTTCGAATCAATCACC-3′) for SPDS2 in combination with a T-DNA left-border (LB) primer (5′-ATAACGCTGCGGACACATCTAC-3′) and a right-border primer (5′-ATCCAGGCTTTGATAGTCA-3′). T-DNA insertion sites were determined by sequencing the resulting positive PCR fragments with the LB or right-border primer. Plant genotyping for spds1-1 and spds2-1 alleles was performed by PCR using the SPDS1 and SPDS2 gene-specific primers and the LB primer described above.
An spds2-2 mutation was found in the searchable database of T-DNA insertion sequences established by the Salk Institute Genome Analysis Laboratory. The T-DNA Express database is accessible at http://signal.salk.edu/cgi-bin/tdnaexpress. DNA flanking the LB of the T-DNA was amplified with a T-DNA LB′ primer (5′-ACGTCCGCAATGTGTTAT-3′) and sequenced. Plant genotyping for the spds2-2 allele was performed by PCR using the SPDS2 gene-specific primers and the LB′ primer described above.
Gene Expression Analyses
Total RNA was extracted from 10-d-old seedlings grown on Murashige and Skoog agar plates according to the SDS-phenol method (Takahashi et al., 1992). RT-PCR was conducted by using the RNA LA PCR Kit (Takara, Kyoto) with 0.5 μg of total RNA. For the first-strand cDNA synthesis, oligo(dT)-adaptor primer (5′-GCGGCCGCT(dT)18-3′) was used. Gene-specific primers for SPDS1 and SPDS2 were as described above. PCR conditions were 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min. As a control, the Actin8 transcript was amplified for 25 cycles using the primers ACT8F (5′-TGAGCCAGATCTTCATCGTC-3′) and ACT8R (5′-TCTCTTGCTCGTAGTCGACA-3′; An et al., 1996).
For RNA gel-blot analysis, 10 μg of each total RNA sample was separated on 1.2% formaldehyde agarose gels and blotted onto Hybond-N+ membranes (Amersham, Buckinghamshire, UK). Gene-specific probes for SPDS1 and SPDS2 were prepared by PCR using Arabidopsis genomic DNA as a template with primer pairs, S1F′ (5′-TTCGCCAAGAAGGTCATTGAGTCA-3′) and S1R′ (5′-TGATGTTAATGTTCTTGGTCTTCGG-3′) for the SPDS1 3′-UTR, and S2F′ (5′-TTTTGCTAAGAAGGTGATTGATTCG-3′) and S2R′ (5′-TATAACGTCAACGTCGACAATACC-3′) for the SPDS2 3′-UTR. These fragments were 32P-labeled by random-primed synthesis (Takara). The blots were hybridized at 42°C for 16 h with a labeled probe in 50% (v/v) formamide, 10% (w/v) dextran sulfate, 1 m NaCl, and 1% SDS and then washed twice with 2 × SSC, 0.1% SDS at 65°C for 30 min, and once with 0.1 × SSC at room temperature for 5 min. The membranes were exposed to x-ray films at −80°C for 72 h.
Microscopy
Seeds of wild-type and spds1-1 spds2-1 double-mutant plants were harvested from siliques, fixed in ethanol:acetic acid (6:1) solution for 4 h and washed successively in 90% and 70% (v/v) ethanol for 30 min each. Then the seeds were cleared with 72.7% (w/v) chloral hydrate in 50% (w/v) glycerol for 4 h prior to microscopy. Embryos were visualized using a Zeiss (Jena, Germany) LSM410 invert Laser Scan Microscope equipped with Nomarski optics. Mature embryos in dry seeds were dissected using fine forceps after the seeds were imbibed.
Polyamine Analysis
In order to extract polyamines, seeds (10 mg) were homogenized in 0.5 mL of 5% (w/v) PCA containing 1 nmol of 1,6-hexanediamine as an internal standard. After centrifugation, the supernatant was preserved and the pellet was resuspended in 5% PCA after several washes with the same solution. Aliquots of acid-soluble and acid-insoluble fractions, containing free plus conjugated polyamines and bound polyamines, respectively, were subjected to hydrolysis in 6 m HCl at 110°C for 18 h to convert the conjugated and bound forms to free form. After the hydrolyzate was taken to dryness in vacuum at 70°C, the residues were dissolved in 500 μL of 5% PCA. Aliquots (100 μL) were added to 200 μL of saturated sodium carbonate and 200 μL of dansyl chloride (5 mg mL−1 acetone). After brief vortexing, the mixture was incubated in darkness at 30°C for 16 h. Excess dansyl reagent was inactivated by the addition of 50 μL of 0.9 m Pro. Dansylated polyamines were extracted in 0.5 mL toluene, dried in Speed-Vac concentrator (Savant, Holbrook, NY), redissolved in 50 μL of methanol, and analyzed by HPLC using a Wakosil-II 5C18 HG reverse phase column (particle size 5 μm; 4.6 × 150 mm; Wako, Osaka). Portions (20 μL) of the polyamine fractions were applied to the column and eluted with a programmed methanol to water solvent gradient, changing from 55% to 85% over 15 min at a flow rate of 0.8 mL min−1. Elution was complete after 15 min. Polyamines were quantified by a fluorescence detector set at excitation and emission wavelengths of 365 and 510 nm, respectively. Conjugated polyamine content was calculated by subtracting free polyamine content from total acid-soluble polyamine content. Results were standardized with equimolar (0.5 nmol) mixtures of dansylated polyamines.
Plasmid Construction and Plant Transformation
A 3,854-bp genomic fragment containing the SPDS1 gene was amplified by PCR using the forward primer cF (5′-CTATCGATGATGCGGATGCG-3′) at 1,049 bp upstream of the SPDS1 ATG start codon and the reverse primer cR (5′-TGGAATTCGGTCTTGCGACAAGGA-3′) at 490 bp downstream of the stop codon. The underlined sequences are endogenous ClaI and engineered EcoRI restriction sites, respectively. The fragment was then cloned into the pBI101.2 vector (CLONTECH, Palo Alto, CA) at ClaI-EcoRI sites, resulting in a cSPDS construct.
To make a hairpin RNA construct for SPDS1, a 0.9-kb fragment (from −564 to +335 relative to the transcriptional initiation site) and a 1.4-kb fragment (from −564 to +881) of the SPDS1 gene were each amplified by PCR from genomic DNA with the following primer pairs: S1Fi (5′-AAGCTTCTTCGTTTAAATGCCTTCC-3′)/S1Ri (5′-GGATCCACCCAGGAATAACAGTG-3′) and S1Fi / S1R'i (5′-GGATCCTACTTTACCGGAGAAGAA-3′), respectively. The underlined sequences are additional HindIII and BamHI restriction sites. These amplified fragments were separately cloned into the pGEM-T Easy vector (Promega, Madison, WI) to generate pSPDS1a and pSPDS1b, respectively. The SpeI-BamHI fragment of pSPDS1a and the BamHI-SacI fragment of pSPDS1b were successively transferred into XbaI-BamHI and BamHI-SacI sites downstream of the CaMV35S promoter of a binary vector, pBI121 (CLONTECH), to remove the β-glucuronidase gene and generate an SPDS1 RNAi construct (Fig. 5A).
Transformation of Arabidopsis was carried out by the floral dip method (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens strain C58C1. The cSPDS construct was introduced into spds1-1/spds1-1 spds2-1/+ plants. Transformants were selected in Murashige and Skoog agar plates supplemented with 60 μg mL−1 kanamycin.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041699.
References
- Alabadí D, Carbonell J (1999) Differential expression of two spermidine synthase genes during early fruit development and in vegetative tissues of pea. Plant Mol Biol 39: 933–943 [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss SC, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bagni N (1966) Aliphatic amines and a growth factor of coconut milk as stimulating cellular proliferation of Helianthus tuberosus (Jerusalem artichoke) in vitro. Experientia 22: 732–733 [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Besford RT, Richardson C, Campos JL, Tiburcio AF (1993) Effect of polyamines on stabilization of molecular complexes in thylakoid membranes of osmotically stressed oat leaves. Planta 189: 201–206 [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J (1999) Polyamines and environmental challenges: recent development. Plant Sci 140: 103–125 [Google Scholar]
- Chattopadhyay MK, Tabor CW, Tabor H (2002) Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe). Proc Natl Acad Sci USA 99: 10330–10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S (1998) A Guide to the Polyamines. Oxford University Press, New York
- Evans PT, Malmberg RL (1989) Do polyamines have roles in plant development? Annu Rev Plant Physiol Plant Mol Biol 40: 235–269 [Google Scholar]
- Fuller DJ, Gerner EW, Russell DH (1977) Polyamine biosynthesis and accumulation during the G1 to S phase transition. J Cell Physiol 93: 81–88 [DOI] [PubMed] [Google Scholar]
- Galston AW, Sawhney RK (1990) Polyamines in plant physiology. Plant Physiol 94: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki N, Galston AW (1990) The polyamines of Xanthium strumarium and their response to photoperiod. Photochem Photobiol 52: 181–186 [DOI] [PubMed] [Google Scholar]
- Hamasaki-Katagiri N, Katagiri Y, Tabor CW, Tabor H (1998) Spermine is not essential for growth of Saccharomyces cerevisiae: identification of the SPE4 gene (spermine synthase) and characterization of a spe4 deletion mutant. Gene 210: 195–201 [DOI] [PubMed] [Google Scholar]
- Hamasaki-Katagiri N, Tabor CW, Tabor H (1997) Spermidine biosynthesis in Saccharomyces cerevisiae: polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase). Gene 187: 35–43 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27: 551–560 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Imai A, Michael AJ, Komeda Y, Takahashi T (2002) Characterization of the spermidine synthase-ralated gene family in Arabidopsis thaliana. FEBS Lett 527: 176–180 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y (2000) ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J 19: 4248–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K (2000) Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271: 559–564 [DOI] [PubMed] [Google Scholar]
- Imai A, Akiyama T, Kato T, Sato S, Tabata S, Yamamoto KT, Takahashi T (2004) Spermine is not essential for survival of Arabidopsis. FEBS Lett 556: 148–152 [DOI] [PubMed] [Google Scholar]
- Korolev S, Ikeguchi Y, Skarina T, Beasley S, Arrowsmith C, Edwards A, Joachimiak A, Pegg AE, Savchenko A (2002) The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat Struct Biol 9: 27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DL, Chang BD, Chen Y, Diegelman P, Alm K, Black AR, Roninson IB, Porter CW (2001) Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res 61: 7754–7762 [PubMed] [Google Scholar]
- Kumar A, Altabella T, Taylor MA, Tiburcio AF (1997) Recent advances in polyamine research. Trends Plant Sci 2: 124–130 [Google Scholar]
- Mackintosh CA, Pegg AE (2000) Effect of spermine synthase deficiency on polyamine biosynathesis and content in mice and embryonic fibroblasts, and the sensitivity of fibroblasts to 1,3-bis-(2-chloroethyl)-N-nitrosourea. Biochem J 351: 439–447 [PMC free article] [PubMed] [Google Scholar]
- Mariani P, D'Orazi D, Bagni N (1989) Polyamines in primary walls of carrot cells: endogenous content and interactions. J Plant Physiol 135: 508–510 [Google Scholar]
- Masgrau C, Altabella T, Farrás R, Flores D, Thompson AJ, Besford RT, Tiburcio AF (1997) Inducible overexpression of oat arginine decarboxylase in transgenic tobacco plants. Plant J 11: 465–473 [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJM (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19: 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noury M, Bassie L, Lepri O, Kurek I, Christou P, Capell T (2000) A transgenic rice cell lineage expressing the oat arginine decarboxylase (adc) cDNA constitutively accumulates putrescine in callus and seeds but not in vegetative tissues. Plant Mol Biol 43: 537–544 [DOI] [PubMed] [Google Scholar]
- Panicot M, Masgrau C, Borrell A, Cordeiro A, Tiburcio AF, Altabella T (2002. a) Effects of putrescine accumulation in tobacco transgenic plants with different expression levels of oat arginine decarboxylase. Physiol Plant 114: 281–287 [DOI] [PubMed] [Google Scholar]
- Panicot M, Minguet EG, Ferrando A, Alcázar R, Blázquez MA, Carbonell J, Altabella T, Koncz C, Tiburcio AF (2002. b) A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell 14: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J, Whitham SA (2001) Gene silencing and DNA methylation processes. Curr Opin Plant Biol 4: 123–129 [DOI] [PubMed] [Google Scholar]
- Schuber F (1989) Influence of polyamines on membrane functions. Biochem J 260: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebela M, Radová A, Angelini R, Tavladoraki P, Frébort II, Pec P (2001) FAD-containing polyamine oxidases: a timely challenge for researchers in biochemistry and physiology of plants. Plant Sci 160: 197–207 [DOI] [PubMed] [Google Scholar]
- Slocum RD, Galston AW (1985) Changes in polyamine biosynthesis associated with postfertilization growth and development in tobacco ovary tissues. Plant Physiol 79: 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H (1984) Polyamines. Annu Rev Biochem 53: 749–790 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Naito S, Komeda Y (1992) Isolation and analysis of the expression of two genes for 81-kilodalton heat shock proteins from Arabidopsis. Plant Physiol 99: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2003) Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ 26: 1917–1926 [Google Scholar]
- Walden R, Cordeiro A, Tiburcio AF (1997) Polyamines: small molecules triggering pathways in plant growth and development. Plant Physiol 113: 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie QW, Tabor CW, Tabor H (1993) Deletion mutations in the speED operon: spermidine is not essential for the growth of Escherichia coli. Gene 126: 115–117 [DOI] [PubMed] [Google Scholar]