Abstract
Cyanobacteria are key contributors to global photosynthetic productivity, and iron availability is essential for cyanobacterial proliferation. While iron is abundant in the earth's crust, its unique chemical properties render it a limiting factor for photoautotrophic growth. As compared to other nonphotosynthetic organisms, oxygenic photosynthetic organisms such as cyanobacteria, algae, and green plants need large amounts of iron to maintain functional PSI complexes in their photosynthetic apparatus. Ferritins and bacterioferritins are ubiquitously present iron-storage proteins. We have found that in the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis 6803), bacterioferritins are responsible for the storage of as much as 50% of cellular iron. Synechocystis 6803, as well as many other cyanobacterial species, have two bacterioferritins, BfrA and BfrB, in which either the heme binding or di-iron center ligating residues are absent. Purified bacterioferritin complex from Synechocystis 6803 has both BfrA and BfrB proteins. Targeted mutagenesis of each of the two bacterioferritin genes resulted in poor growth under iron-deprived conditions. Inactivation of both genes did not result in a more severe phenotype. These results support the presence of a heteromultimeric structure of Synechocystis bacterioferritin, in which one subunit ligates a di-iron center while the other accommodates heme binding. Notably, the reduced internal iron concentrations in the mutant cells resulted in a lower content of PSI. In addition, they triggered iron starvation responses even in the presence of normal levels of external iron, thus demonstrating a central role of bacterioferritins in iron homeostasis in these photosynthetic organisms.
Iron serves as a cofactor in a multitude of cellular processes. As such, iron accumulation and storage processes are essential for the survival of all organisms. However, the same redox properties that make iron a valuable cofactor also lead to oxidative interactions resulting in the formation of harmful radicals. Therefore, iron accumulation in the cells is tightly regulated to ensure that very little free iron is present (Kakhlon and Cabantchik, 2002).
While iron is abundant in the earth's crust, the bioavailability of iron in the current oxidative terrestrial environment is limited. This is due to the fact that in the presence of dioxygen at neutral pH, iron precipitates as Fe(OH)3. As a consequence, the bioavailability of iron limits the proliferation of photosynthetic organisms. Indeed, studies conducted in the Southern Ocean have indicated that iron is a limiting factor for primary photosynthetic productivity (Falkowski et al., 1998). In many of the ecological niches occupied by photosynthetic organisms, iron concentrations are limited to nanomolar levels (Morel and Price, 2003). Surges in the iron concentration due to aeolian dust deposition are transient (Falkowski et al., 1998). To accommodate such an infrequent supply of iron, photosynthetic organisms must have efficient iron storage mechanisms, the molecular natures of which are poorly understood.
Here, we report on our studies on iron homeostasis in Synechocystis sp. PCC 6803 (Synechocystis 6803), a model organism representing an abundant group of oxygenic photosynthetic organisms, the cyanobacteria. Synechocystis 6803 is a unicellular cyanobacterium in which targeted gene replacement can be easily accomplished. As in all other gram-negative bacteria, iron is transported through the outer and then the inner membrane. However, unlike other bacteria, cyanobacteria contain intracellular photosynthetic thylakoid membranes (Gantt, 1994) in which many iron-dependent enzymes function.
Iron plays a key role in photosynthetic electron transfer. PSII contains two cytochromes and one nonheme iron (Zouni et al., 2001; Kamiya and Shen, 2003). The cytochrome b6f complex has four hemes and one Fe2-S2 cluster (Kurisu et al., 2003; Stroebel et al., 2003). However, the largest sink for iron in the photosynthetic system is PSI, which contains 12 iron atoms in three Fe4-S4 clusters (Jordan et al., 2001). Adaptations of the photosynthetic apparatus to iron limitation include the induction of a specific PSI antenna complex, CP43′ (Boekema et al., 2001) and a flavodoxin protein that can serve as an alternative to ferredoxin in the electron transport chain (Laudenbach et al., 1988).
Once iron is transported inside the cell, it needs to be stored in a way that prevents its interaction with various redox active components. The Synechocystis 6803 genome encodes two iron storage proteins that are members of the bacterioferritin (Bfr) family (Bertani et al., 1997). Bfrs, like other ferritin family proteins, store iron in a cavity at the center of their 24-mer ultrastructure. Iron enters the Bfr complex as Fe2+ and is oxidized on its way to the central cavity (Carrondo, 2003). This ferroxidase activity is carried out by the di-iron center. In addition to the ferroxidase center, which is found in all ferritin family proteins, Bfrs contain a heme moiety ligated at the interface between two subunits. The role of this heme remains poorly defined (Carrondo, 2003).
Synechocystis 6803 bfr genes belong to a subfamily of Bfr genes in which one gene codes for a protein with a conserved heme ligand and the other codes for a protein with conserved di-iron center ligands (Bertani et al., 1997). In this work, we present a functional analysis of both Bfr proteins in Synechocystis 6803. A deletion of either one of the bfr genes resulted in a substantial loss in intracellular iron content and directly affected the photosynthetic apparatus. We propose a mechanistic model for the role of the two Bfr proteins and discuss the implications of our findings for a wide range of cyanobacterial species.
RESULTS
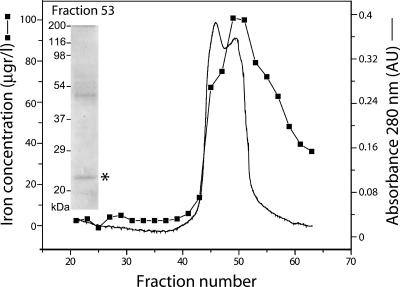
To study the protein composition of Bfr from Synechocystis 6803, we isolated Bfr complexes from wild-type cells (Fig. 1). The heavy fraction collected from broken cells by gradient centrifugation was applied to a size exclusion column. Fraction 53, eluted from the column at the 400-kD molecular mass range, corresponded to the peak in the iron profile. Mass spectroscopic analysis of the peak fraction revealed that two Bfr proteins, Sll1341 (BfrA) and Slr1890 (BfrB), comigrated on an SDS-PAGE gel at an apparent molecular mass of approximately 20 kD (Fig. 1 Insert). It is noteworthy that previous analysis of Bfr from Synechocystis 6803 by N-terminal sequencing had detected only the BfrA protein (Laulhere et al., 1992). In Neisseria gonorrhoeae, a bacterium with a similar bfr gene composition, both subunits were found in isolated Bfr (Chen and Morse, 1999). The additional band at 50 kD contains the periplasmic urea transport UrtA protein. This is most probably a result of cross contamination from periplasmic material that precipitates with the heavy fraction as well.
Figure 1.
Purification of Bfr from Synechocystis 6803 cells. Elution profile of partially purified Bfr from wild-type cells on a Sephacryl S-300 column. Solid line, A280; squares, iron concentration in the fractions. Fraction 53 was concentrated and fractionated by SDS-PAGE (insert, Coomassie R-250-stained gel). Mass spectroscopic analysis reveals the presence of Bfr proteins in the band marked by an asterisk. BfrA and BfrB proteins were identified with MOWSE scores (as defined at http://prospector.ucsf.edu/) of 2.45 × 106 and 1.31 × 103, respectively. The additional band at approximately 50 kD contains the UrtA protein (MOWSE score 5.58 × 104).
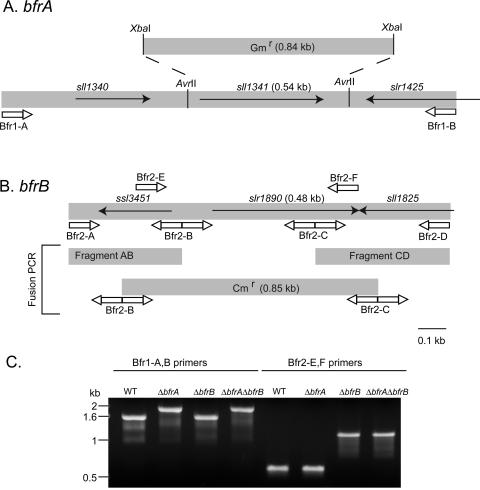
To examine the role of the Bfr genes in cyanobacterial iron homeostasis, we have produced two knockout constructs targeted for each of the Synechocystis 6803 bfr genes, respectively. Using these constructs, three mutant strains were produced: lacking the sll1341 gene (bfrA), slr1890 gene (bfrB), or both genes (Fig. 2). High molecular mass-assembled Bfr complexes could not be found in the heavy fraction collected from ΔbfrA or ΔbfrB cells (data not shown).
Figure 2.
Construction of bfr disruption mutants. A, Strategy for cloning and replacing the sll1341 (bfrA) gene with a gentamycin (Gm) resistance gene. B, Strategy for cloning and replacing the slr1890 (bfrB) gene with a chloramphenicol (Cm) resistance gene. C, Segregation of the mutant strains tested by genomic PCR. Supplemental Table I contains the primer sequences.
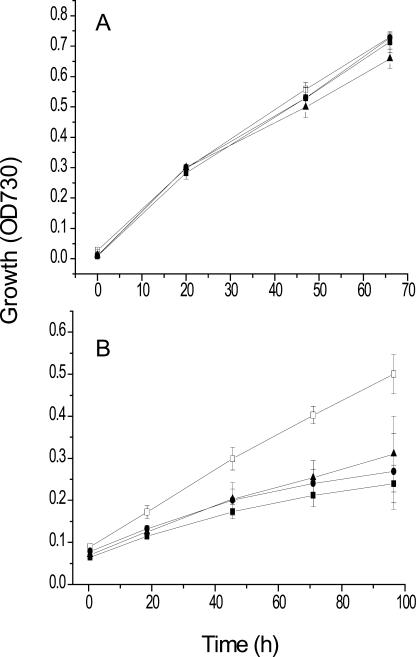
In the BG11 medium, the growth rates of the three mutant strains were virtually indistinguishable from that of the wild type (Fig. 3A). By contrast, in the BG11-Fe medium, the two deletion strains grew at half the rate of the wild-type cells (Fig. 3B). Interestingly, the double deletion strain grew as well as the single deletion strains. It is important to note that the iron concentrations that cyanobacteria encounter in nature are in the nanomolar range (Morel and Price, 2003). This concentration is closer to the iron concentration in BG11-Fe rather than to that in BG11 that contains 30 μm iron.
Figure 3.
Growth of wild type and bfr mutants on BG11 ± Fe media. The four strains were grown in BG11. The cells were washed free of excess iron (see “Materials and Methods”) and resuspended in BG11 (A) or BG11-Fe (B). sd was based on measurements of three replicate cultures. □, Wild type; ▪, ΔbfrA; •, ΔbfrB; ▴, ΔbfrAΔbfrB.
Iron limitation did not halt the growth of cyanobacteria completely (Fig. 3). Cyanobacteria acclimatize to low iron concentrations and are able to sustain slow rates of growth in spite of limited iron availability (Sherman and Sherman, 1983). One of the adaptations to iron stress is the synthesis of the CP43′ pigment protein complex (Boekema et al., 2001). The CP43′ protein is homologous to the PSII light-harvesting antenna protein CP43. However, CP43′ is associated with PSI (Bibby et al., 2003).
Transcription of the isiA gene that codes for the CP43′ protein is strongly correlated to iron availability (Singh et al., 2003). In addition to iron, isiA transcription was found to be regulated by salt and oxidative stresses (Vinnemeier et al., 1998; Yousef et al., 2003, respectively).
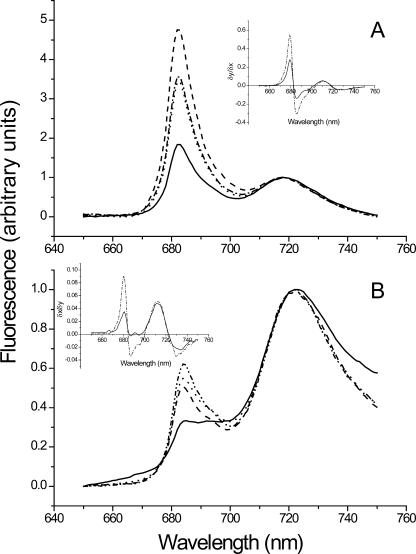
Fluorescence emission spectra at 77 K were used to test for the presence of the CP43′ antenna complex (Fig. 4). The fluorescence from CP43′ peaks at 682 nm, PSII fluorescence peaks at 685 and 695 nm, and PSI fluorescence peaks at 720 nm. Under limiting iron concentrations (Fig. 4A), a strong CP43′ fluorescence peak could be detected in all strains. However, the intensity of this band was higher in the mutants than in the wild-type cells (Table I). When grown under iron-sufficient conditions (Fig. 4B), the intensity of the 682-nm peak was lower in all of the strains (Table I). Nevertheless, it was still detectable in the three bfr deletion strains, while in the wild-type cell the PSII peak at 685 nm could be observed (Fig. 4B, insert). The CP43′ fluorescence peak could be observed only in late exponential phase and not early exponential phase cells (Table I). These results indicate that, even under iron-sufficient conditions, the mutants experienced iron limitation. The induction of CP43′ was tested during exposure of the cells to 0.7 m NaCl or 5 mm H2O2 as well. These conditions were reported to induce isiA transcription (Vinnemeier et al., 1998; Yousef et al., 2003). However, no differential effect on CP43′ fluorescence in the mutant cultures could be observed in these experiments (Table I).
Figure 4.
77 K fluorescence emission spectra from wild type and bfr mutants. Chlorophyll fluorescence spectra at 77 K from cells grown in BG11-Fe (A) and BG11 (B). Excitation was at 420 nm ± 5 nm. The spectra were normalized to the emission intensity at 720 nm. The samples were in the range of 1 to 2 × 109 cells/mL. Solid lines, wild type; dashed lines, ΔbfrA; dotted lines, ΔbfrB; dot-dashed lines, ΔbfrAΔbfrB. Inserts display the first derivative of the fluorescence curves. For the sake of clarity, only wild-type and ΔbfrAΔbfrB derivatives are shown. The experiment was repeated three times for each growth condition. Representative data are presented.
Table I.
Relative CP43′ content
| Wild Type | ΔbfrA | ΔbfrB | ΔbfrAΔbfrB | |
|---|---|---|---|---|
| Early log cells | 0.17 | 0.23 | 0.19 | 0.19 |
| Late log cells* | 0.27 | 0.55 | 0.49 | 0.68 |
| Low iron* | 1.84 | 3.43 | 3.56 | 4.76 |
| 5 mm H2O2 | 0.28 | 0.56 | 0.52 | 0.55 |
| 0.7 m NaCl | 0.44 | 0.44 | 0.45 | 0.46 |
The relative CP43′ content was calculated as the ratio of the 77 K fluorescence intensity at 682 nm (CP43′) to the intensity at 720 nm (PSI). The fluorescence traces for rows marked with an asterisk are presented in Figure 4. Early exponential phase cells were in the 7.2 to 9.2 × 107 cells/mL range. Late exponential phase cells were in the 1 to 2 × 109 cells/mL range. For low iron conditions, cells were grown in BG11-Fe. For oxidative stress, 5 mm H2O2 were applied for 30 min (culture concentrations 3.3–4.6 × 108 cells/mL). For salt stress conditions cells (3.3–4.6 × 108 cells/mL) were harvested, resuspended in BG11 media containing 0.7 m NaCl, and incubated for 3 d. Experiments marked by an asterisk were repeated three times; all other experiments were repeated two times. Representative data are presented.
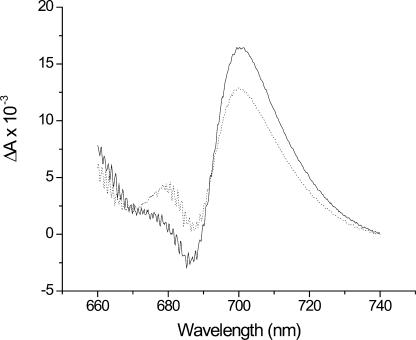
In late exponential phase ΔbfrAΔbfrB cells, the PSI content, determined by measuring the absorbance of the reaction center P700 chlorophylls (Sonoike and Katoh, 1989), was lower then that of the wild type (Fig. 5). On an equal chlorophyll basis, the ΔbfrAΔbfrB strain contained only 75% PSI, as compared to wild-type cells (189 and 145 Chl/P700, respectively), indicating a lower PSI to PSII ratio. The chlorophyll concentration in both wild-type and mutant cells was 2.16 × 10−14 g Chl/cell or 1.4 × 107 Chl molecules/cell. Hence, the wild-type and mutant cells contained 9.6 × 104 and 7.3 × 104 PSI units, respectively. Considering that each PSI unit contains 12 iron atoms, these numbers correspond to 1.2 × 106 and 8.8 × 105 atoms of iron in PSI complexes per cell, respectively.
Figure 5.
Spectroscopic determination of the P700 content of wild-type and ΔbfrAΔbfrB mutant strains. The reduced minus oxidized (ascorbate minus ferricyanide) absorption spectra were measured for thylakoid membranes isolated from the wild type and ΔbfrAΔbfrB mutant. The absorption at 701 nm was used for calculating the ratio of P700 to chlorophyll (Chl). Wild type and ΔbfrAΔbfrB were harvested at 9.1 × 108 and 9.4 × 108 cells/mL; 19.6 and 20.3 μg/mL Chl, respectively. An equal amount of Chl was used in both spectra presented (20 μgr Chl/mL). Solid line, wild type; dashed line, ΔbfrAΔbfrB. The vertical gray line marks the absorption maxima for P700. The experiment was repeated on membranes from two cultures for every strain. Each measurement was repeated twice. On average, P700 content in the mutant strain was 75% ± 5% of the wild-type strain.
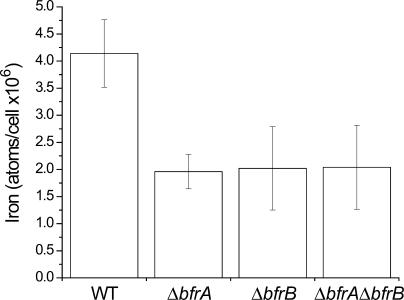
Direct measurements of the intracellular iron content using atomic absorption spectrometry revealed that in early exponential phase cells, the iron content in all three mutant strains was approximately 70% of that in wild-type cells (wild-type content: 9.3 × 106 ± 7.5 × 105 atoms/cell). Synechocystis 6803 cells are extremely efficient accumulators of iron. The amount of iron detected in these cells was approximately two orders of magnitude larger than that reported for Escherichia coli cells (Finney and O'Halloran, 2003). In early exponential phase cells, no iron stress could be detected (Table I). As the density of the cells increased, the stored iron in the cells was utilized and its concentration decreased (Fig. 6). By late exponential phase, the internal iron concentration of wild-type cells was twice as much as that in each of the three mutant strains (Fig. 6). Under these conditions, iron stress response was evident in all three mutant strains (Fig. 4).
Figure 6.
Iron content of wild-type and bfr mutant cells. Concentration of iron was measured in cells grown in BG11 medium. Late exponential phase cells were harvested at 6 to 22 × 108 cells/mL and EDTA treated to remove external iron. Error bars, sd, based on two experiments, each measured in three atomic absorption replicates. In a paired t test between wild type and each of the mutants, wild-type values were found to be significantly higher, with a confidence level P < 0.05.
The large amount of iron accumulated from fresh media during early growth stages of the culture is used later as the culture becomes dense and iron limiting. This process is, in many respects, similar to that described for manganese accumulation in Synechocystis 6803 cells (Keren et al., 2002). Mn2+ accumulates externally to the inner membrane and can be removed by an EDTA wash. The accumulation of an EDTA-washable fraction of iron could not be tested because the iron in the media sedimented when cells were centrifuged. Therefore, it was not possible to distinguish bound iron, removed by EDTA, from the free iron in the medium.
DISCUSSION
The data presented in this article reveal the significance of Bfrs in iron homeostasis in cyanobacteria. While in a number of nonphotosynthetic bacteria, no direct role for Bfrs in iron storage could be found (Andrews et al., 2003), we show here that in Synechocystis 6803, roughly 50% of the intracellular iron is accumulated in association with Bfrs. The reduced iron content, the impaired growth on media containing low iron concentrations, and the induction of CP43′ synthesis were observed in the two single deletion strains and, to the same extent, in the double deletion strain.
The cellular iron quota affects the photosynthetic apparatus directly. In late exponential phase wild-type cells, roughly one-fourth of the iron is in PSI complexes. In late exponential phase ΔbfrAΔbfrB cells that contain only half the iron quota of wild-type cells, the PSI concentration was lower. However, in this case the iron in PSI represents a much larger fraction of the intracellular iron pool, and therefore much less iron is available for nonphotosynthetic processes. The limiting iron quota of the mutants is reflected in the induction of the CP43′ antenna complex. While the exact role of CP43′ is still under investigation, spectroscopic data suggest that it can transfer excitation energy efficiently to PSI (Melkozernov et al., 2003) and compensate for the decreased PSI to PSII ratio.
The induction of CP43′ synthesis, as well as other iron stress-responsive proteins, has been studied in detail and is known to be regulated by the Fur family of transcriptional regulators (Ghassemian and Straus, 1996). However, it is important to note that in the case of the Bfr mutants, iron stress response was triggered when iron was abundant in the medium, conditions under which wild-type cells exhibited no sign of stress. Therefore, the sensing of iron concentration in these cells takes place internally, regardless of external iron concentrations. It is possible that an iron-loaded Bfr complex serves as a signal reporting the iron status inside the cell.
Why would the two Bfr proteins be necessary for iron accumulation? Comparing the sequences of the two cyanobacterial proteins to that of E. coli, Bfr reveals striking differences (Supplemental Fig. 2). While E. coli Bfr contains all of the necessary residues to form an active di-iron-binding site as well as a heme-binding site, the cyanobacterial BfrB protein is missing most of the residues required for di-iron cluster formation. Most notably, the conserved Glu-51 residue in the E. coli sequence is replaced by an Ala in BfrB. Therefore, it is unlikely that a BfrB dimer would have a functional di-iron cluster. BfrA, on the other hand, retains all of the conserved di-iron ligands, but is missing the Met residue at position 52 that serves as a heme ligand in E. coli Bfr. A BfrA-BfrB heterodimer would be able to bind heme and also contain active di-iron centers. Therefore, both proteins would be necessary in order to bind an active di-iron center as well as heme, as suggested by Andrews (1998). As a consequence, the absence of either of them is expected to result in a nonfunctional Bfr structure.
This hypothesis is strengthened by the observation that no assembled Bfr could be found in either of the single deletion strains. This, however, does not imply that both units are required in stoichiometric amounts or that the ratio of the two subunits is fixed.
Multiple bfr genes are not unique to Synechocystis 6803. Recent advents in cyanobacterial genome sequencing have enabled us to gain insights into iron-storage in a variety of cyanobacterial species by comparing their ferritin family protein sequences (Table II). In the ground-dwelling Gloeobacter violaceus PCC 7421 species, one bimodal bfr gene as well as a gene for a nonheme ferritin-like protein were found. Interestingly, the G. violaceus genome encodes two additional ferritins in which neither heme nor di-iron cluster-binding residues are conserved. G. violaceus lacks thylakoid membranes, and its photosynthetic apparatus resides in its plasma membrane. It is considered to be a member of an early branching lineage of cyanobacteria. A similar composition of ferritin family genes was found in Thermosynechococcus elongatus BP-1, a thermophilic cyanobacterium. In the nitrogen-fixing, filamentous Anabaena sp. PCC 7120 species, only nonheme-binding ferritin family genes were identified. However, in Synechococcus sp. PCC 7002 as well as in Prochlorococcus marinus MED4, two ferritin family genes with either heme- or di-iron center-binding conserved residues, can be found, a situation similar to that in Synechocystis 6803. Synechococcus 7002 represents a family of coastal cyanobacteria, while P. marinus MED4 represents a family of open ocean cyanobacteria that are major contributors to the primary productivity in the oceans (Rocap et al., 2003).
Table II.
Ferritin homologs in cyanobacterial species
| Organism | GenBank ORF Name | Heme-Binding Met | Di-Iron Binding |
|---|---|---|---|
| Synechocystis sp. PCC 6803 | Sll1341 (BfrA) | − | + |
| Slr1890 (BfrB) | + | − | |
| Synechococcus sp. PCC 7002 | BfrA | − | + |
| BfrB | + | − | |
| P. marinus MED4 | NP_892922 | + | − |
| NP_875274 | − | + | |
| T. elongatus BP-1 | NP_682531 | + | + |
| NP_681404 | − | + | |
| G. violaceus PCC 7421 | Gll3486Gll2819 | + | + |
| Gll0337 | − | + | |
| Glr2566 | − | − | |
| − | − | ||
| Anabaena sp. PCC 7120 | All0458 | − | + |
| Alr3808 | − | + | |
| All4145 | − | + | |
| All1173 | − | + |
The presence of genes encoding ferritin family proteins in the genomes of a number of cyanobacterial species was examined by scanning all of the translated ORFs with a PFAM for ferritin-like domain (see “Materials and Methods”). All ORFs that scored at E-values above 1 × 10−3 are shown. A multiple sequence alignment (Supplemental Fig. 1) of these proteins was generated, and the existence of conserved functional residues involved in heme or di-iron binding was assessed (Supplemental Table II).
What might be the adaptive advantage of having two different bfr genes? An answer may be found in the function of the heme cofactor. Andrews and co-workers (Andrews et al., 1995) have reported that an E. coli hemeless Bfr accumulates 4 times more iron than a Bfr that binds heme, in vitro. This suggests that while the di-iron center is needed for iron acquisition, the heme may be needed for iron extraction from the Bfr structure. In this case, splitting the iron input and output functions between two genes may form the basis for a mechanism for regulating iron homeostasis by changing the subunit composition in the Bfr ultrastructure. Efficient control of iron storage is essential for cyanobacterial proliferation in an environment in which available iron abundance is rare and short lived.
MATERIALS AND METHODS
Isolation and Characterization of Bfr
Bfr from Synechocystis 6803 cells was purified as described by Laulhere and co-workers with the following modifications (Laulhere et al., 1992). Cells from a 2.5-L culture (2 × 108 cell/mL) were broken with glass beads. The broken cells were incubated in 50 mm Tris/Malate, pH 7.5, 0.1% Triton X-100 and 1.1 g/mL CsCl for 30 min. Unbroken cells were discarded after centrifugation for 5 min at 16,000g. The broken cells were loaded on a cesium chloride step gradient (1.1 g/mL to 1.2 g/mL, each fraction 8 mL). Following centrifugation for 3 h at 350,000g, the pellet was resuspended in 50 mm Tris/Malate, pH 7.5, buffer containing 0.1% Triton X-100. The resuspended material was fractionated by gel filtration on a Sephacryl S-300 column (Amersham, Piscataway, NJ). The moving phase was 20 mm Tris/Malate, pH 7.5, buffer containing 0.1% Triton X-100, the flow rate was 1.5 mL/min, and the column dimensions were 55 cm length and 26 mm i.d. The iron concentrations in the fractions were determined by atomic absorption spectroscopy (see below). The fraction correlating to the peak in the iron profile was concentrated using a Biomax 30 K spin filter (Millipore, Bedford, MA) and fractionated on an SDS-PAGE gel (3 m Urea, 16%). Protein bands were excised and reduced with 10 mm dithiothreitol for 30 min, followed by alkylation with 55 mm iodoacetamide for 30 min. The treated bands were trypsin digested at 37°C for 12 h (6 ng/mL trypsin, 50 mm NH4HCO3). Digested peptides were eluted from the gel into a 1% formic acid, 2% acetonitrile solution and analyzed on a MALDI-TOF Voyager DE STR instrument (Applied Biosystems, Foster City, CA). The Protein Prospector software was used to identify hits in translated open reading frames (ORF) of Synechocystis 6803, with the maximum miss cleavage of 1 and mass tolerance of 35 ppm (mass spectrometric parameters are defined in http://prospector.ucsf.edu/).
Construction of Mutants
The region surrounding the sll1341 gene (bfrA) was amplified from genomic DNA (Fig. 2A; Supplemental Table I) and cloned into the Zero-Blunt TOPO plasmid (Invitrogen, Carlsbad, CA). A deletion of the gene was created by introducing a gentamycin resistance cassette between the two AvrII sites flanking the gene (Fig. 2A). The region surrounding slr1890 (bfrB) was amplified from genomic DNA (Fig. 2B; Supplemental Table I). Replacement of the gene with a chloramphenicol resistance cassette was achieved by a fusion PCR technique (Wang et al., 2002). The double Δsll1341Δslr1890 (ΔbfrAΔbfrB) mutant was generated by transforming the Δslr1890 strain with the Δsll1341 construct. Segregation was tested by genomic PCR (Fig. 2C).
Cell Growth and Sample Preparation
Cultures were grown in BG11 mineral medium (Allen, 1968) at 30°C with constant shaking. Illumination was at 30 μmol photons m−2 s−1. For growth experiments, BG11-Fe was prepared. No iron could be detected in BG11-Fe by atomic absorption spectroscopy (for details, see below). Considering that the lower sensitivity limit was 2 μg Fe/L, the iron concentration in BG11-Fe could not be higher then 36 nm. BG11 medium was prepared by supplementing BG11-Fe with 30 μm Fe(NO3)3. Prior to the experiment, cells were incubated in 20 mm MES, 10 mm EDTA, pH 5.0, for 15 min, spun down, and resuspended in BG11-Fe. This treatment is designed to remove insoluble iron from the media. In order to ensure that all of the extracellular iron is removed, cells (1.8 × 109 cells/mL) were subjected to two rounds of the wash procedure. No additional iron was released in the second round of washing.
Growth experiments were carried out in 12-well microtiter plates with constant shaking. Growth rate was determined by measuring absorption at 730 nm on a μQuant plate reader (Bio-Tek Instruments, Winooski, VT). Since centrifugations and the EDTA treatment inhibited growth for the first 24 h, sample collection started immediately after that period.
For iron concentration measurements, 5-mL samples were treated with the EDTA wash solution. The samples were digested in 80% nitric acid in a microwave oven (CEM, Matthews, NC). After digestion, the nitric acid was evaporated and the samples were reconstituted in a fresh 5% nitric acid solution.
Spectroscopy
Cell density and chlorophyll concentration were measured spectroscopically as previously described (Keren et al., 2002). P700 measurements were performed according to Sonoike and Katoh (1989). 77 K fluorescence spectra were obtained on a FluoroMax-2 fluorometer (Jobin Ivon, Longjumaeu, France). Metal concentrations were measured on an AA600 atomic absorption spectrometer (Perkin-Elmer, Ueberlingen, Germany).
Genomic Analysis
The genome sequences of a number of cyanobacterial species were examined for the presence of ferritin-like genes by scanning all of the translated ORFs in these genomes with the PFAM for the ferritin-like domain (PF00210; http://pfam.wustl.edu). The genome sequence of the Synechococcus sp. PCC 7002 genome is not finished, and the two Bfr genes that were included in the analysis were found on the contig gnl|jmarq_32049|Contig051302. To identify whether any ORF is predicted to bind a heme or a di-iron center, we generated a multiple sequence alignment (ClustalW 1.81; Chenna et al., 2003) that includes Bfrs whose crystal structures are available (E. coli-PDB identifier 1BFC, Rhodobacter capsulatus-1JGC, and Desulfovibrio desulfuricans-1NFV) to evaluate the secondary structures and thus determine whether the functional residues were in registration (Supplemental Tables II and III; Supplemental Fig. 1).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the accession numbers listed in Table II.
Supplementary Material
Acknowledgments
We thank Dr. Thomas J. Smith and his lab members for their kind assistance as well as Dr. Sixue Chen from the mass spectrometry facility at the Danforth Plant Science Center for the MALDI analysis.
This work was supported by grants from the U.S. Department of Energy, (USDA-NRI grant no. 00003348) and by the Department of Biology, Washington University, from funds provided by the Danforth Foundation. N.K. was partially supported by the International Human Frontier Science Program and The European Molecular Biology Organization.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042770.
References
- Allen MM (1968) Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Andrews SC (1998) Iron storage in bacteria. Adv Microb Physiol 40: 281–351 [DOI] [PubMed] [Google Scholar]
- Andrews SC, Le Brun NE, Barynin V, Thomson AJ, Moore GR, Guest JR, Harrison PM (1995) Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J Biol Chem 270: 23268–23274 [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237 [DOI] [PubMed] [Google Scholar]
- Bertani LE, Huang JS, Weir BA, Kirschvink JL (1997) Evidence for two types of subunits in the bacterioferritin of Magnetospirillum magnetotacticum. Gene 201: 31–36 [DOI] [PubMed] [Google Scholar]
- Bibby TS, Mary I, Nield J, Partensky F, Barber J (2003) Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424: 1051–1054 [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK, Kruip J (2001) A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412: 745–748 [DOI] [PubMed] [Google Scholar]
- Carrondo MA (2003) Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J 22: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Morse SA (1999) Neisseria gonorrhoeae bacterioferritin: structural heterogeneity, involvement in iron storage and protection against oxidative stress. Microbiology 145: 2967–2975 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Barber RT, Smetacek VV (1998) Biogeochemical controls and feedbacks on ocean primary production. Science 281: 200–207 [DOI] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV (2003) Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300: 931–936 [DOI] [PubMed] [Google Scholar]
- Gantt E (1994) Supramolecular membrane organization. In DA Bryant, ed, The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 119–138
- Ghassemian M, Straus NA (1996) Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology 142: 1469–1476 [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kakhlon O, Cabantchik ZI (2002) The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radic Biol Med 33: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Shen JR (2003) Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-A resolution. Proc Natl Acad Sci USA 100: 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Kidd MJ, Penner-Hahn JE, Pakrasi HB (2002) A light-dependent mechanism for massive accumulation of manganese in the photosynthetic bacterium Synechocystis sp. PCC 6803. Biochemistry 41: 15085–15092 [DOI] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, Cramer WA (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Laudenbach DE, Reith ME, Straus NA (1988) Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J Bacteriol 170: 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulhere JP, Laboure AM, Van Wuytswinkel O, Gagnon J, Briat JF (1992) Purification, characterization and function of bacterioferritin from the cyanobacterium Synechocystis P.C.C. 6803. Biochem J 281: 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkozernov AN, Bibby TS, Lin S, Barber J, Blankenship RE (2003) Time-resolved absorption and emission show that the CP43′ antenna ring of iron-stressed Synechocystis sp. PCC6803 is efficiently coupled to the photosystem I reaction center core. Biochemistry 42: 3893–3903 [DOI] [PubMed] [Google Scholar]
- Morel FM, Price NM (2003) The biogeochemical cycles of trace metals in the oceans. Science 300: 944–947 [DOI] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al (2003) Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424: 1042–1047 [DOI] [PubMed] [Google Scholar]
- Sherman DM, Sherman LA (1983) Effect of iron deficiency and iron restoration on ultrastructure of Anacystis nidulans. J Bacteriol 156: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA (2003) Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132: 1825–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K, Katoh S (1989) Simple estimation of the differential absorption coefficient of P-700 in detergent-treated preparations. Biochim Biophys Acta 976: 210–213 [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D (2003) An atypical haem in the cytochrome b6f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Vinnemeier J, Kunert A, Hagemann M (1998) Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol Lett 169: 323–330 [DOI] [PubMed] [Google Scholar]
- Wang HL, Postier BL, Burnap RL (2002) Polymerase chain reaction-based mutageneses identify key transporters belonging to multigene families involved in Na+ and pH homeostasis of Synechocystis sp. PCC 6803. Mol Microbiol 44: 1493–1506 [DOI] [PubMed] [Google Scholar]
- Yousef N, Pistorius EK, Michel KP (2003) Comparative analysis of idiA and isiA transcription under iron starvation and oxidative stress in Synechococcus elongatus PCC 7942 wild-type and selected mutants. Arch Microbiol 180: 471–483 [DOI] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.