Abstract
Salt cress (Thellungiella halophila), a halophyte, is a genetic model system with a small plant size, short life cycle, copious seed production, small genome size, and an efficient transformation. Its genes have a high sequence identity (90%–95% at cDNA level) to genes of its close relative, Arabidopsis. These qualities are advantageous not only in genetics but also in genomics, such as gene expression profiling using Arabidopsis cDNA microarrays. Although salt cress plants are salt tolerant and can grow in 500 mm NaCl medium, they do not have salt glands or other morphological alterations either before or after salt adaptation. This suggests that the salt tolerance in salt cress results from mechanisms that are similar to those operating in glycophytes. To elucidate the differences in the regulation of salt tolerance between salt cress and Arabidopsis, we analyzed the gene expression profiles in salt cress by using a full-length Arabidopsis cDNA microarray. In salt cress, only a few genes were induced by 250 mm NaCl stress in contrast to Arabidopsis. Notably a large number of known abiotic- and biotic-stress inducible genes, including Fe-SOD, P5CS, PDF1.2, AtNCED, P-protein, β-glucosidase, and SOS1, were expressed in salt cress at high levels even in the absence of stress. Under normal growing conditions, salt cress accumulated Pro at much higher levels than did Arabidopsis, and this corresponded to a higher expression of AtP5CS in salt cress, a key enzyme of Pro biosynthesis. Furthermore, salt cress was more tolerant to oxidative stress than Arabidopsis. Stress tolerance of salt cress may be due to constitutive overexpression of many genes that function in stress tolerance and that are stress inducible in Arabidopsis.
Abiotic stresses greatly affect plant growth and crop production. To survive against these stresses, plants respond and adapt with complex mechanisms, including developmental, morphological, physiological, and biochemical strategies. High salinity is a major abiotic stress causing both osmotic and ionic stress. The adaptive strategy to osmotic stress is the accumulation of osmoprotectants such as Pro, glycinebetaine, mannitol, and the raffinose family oligosaccharides. This strategy has been shown to be important in improving stress tolerance in plants by manipulating genes encoding key enzymes of the osmoprotectant synthesis or degradation pathway (Tarczynski et al., 1993; Kavi-Kishor et al., 1995; Thomas et al., 1995; Nanjo et al., 1999a, 1999b; Taji et al., 2002). A high level of Na+ is toxic to plants because it disturbs cytoplasmic K+/Na+ homeostasis. To prevent Na+ accumulation in the cytoplasm, plants use the following three strategies: reducing Na+ influx, vascuolar compartmentation of Na+, and excretion of Na+ via plasma membrane Na+/H+ antiporters (Ward et al., 2003). Overexpression of the vacuolar Na+/H+ antiporter NHX1 has been shown to confer salt tolerance in Arabidopsis and tomato, suggesting the utility of this vascuolar compartmentation of Na+ (Apse et al., 1999; Zhang and Blumwald, 2001). Also, overexpression of a plasma membrane Na+/H+ antiporter gene in a freshwater cyanobacterium conferred extreme salt tolerance and allowed growth in seawater, indicating the effectiveness of excreting Na+ via plasma membrane Na+/H+ antiporters for salt tolerance (Waditee et al., 2002). Furthermore, very recently, overexpression of SOS1, a plasma membrane Na+/H+ antiporter, was shown to confer salt tolerance by retrieving Na+ from the xylem of transgenic plants (Shi et al., 2003).
Although there have been many reports on salt tolerance, most of the studies have been on a typical glycophyte, Arabidopsis, and not on halophytes. This is because the technological advantages of this model plant are too significant, and halophytic plants have not been shown to be a suitable genetic and genomic model system. Recently, the halophytic plant species salt cress (Thellungiella halophila) was reported to grow in high salinity coastal areas in eastern China. Salt cress is thought to be a good model plant for the analysis of high salinity tolerance because it is closely related to Arabidopsis and has good genetic features such as similar morphology, small size, short life cycle, high seed number, and the ability to self-pollinate. Furthermore, salt cress has a gene composition with >90% nucleotide identity with that of Arabidopsis, and it can be transformed in planta according to the Arabidopsis protocol (Bressan et al., 2001; Zhu, 2001).
Although Arabidopsis is a typical glycophyte that is not particularly salt tolerant, a number of recent studies suggest that it may contain most, if not all, of the salt tolerance genes that one might find in halophytes (Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2000; Shinozaki et al., 2003). It is now hypothesized that a substantial percentage of halophytes use the mechanisms of salt tolerance found in glycophytes and that subtle differences in regulation result in large variations in tolerance or sensitivity between glycophytes and halophytes. This hypothesis can be tested by applying comparative genomics using microarray for genome-scale gene expression. Fortunately, since expressed sequence tag (EST) analyses of several hundred salt cress clones revealed 90% to 95% identities between salt cress and Arabidopsis cDNA sequences (Bressan et al., 2001), we assumed that a full-length cDNA microarray composed of 7,000 Arabidopsis genes could be applied in this case (Seki et al., 2002b). Recently, a microarray containing 8,987 cDNAs of rice EST was used to analyze the gene expression profile in barley roots during Fe-deficiency stress (Negishi et al., 2002). Genes of cereal crops tend to be highly conserved at the DNA sequence level (Devos and Gale, 2000), and this conservation allows the use of heterologous probes to identify orthologous DNA sequences among different species in DNA hybridization experiments.
In this study, we applied the full-length Arabidopsis cDNA microarray to reveal the differences in the regulation of salt tolerance mechanisms between a glycophyte, Arabidopsis, and a halophyte, salt cress. Furthermore, we compared the expression profiling data of salt cress with our previous Arabidopsis expression profiling data of genes that included not only those obtained by various abiotic stress treatments but also those obtained by biotic stress-related treatments using the same full-length Arabidopsis cDNA microarray. Salt stress tolerance of salt cress is discussed by comparison of Arabidopsis based on gene expression profiling of stress-inducible genes.
RESULTS
Salt Cress Is Extremely Salt Tolerant
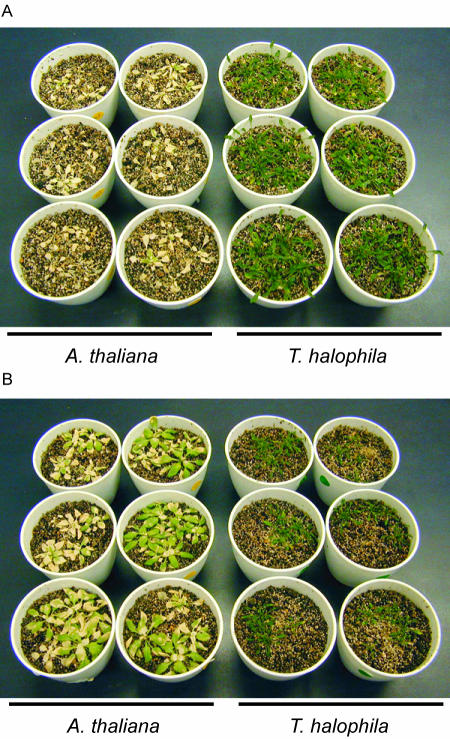
The germination rate of salt cress in the absence of 4°C stratification was very low and not uniform. Stratification treatment at 4°C for 1 week greatly improved the low germination rate. Growth of the salt cress seedling was slower than that of Arabidopsis. Thus, it was necessary to examine the salt stress tolerance of salt cress and Arabidopsis either with seedlings of the same size or at the same age. Three-week-old Arabidopsis and 4-week-old salt cress plants were exposed to 500 mm NaCl solution to investigate the salt tolerance with seedlings of the same size. The salt tolerance of seedlings at the same age was examined using 3-week-old Arabidopsis and the salt cress plants exposed to 500 mm NaCl solution. After 3 weeks of treatment, complete chlorosis was observed in all the Arabidopsis plants of the same size and same age. By comparison, the salt cress plants were not affected by either condition (Fig. 1, A and B).
Figure 1.
Survivability of salt cress and Arabidopsis under high-salt stress. A, High-salt stress tolerance of salt cress and Arabidopsis of the same size. Three-week-old Arabidopsis and 4-week-old salt cress plants that were the same in size were exposed to 500 mm NaCl solution for 3 weeks. B, High salt stress tolerance of salt cress and Arabidopsis of the same age. Three-week-old Arabidopsis and salt cress plants that were the same in size were exposed to 500 mm NaCl solution for 3 weeks.
Salt Tolerance in the Hydroponic Culture System and Differences in NaCl Uptake
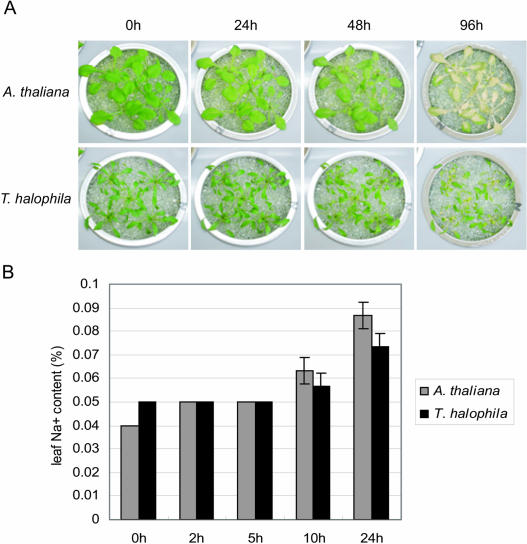
The demonstration of extreme salt tolerance of salt cress plants implies the existence of important mechanisms for salt tolerance in the root. It was necessary to gather not only the rosette leaves but also the roots. However, we could not harvest the root of the soil-grown Arabidopsis and salt cress plants. So we adopted the hydroponic culture system with glass beads (Nanjo et al., 1999a). This method enabled us to collect the roots easily and to change the salinity condition of the growth media reversibly. To check the salt tolerance on the system, we exposed 3-week-old Arabidopsis and 4-week-old salt cress plants of the same size to 250 mm NaCl solution (Fig. 2A). After a 24-h treatment, salt cress plants were not affected by the salinity stress, whereas significant chlorosis was observed in Arabidopsis. After a 96-h treatment, Arabidopsis plants showed complete chlorosis, conversely one-half of the salt cress plants survived. Consequently, we also confirmed the salt tolerance of salt cress in the hydroponic system.
Figure 2.
Evaluation of salt tolerance and Na+ uptake in hydroponic culture system. A, High salinity stress tolerance in hydroponic culture system. Three-week-old Arabidopsis and 4-week-old salt cress plants that were the same in size were exposed to 250 mm NaCl solution for 24, 48, and 96 h. B, Accumulation of NaCl in leaves of Arabidopsis and salt cress during high salt stress in hydroponic culture system. Three-week-old Arabidopsis and 4-week-old salt cress plants that were the same size were exposed to 250 mm NaCl solution for 2, 5, 10, and 24 h.
To investigate the importance of salt cress roots in the tolerance to salt stress, we measured the accumulation of NaCl in the leaves of Arabidopsis and salt cress plants during high salinity stress in the hydroponic culture system. Three-week-old Arabidopsis and 4-week-old salt cress plants were exposed to 250 mm NaCl solution for 2, 5, 10, and 24 h. After exposure for 10 or 24 h, NaCl uptake of salt cress plants became slower than that of Arabidopsis (Fig. 2B). This suggested that salt cress roots possess special mechanisms to prevent inflow of NaCl.
Salt Stress-Inducible Genes in Salt Cress
To analyze the molecular mechanisms of salt tolerance in salt cress, we compared gene expression profiles in salt cress with those of Arabidopsis by using a full-length cDNA microarray (approximately 7,000 Arabidopsis genes). The Arabidopsis full-length cDNA microarray is expected to hybridize mRNA of salt cress more efficiently than Arabidopsis oligonucleotide microarray because the oligonucleotide microarray needs strict identity between mRNAs in comparison with the full-length cDNA microarray. In addition, since salt cress genes share >90% nucleotide identity with Arabidopsis genes, efficient cross-hybridization was expected.
To identify salt stress-inducible genes in salt cress and Arabidopsis, we treated plants with salt at 250 mm NaCl for 2 h. The reasons for using this condition are as follows: (1) genes rapidly inducible should be important for salt tolerance; (2) significant chlorosis is not observed within 2 h in control Arabidopsis; and (3) NaCl is taken up more efficiently at 10 h of salt treatment in Arabidopsis. The salt stress-inducible genes in salt cress and Arabidopsis were identified by microarray analyses that hybridized with Cy3 and Cy5 fluorescence-labeled probe pairs of salt-stressed plants plus unstressed plants. Subsequently, we investigated the differences in salt-inducible genes between salt cress and Arabidopsis by comparing their microarray data. Down-regulated genes could not be evaluated precisely because of lower hybridization signals. These genes might include not only down-regulated genes but also ortholog genes with low homology. Therefore, we concluded that the expression data obtained with heterologous microarray analysis can be used for the identification of up-regulation of genes.
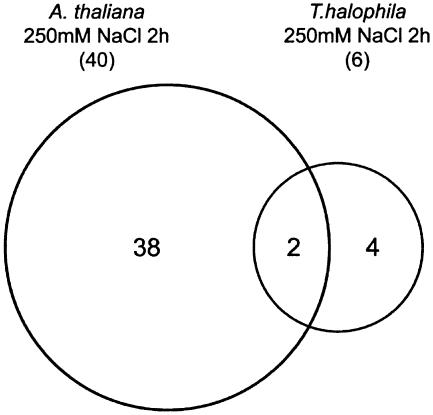
Tables I and II show the genes that are up-regulated (ratio = log2 Cy5/Cy3 ≥ 1.5) by salt stress with a signal value greater than 3,000 in salt cress and Arabidopsis, respectively. We identified only 6 salt stress-inducible genes whose log2 ratios of salt stress versus nonstress were ≥1.5 in salt cress, whereas we showed 40 such salt stress-inducible genes in Arabidopsis. Figure 3 shows the classification of salt stress-inducible genes in salt cress and Arabidopsis. These genes were categorized into three groups: (1) genes induced in Arabidopsis; (2) genes induced in salt cress; and (3) genes induced in both Arabidopsis and salt cress. The genes induced by salt stress in salt cress include myoinositol-1-phosphate synthase (INPS) gene. In the halophyte common ice plant (Mesembryanthemum crystallinum), known to accumulate methyl inositol, d-ononitol, and d-pinitol, which serves as an osmoprotectant, INPS RNA levels are up-regulated and levels of free myoinositol accumulate during salinity stress (Ishitani et al., 1996). By contrast, Arabidopsis does not show up-regulation of INPS or increased levels of myoinositol under salinity stress (Ishitani et al., 1996). Thus, salt cress may accumulate myoinositol, d-ononitol, and d-pinitol during salt stress as in the ice plant. Furthermore, galactinol synthase gene AtGolS2 is up-regulated in salt cress under salt stress. Galactinol synthase catalyzes the first step in the biosynthesis of the raffinose family oligosaccharides. The transgenic plants that overaccumulate galactinol and raffinose have improved tolerance to drought or freezing (Taji et al., 2002; Pennycooke et al., 2003). Myoinositol is also used as a substrate of raffinose. Consequently, salt cress may also accumulate more raffinose and other raffinose family oligosaccharides than Arabidopsis during salt stress.
Table I.
Genes up-regulated in salt cress under 250 mM NaCl stress for 2h
| Clone Namea | MIPSb | Genec | Log2 Ratiod | SDe |
|---|---|---|---|---|
| RAFL09-15-K07 | At4g39800 | Myoinositol-1-phosphate synthase | 2.48 | 0.13 |
| RAFL09-14-B02 | At2g22240 | Putative myoinositol 1-phosphate synthase | 2.09 | 0.09 |
| RAFL08-08-L20 | At1g56600 | Galactinol synthase, AtGolS2 | 1.72 | 0.32 |
| RAFL08-16-M12 | At2g33380 | Putative calcium-binding EF-hand protein, responsive to desiccation, RD20 | 1.68 | 0.65 |
| RAFL08-11-C23 | At5g06760 | Late embryogenesis abundant protein LEA-like | 1.67 | 0.15 |
| RAFL05-13-C23 | At5g53450 | Protein kinase family | 1.51 | 0.07 |
RIKEN Arabidopsis full-length (RAFL) cDNA.
MIPS protein code.
Indicates the putative functions of the gene products that are expected on the basis of sequence similarity.
Ratio = log2 Cy5/Cy3.
Standard deviation.
Table II.
Genes up-regulated in Arabidopsis under 250 mM NaCl stress for 2h
| Clone Namea | MIPSb | Genec | Log2 Ratiod | SDe |
|---|---|---|---|---|
| RAFL07-11-M21 | At5g52310 | Hydrophilic protein, responsive to desiccation, RD29A | 3.77 | 0.09 |
| RAFL08-16-M12 | At2g33380 | Putative calcium-binding EF-hand protein, responsive to desiccation, RD20 | 3.52 | 0.15 |
| RAFL05-21-C17 | At4g27410 | NAM-like protein and ATAF-like protein, responsive to desiccation, RD26 | 2.28 | 0.39 |
| RAFL04-12-G16 | At2g43570 | Putative endochitinase | 2.25 | 0.25 |
| RAFL05-12-M18 | At4g09010 | Ascorbate peroxidase, putative | 2.13 | 0.1 |
| RAFL08-12-H04 | At2g26690 | Putative nitrate transporter | 2.12 | 0.39 |
| RAFL05-20-O23 | At2g39800 | Putative Δ-1-pyrroline 5-carboxylase synthetase, AtP5CS1 | 2 | 0.29 |
| RAFL05-10-D11 | At1g73480 | Lysophospholipase homolog, putative | 1.96 | 0.15 |
| RAFL08-11-P07 | At5g17460 | Unknown protein | 1.95 | 0.14 |
| RAFL09-66-C10 | At3g57260 | β-1,3-glucanase 2, BG2 | 1.93 | 0.07 |
| RAFL05-21-F13 | At1g16850 | Unknown protein | 1.9 | 0.42 |
| RAFL05-21-C06 | At2g24850 | Putative tyrosine aminotransferase | 1.89 | 0.39 |
| RAFL05-05-G20 | At3g02480 | Unknown protein | 1.86 | 0.36 |
| RAFL05-14-E16 | At1g62570 | Similar to glutamate synthase | 1.84 | 0.23 |
| RAFL05-08-P17 | At1g20450 | Group II LEA protein, early responsive to desiccation, ERD10 | 1.84 | 0.08 |
| RAFL07-15-N09 | At4g04020 | Plastid-lipid associated protein PAP1 | 1.76 | 0.26 |
| RAFL06-09-E13 | NDf | Unknown protein | 1.76 | 0.14 |
| RAFL05-10-J09 | At1g78070 | Unknown protein | 1.73 | 0.03 |
| RAFL05-11-A18 | At4g37590 | Phototropic response protein family | 1.69 | 0.27 |
| RAFL05-16-I09 | At5g20830 | Suc-UDP glucosyltransferase | 1.67 | 0.3 |
| RAFL05-16-H23 | At4g28140 | AP2 domain transcription factor, putative | 1.66 | 0.24 |
| RAFL09-09-I16 | At5g13630 | Cobalamin biosynthesis protein | 1.65 | 0.04 |
| RAFL08-08-L20 | At1g56600 | Galactinol synthase, AtGolS2 | 1.63 | 0.81 |
| RAFL05-14-A12 | ND | Putative myosin heavy chain | 1.62 | 0.17 |
| RAFL08-11-H16 | At3g14440 | 9-cis-epoxycarotenoid dioxygenase, AtNCED3 | 1.62 | 0.34 |
| RAFL08-09-C23 | At1g54100 | Aldehyde dehydrogenase, putative | 1.6 | 0.05 |
| RAFL05-17-I08 | At2g43020 | Putative amine oxidase | 1.59 | 0.19 |
| RAFL07-07-G15 | At1g01720 | NAC domain protein, putative | 1.59 | 0.82 |
| RAFL04-16-P21 | At4g37370 | Cytochrome P450-like protein | 1.58 | 0.41 |
| RAFL05-11-I09 | At5g52300 | Hydrophilic protein, responsive to desiccation, RD29B | 1.58 | 0.66 |
| RAFL05-21-L12 | At3g22830 | Heat shock transcription factor-like protein | 1.57 | 0.53 |
| RAFL07-07-J02 | At4g24960 | ABA-induced like protein, AtHVA22d | 1.57 | 0.43 |
| RAFL05-19-I05 | At1g01720 | Strong similarity to OsNAC6 protein from Oryza sativa | 1.56 | 0.27 |
| RAFL06-07-B19 | At3g11410 | Protein phosphatase 2C, PP2C | 1.55 | 0.18 |
| RAFL05-19-O22 | ND | Unknown protein | 1.54 | 0.83 |
| RAFL09-06-L09 | At3g12580 | Heat shock protein 70 | 1.52 | 0.16 |
| RAFL05-03-A05 | At2g42540 | Cold-regulated protein, cor15a | 1.51 | 0.3 |
| RAFL05-15-H13 | At5g25610 | Responsive to desiccation, RD22 | 1.51 | 0.26 |
| RAFL05-14-J01 | At2g29450 | Glutathione S-transferase | 1.51 | 0.26 |
| RAFL06-12-H12 | At5g02020 | Unknown protein | 1.51 | 0.26 |
| RAFL05-15-A16 | At5g53120 | Spermidine synthase | 1.5 | 0.26 |
| RAFL05-15-E19 | At3g05640 | Putative protein phosphatase 2C, PP2C | 1.5 | 0.09 |
RIKEN Arabidopsis full-length (RAFL) cDNA.
MIPS protein code.
Indicates the putative functions of the gene products that are expected on the basis of sequence similarity.
Ratio = log2Cy5/Cy3.
Standard deviation.
ND, No data.
Figure 3.
Classification of salt stress-inducible genes in salt cress and Arabidopsis. The salt stress-inducible genes identified were categorized into three groups: (1) genes induced in Arabidopsis; (2) genes induced in salt cress; and (3) genes induced in both Arabidopsis and salt cress. The genes with expression log2 ratios (salt stressed/unstressed) greater than 1.5 times the average of the three experimental sets were regarded as salt stress-inducible genes.
Genes Up-Regulated in Salt Cress Compared with Arabidopsis under Normal Growth Conditions
To compare expression profiles under normal growth conditions between salt cress and Arabidopsis, we hybridized the full-length Arabidopsis cDNA microarrays with Cy3 and Cy5 fluorescence-labeled probe pairs of nonstressed salt cress plus nonstressed Arabidopsis plants. Interestingly, a number of abiotic or biotic stress-inducible genes were expressed under normal growth conditions in salt cress (Table III; Fig. 4). Especially, the tendency observed in the genes up-regulated to high levels, and five of the eight genes with log2 ratios greater than 2.5 are known to be important genes in abiotic stress or biotic stress tolerance. The up-regulated genes are Fe-superoxide dismutase (SOD); 9-cis-epoxycarotenoid dioxygenase (AtNCED2); chitinase; plant defensin1.2 (PDF1.2); Δ-1-pyrroline-5-carboxylate synthetase (AtP5CS); plasma membrane Na+/H+ antiporter (SOS1); P-protein associated with nitric oxide (NO) production; and β-glucosidase.
Table III.
Genes up-regulated in salt cress grown at normal conditions compared with Arabidopsis
| Clone Namea | MIPSb | Genec | Log2 Ratiod | sde | Groupf |
|---|---|---|---|---|---|
| RAFL02-01-K01 | At4g25100 | Superoxide dismutase, SODg | 4.10 | 0.45 | V |
| RAFL09-13-C16 | At1g32900 | Granule-bound starch synthase-like protein | 3.09 | 0.63 | V |
| RAFL08-17-C21 | At1g54010 | Myrosinase-associated protein | 2.89 | 0.25 | II |
| NDh | At4g19170 | 9-cis-epoxycarotenoid dioxygenase, AtNCED2g | 2.87 | 0.47 | V |
| RAFL04-12-G16 | At2g43570 | Putative chitinaseg | 2.79 | 0.13 | III |
| RAFL06-82-G15 | At5g44420 | Antifungal protein-like, PDF1.2g | 2.75 | 0.64 | III |
| RAFL04-12-I10 | At3g06660 | Hypothetical protein | 2.61 | 0.56 | IV |
| RAFL05-20-O23 | At2g39800 | Putative Δ-1-pyrroline 5-carboxylase synthetase, AtP5CS1g | 2.47 | 0.26 | I |
| RAFL07-13-P10 | At5g58540 | Ser/Thr-specific protein kinase NPK15, Nicotiana tabacum | 2.31 | 0.13 | IV |
| RAFL07-12-I18 | At2g45290 | Putative transketolase precursor | 2.27 | 0.24 | V |
| RAFL05-18-M07 | At4g02280 | Putative Suc synthetase | 2.24 | 0.46 | I |
| RAFL09-06-E16 | At2g26080 | Putative Gly dehydrogenase | 2.22 | 0.16 | V |
| RAFL04-15-P13 | At4g17260 | Lactate dehydrogenase, LDH1 | 2.18 | 0.1 | III |
| RAFL07-18-C20 | At2g21330 | Fructose bisphosphate aldolase-like protein | 2.16 | 0.09 | V |
| RAFL08-16-N03 | At1g09970 | Leu-rich repeat receptor-like kinase | 2.16 | 0.19 | III |
| RAFL07-10-O06 | At4g33010 | P-proteing | 2.16 | 0.31 | V |
| RAFL09-09-I16 | At5g13630 | Cobalamin biosynthesis protein | 2.08 | 0.21 | V |
| RAFL05-05-K17 | At3g03780 | Met synthase-like protein | 2.08 | 0.15 | II |
| RAFL07-13-A16 | At5g67030 | Zeaxanthin epoxidase | 2.02 | 0.13 | I |
| RAFL04-13-A11 | At2g29340 | Tropinone reductase-like protein | 2.02 | 0.19 | III |
| RAFL05-11-A02 | At4g14030 | Selenium-binding protein-like | 2.02 | 0.03 | III |
| RAFL08-18-C10 | At2g21330 | Fructose bisphosphate aldolase-like protein | 2.00 | 0.07 | V |
| RAFL04-15-M13 | At5g56030 | HEAT SHOCK PROTEIN 81-2, HSP81-2 | 1.96 | 0.11 | III |
| RAFL07-08-F20 | At3g24800 | E3 ubiquitin ligase, PRT1 | 1.96 | 0.64 | IV |
| RAFL05-17-G21 | At5g58330 | NADP-dependent malate dehydrogenase | 1.95 | 0.4 | V |
| RAFL06-09-C11 | At4g23190 | Ser/Thr kinase-like protein | 1.95 | 0.3 | III |
| RAFL09-15-F15 | At4g30190 | H+-transporting ATPase type 2, plasma membrane | 1.94 | 0.62 | II |
| RAFL11-07-C13 | At4g27470 | Putative RING zinc finger protein | 1.91 | 0.25 | IV |
| RAFL05-11-L07 | At3g27690 | Putative chlorophyll a- b-binding protein | 1.9 | 0.34 | V |
| RAFL09-12-L07 | At4g01950 | Predicted protein of unknown function | 1.9 | 0.17 | III |
| RAFL02-01-D07 | At3g14990 | 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative | 1.89 | 0.15 | III |
| RAFL09-17-L12 | At1g62180 | Putative adenosine-5′-phosphosulfate reductase | 1.88 | 0.55 | III |
| RAFL09-15-M21 | At5g17310 | UDP-glucose pyrophosphorylase | 1.86 | 0.16 | IV |
| RAFL09-06-L09 | At3g12580 | Heat shock protein 70 | 1.86 | 0.61 | III |
| RAFL05-18-L03 | At1g06430 | Cell division protease FtsH, putative | 1.86 | 0.25 | I |
| RAFL04-16-H01 | At1g14960 | Major latex protein, putative | 1.84 | 0.33 | V |
| RAFL05-21-D08 | At3g18490 | Putative chloroplast nucleoid DNA-binding protein | 1.81 | 0.4 | V |
| RAFL05-16-C09 | At3g54400 | Nucleoid DNA-binding-like protein | 1.81 | 0.26 | II |
| RAFL08-13-D07 | At1g66280 | β-glucosidase, putativeg | 1.8 | 0.33 | II |
| RAFL09-06-M16 | At2g01980 | Na+/H+ antiporter, SOS1g | 1.78 | 0.61 | III |
| RAFL09-07-L16 | At2g39210 | Nodulin-like protein | 1.78 | 0.5 | III |
| RAFL08-18-C03 | At5g63640 | Unknown protein | 1.77 | 0.82 | III |
| RAFL04-09-L18 | At3g60900 | GPI-anchored protein, Fla10 | 1.77 | 0.29 | V |
| RAFL05-09-D06 | At5g13490 | Adenosine nucleotide translocator | 1.77 | 0.37 | III |
| RAFL04-16-G14 | At1g44446 | Chlorophyll a oxygenase | 1.77 | 0.6 | V |
| RAFL07-12-M09 | At2g21330 | Fructose bisphosphate aldolase-like protein | 1.76 | 0.21 | V |
| RAFL09-13-P07 | At3g53130 | Cytochrome P450-like protein | 1.75 | 0.33 | V |
| RAFL07-15-N09 | At4g04020 | Putative fibrillin | 1.75 | 0.17 | I |
| RAFL09-13-N23 | At2g21410 | Putative vacuolar proton-ATPase subunit | 1.71 | 0.4 | III |
| RAFL06-08-O12 | At5g15780 | Pro-rich protein | 1.69 | 0.28 | V |
| RAFL09-10-F11 | At5g13630 | Cobalamin biosynthesis protein | 1.69 | 0.45 | V |
| RAFL11-03-D07 | At5g56000 | Heat shock protein | 1.67 | 0.35 | III |
| RAFL11-11-M23 | At4g12470 | PEARLI 1-like protein | 1.66 | 0.42 | II |
| RAFL09-06-A02 | At3g59290 | Epsin-like protein | 1.66 | 0.38 | III |
| RAFL09-07-O19 | At1g58270 | Unknown protein | 1.66 | 0.57 | I |
| RAFL08-13-B17 | At1g53280 | 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative | 1.66 | 0.2 | III |
| RAFL09-09-M16 | At1g53310 | Putative phosphoenolpyrovate carboxylase | 1.64 | 0.47 | III |
| RAFL04-10-D23 | At5g23660 | MtN3-like protein | 1.62 | 0.05 | III |
| RAFL04-20-P19 | At3g49110 | Peroxidase | 1.61 | 0.01 | III |
| RAFL09-12-F24 | At5g64940 | ABC transporter-like | 1.61 | 0.54 | V |
| RAFL09-06-M11 | At2g32950 | Photomorphogenesis repressor, COP1 | 1.6 | 0.57 | IV |
| RAFL09-16-J07 | AT5g06600 | Ubiquitin carboxyl-terminal hydrolase | 1.59 | 0.46 | III |
| RAFL08-17-N09 | At3g52640 | Unknown protein | 1.58 | 0.51 | IV |
| RAFL04-17-N08 | At1g63900 | Putative RING zinc finger protein | 1.58 | 0.08 | IV |
| RAFL09-10-N21 | At3g56200 | Amino acid transporter family | 1.58 | 0.42 | III |
| RAFL05-21-G06 | At2g18960 | Plasma membrane proton ATPase, PMA | 1.58 | 0.17 | III |
| RAFL08-16-K20 | AT4g21990 | 5′-adenylylsulfate reductase, APR3 | 1.57 | 0.6 | III |
| RAFL07-08-K14 | At3g09440 | Heat-shock protein, At-hsc70-3 | 1.56 | 0.05 | III |
| RAFL05-16-I09 | At5g20830 | Sucrose-UDP glucosyltransferase | 1.56 | 0.58 | I |
| RAFL09-13-F15 | At1g71270 | ARE1-like protein | 1.55 | 0.47 | IV |
| RAFL05-02-M09 | At3g61820 | Unknown protein | 1.54 | 0.12 | V |
| RAFL06-09-I16 | At5g16570 | Gln synthetase | 1.54 | 0.52 | II |
| RAFL11-13-K15 | At1g60470 | Galactinol synthase, AtGolS4 | 1.54 | 0.76 | I |
| RAFL04-20-A09 | At3g16370 | Putative APG protein | 1.54 | 0.11 | V |
| RAFL09-09-D18 | At3g17650 | Unknown protein | 1.53 | 0.15 | II |
| RAFL05-07-K16 | At4g28250 | Putative β-expansin/allergen protein | 1.53 | 0.38 | V |
| RAFL07-15-O10 | At2g39210 | Nodulin-like protein | 1.53 | 0.42 | III |
| RAFL09-14-N06 | At2g04030 | Heat shock-like protein | 1.52 | 0.18 | V |
| RAFL07-10-M10 | At4g11420 | Initiation factor 3a-like | 1.52 | 0.37 | III |
| RAFL08-18-M13 | At2g38110 | Phospholipid/glycerol acyltransferase family | 1.51 | 0.75 | III |
| RAFL04-19-A22 | At5g08650 | GTP-binding protein LepA homolog | 1.5 | 0.33 | V |
| RAFL05-11-D11 | At1g55490 | Rubisco subunit binding-protein β subunit | 1.5 | 0.11 | V |
| RAFL08-10-G22 | At4g02280 | Putative sucrose synthetase | 1.5 | 0.76 | I |
RIKEN Arabidopsis full-length (RAFL) cDNA.
MIPS protein code.
Indicates the putative functions of the gene products that are expected on the basis of sequence similarity.
Ratio = log2Cy5/Cy3.
Standard deviation.
Five groups: I: abiotic stress inducible genes, II: biotic stress inducible genes, III: both abiotic and biotic stress inducible genes, IV: abiotic and biotic stress non-inducible genes, V: abiotic and biotic stress supressible genes (see Fig. 4).
genes discussed in manuscript.
ND, No data.
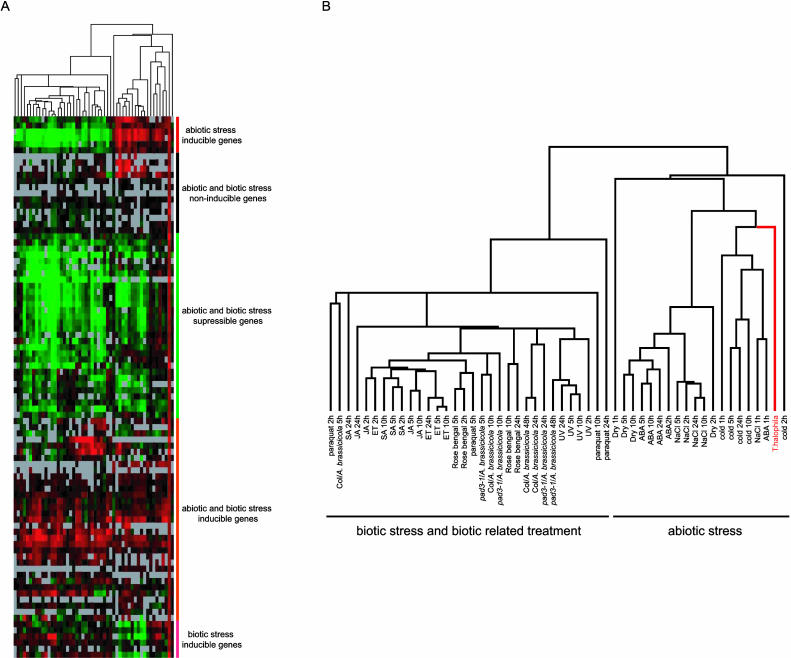
Figure 4.
Hierarchical clustering among the genes highly expressed in salt cress and the results of microarray analyses that included various abiotic stress treatments or various biotic stress and biotic-related treatments of Arabidopsis using this Arabidopsis cDNA microarray. A, Overview of the hierarchical cluster display. The fold change values for each sample, relative to untreated control samples, were log2 transformed and subjected to complete linkage hierarchical clustering. Expression values higher and lower than those of the control are shown in red and green, respectively. As absolute value of fold difference increased, the color intensity increased. The bars at the right end indicate five rough groups: abiotic, biotic, both abiotic and biotic stress-inducible genes, abiotic and biotic stress-suppressible genes, and abiotic and biotic stress-noninducible genes. B, The expansion figure of A. The relationship among experiments across all of the genes included in the cluster analysis and the type of each experiment are indicated (for details, see Seki et al., 2002a, 2002b; Narusaka et al., 2003).
We have performed many kinds of microarray analyses to analyze gene expression profiles in response to various abiotic, biotic stress, and oxidative stress in Arabidopsis using this full-length Arabidopsis cDNA microarray (Seki et al., 2002a, 2002b; Narusaka et al., 2003). Using the database of those microarray analyses, we analyzed the expression of the genes in Table III that were highly expressed (ratio; ≥1.5) in salt cress under normal growth conditions with Arabidopsis genes induced by various abiotic, biotic, and oxidative stress treatments. The abiotic stresses include drought, salt (NaCl), cold, and abscisic acid (ABA) treatment. The biotic and oxidative stress treatments include inoculation with a fungal pathogen, Alternaria brassicicola, to wild-type Arabidopsis (Columbia) and pad3-1 mutant (Glazebrook and Ausubel, 1994), signal molecules (salicylic acid [SA], jasmonic acid [JA], and ethylene [ET]), reactive oxygen species-inducing compounds (paraquat and rose Bengal), and UV-C treatment, which causes cell death (Narusaka et al., 2003). Figure 4 shows the results of cluster analysis of the resultant expression profiles. The expression profiles include five groups, i.e. abiotic stress-inducible genes, biotic stress-inducible genes, both abiotic and biotic inducible genes, abiotic and biotic noninducible genes, and abiotic and biotic suppressible genes (Table III). Interestingly, one-half of the genes belonged to the groups that were abiotic, biotic, or both abiotic and biotic stress-inducible genes in Arabidopsis (Fig. 4, A and B). The fact that many stress-inducible genes were expressed under normal growth conditions in salt cress might account for the relatively fewer salt stress-inducible genes compared to those of Arabidopsis with similar ratios.
Verification of Expression Profiles by RNA Gel-Blot Analysis
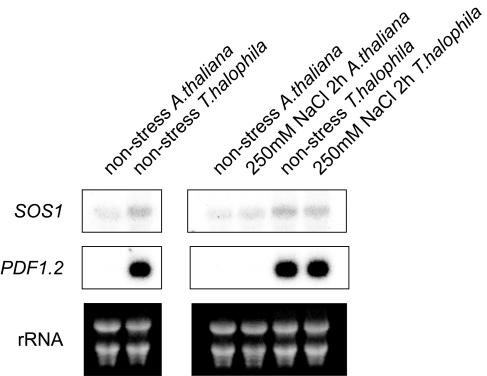
We performed RNA gel-blot analysis on expression profiles of SOS1 and PDF1.2 genes to confirm the validity of the microarray analyses (Fig. 5). The expression levels of SOS1 and PDF1.2 in salt cress were higher than those of Arabidopsis by RNA gel-blot analysis as well as the microarray analysis. Especially, the expression level of PDF1.2 was very high in salt cress under a normal growth condition or salinity stress, whereas that in Arabidopsis was not detectable under these conditions. Therefore, the expression data obtained by microarray analysis were in good agreement with those obtained by RNA gel-blot analysis. When we performed RNA gel-blot analysis of other genes, such as P5CS using ThP5CS cDNA (AtP5CS ortholog in salt cress [accession no. BM985832]), the expression of P5CS was higher in salt cress. However, when we used Arabidopsis P5CS cDNA as a probe, we could not detect higher expression of P5CS in salt cress (data not shown). The disagreement between the microarray analysis and the RNA gel-blot analysis may have arisen because of the difference in their hybridization efficiency and gene family.
Figure 5.
RNA gel-blot analysis of AtP5CS, ThP5CS, SOS1, and PDF1.2 genes in salt cress and Arabidopsis. Total RNA was prepared from 3-week-old Arabidopsis and 4-week-old salt cress plants that were the same in size during nonstress or 250 mm NaCl stress using the microarray analyses. Each lane was loaded with 10 μg of total RNA. The RNA was fractionated on a 1% agarose gel, blotted onto a nylon membrane, and hybridized with 32P-labeled AtP5CS, ThP5CS, SOS1, and PDF1.2 DNA fragments as probes.
ABA and Pro Contents in Salt Cress
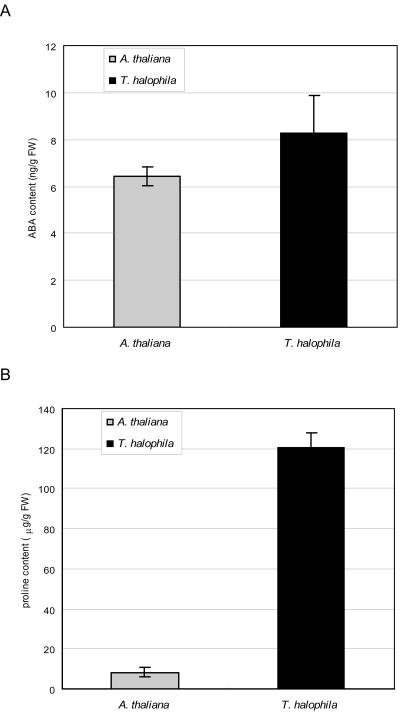
AtNCED2 and AtP5CS genes were expressed at higher levels in nonstressed salt cress. NCED and P5CS encode key enzymes of ABA and Pro biosynthesis, respectively. Thus, we measured the accumulation of endogenous levels of ABA and Pro in salt cress and Arabidopsis under normal growth conditions in a hydroponic culture. The ABA content of salt cress was slightly higher than that of Arabidopsis (Fig. 6A). The Pro content in salt cress was markedly higher than that of Arabidopsis during normal growth conditions in a hydroponic culture. Therefore, the extreme stress tolerance to high salinity characteristic of salt cress is due in part to the overaccumulation of Pro under unstressed conditions.
Figure 6.
ABA and Pro contents in salt cress and Arabidopsis. A, Accumulation of ABA in the entire plant of salt cress and Arabidopsis during normal growth conditions in hydroponic culture. Three-week-old Arabidopsis and 4-week-old salt cress plants grown under normal conditions were used for the analysis. B, Pro content in the entire plant of salt cress and Arabidopsis during normal growth conditions in hydroponic culture. Three-week-old Arabidopsis and 4-week-old salt cress plants grown under normal conditions were used for the analysis.
Salt Cress Is Tolerant to Oxidative Stress
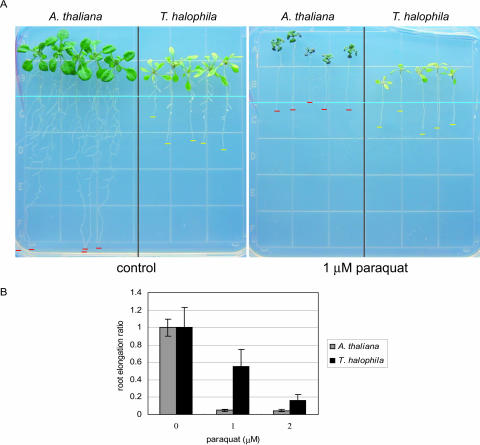
The SOD gene was expressed at much higher levels in nonstressed salt cress, suggesting that salt cress is tolerant to oxidative stress. SOD is one of the key enzymes in the protective system against oxidative stress since it catalyzes the dismutation of the superoxide radical to O2 and H2O2, a reaction that constitutes the first cellular defense against many oxidative stress situations. To analyze the oxidative tolerance in salt cress, we examined the paraquat tolerance of salt cress and Arabidopsis. Paraquat is known to cause the formation of a superoxide anion and a singlet oxygen (Lin and Culotta, 1995), hence producing oxidative stress in plant cells. Arabidopsis and salt cress plants were germinated and grown vertically on normal Murashige and Skoog (MS) agar medium without paraquat for 7 d and then transferred onto MS agar medium either with or without application of 1 μm paraquat. Without paraquat application, salt cress roots grew more slowly than Arabidopsis roots. However, in the presence of 1 μm paraquat for 7 d, the elongation of Arabidopsis roots stopped and the resultant seedling accumulated a large amount of anthocyanin, whereas the roots of salt cress continued elongation and the seedlings did not accumulate anthocyanin (Fig. 7, A and B). Therefore, compared to Arabidopsis, salt cress was more tolerant not only to high salinity but also to oxidative stress. Oxidative tolerance of salt cress may be in part due to the overexpression of SOD genes under normal growth conditions.
Figure 7.
Oxidative stress tolerance of salt cress. A, Root elongation assay with paraquat. Salt cress and Arabidopsis seedlings were grown on vertical MS plates containing 1.2% (w/v) agar and 3% Suc. One week after germination, the seedlings were transferred onto either the control MS agar plates (left) or MS agar plates containing 1 μm paraquat (right). The root tips of the seedlings were arranged uniformly along the light-blue line to ensure accurate measurements. Red and yellow bars show root ends of salt cress and Arabidopsis. The pictures were taken 10 d after the seedlings were transferred. B, Root elongation ratio in salt cress (black bar) and Arabidopsis (gray bar) during the exogenous paraquat treatment. The sd of 10 replicated measurements is indicated.
DISCUSSION
It is now hypothesized that the mechanisms of salt tolerance in halophytes are substantially the same as those known to exist in glycophytes and that subtle differences in regulation result in large variations in tolerance or sensitivity (Zhu, 2001). In this study, we investigated the strategy used for salt tolerance in a halophyte, salt cress, which is a close relative of Arabidopsis, by using the genomic information of Arabidopsis. Comparison of EST sequences from Arabidopsis and salt cress revealed a high DNA sequence identity (90%–95%) for the majority of transcripts, which indicates that the full-length cDNA microarray of Arabidopsis can be used for expression profiling of salt cress genes. As a result of microarray analyses, only 6 genes were shown to be strongly induced in response to high salinity stress in salt cress, whereas 40 genes were identified as salt stress-inducible genes in Arabidopsis (Tables I and II; Fig. 3). Many abiotic or biotic stress-inducible genes with various functions were expressed under normal growth conditions in salt cress than in Arabidopsis. This may be due to only six inducible genes detected in salt cress. Especially, the tendency observed in the genes up-regulated to high levels, and five of the eight genes with log2 ratios greater than 2.5 were known to be important genes in abiotic stress or biotic stress tolerance. As a result of hierarchical clustering among the genes highly expressed in salt cress without stress and the stress-inducible genes in Arabidopsis under different stress conditions, it was revealed that one-half of the genes highly expressed in salt cress belong to groups of genes that were abiotic, biotic, or abiotic and biotic stress-inducible in Arabidopsis (Fig. 4A). Furthermore, the expression profiles of genes highly expressed in salt cress under normal growth conditions resembled those of Arabidopsis genes induced under abiotic stresses rather than biotic stresses (Fig. 4B). However, some important genes involved in biotic stress tolerance were also observed in genes highly expressed in salt cress without stress. These observations suggest that salt cress is tolerant to biotic stresses as well as abiotic stresses because of constitutive overexpression of stress-related genes under unstressed conditions. These results indicate that salt cress does not immediately respond to a variety of environmental stresses, including high salinity at the transcriptional level, but instead adapted to them by expressing a number of genes that are known components of abiotic or biotic stress defense and survival machinery even in the absence of these stresses (Table III; Fig. 4). The genes overexpressed in the salt cress under normal conditions include Fe-SOD, P5CS, PDF1.2, AtNCED, P-protein, β-glucosidase, and SOS1 (see Table III). Possible functions of their gene products in stress tolerance of salt cress are discussed in the following.
Pro, the most common osmoprotectant, accumulates in many organisms, including higher plants exposed to environmental stresses such as high salinity, drought, and freezing. Some reports have even indicated a positive correlation between Pro accumulation and the acquisition of stress tolerance (Kavi Kishor et al., 1995; Nanjo et al., 1999a, 1999b). Since the Pro synthesis gene AtP5CS was up-regulated in salt cress under normal conditions, the Pro content of salt cress and Arabidopsis plants was measured under normal conditions to confirm the accuracy of the microarray analysis. Salt cress was found to accumulate significantly more Pro than Arabidopsis. The amount of Pro in salt cress was much higher than that in Arabidopsis under high salt stress, and equal to that in antisense transgenic Arabidopsis plants known to exhibit improved salt tolerance with an AtProDH cDNA encoding Pro dehydrogenase, which catalyzes Pro degradation (Nanjo et al., 1999a). These results suggest that the result of microarray analysis that AtP5CS was up-regulated is in good agreement with the phenomenon of excess accumulation of Pro. An external supply or extreme accumulation of Pro has also been reported to be toxic to plants in spite of its protective functions under stress (Hellmann et al., 2000; Deuschle et al., 2001; Mani et al., 2002; Nanjo et al., 2003). Growth of the salt cress seedling is slower than that of Arabidopsis. This phenomenon may be induced, in part, by the overaccumulation of Pro under normal growth conditions because a high level of Pro has a negative effect on plant growth (Nanjo et al., 2003).
The Fe-SOD gene was overexpressed in salt cress under normal conditions, and salt cress showed a higher tolerance to paraquat treatment (Fig. 6). SODs are essential components in almost all plant antioxidant defense mechanisms. The SOD isoenzymes can be classified into Cu/Zn, Mn, and Fe types according to their metal cofactor. Plants generally contain Fe-SOD and/or Cu/Zn-SOD in the chloroplasts (Holmberg and Bulow, 1998). In Arabidopsis ecotype Cvi, a new chloroplastic Cu/Zn-SOD isoenzyme, which was encoded by a Cvi-specific allele, has been identified. Paraquat treatments of Arabidopsis ecotype Landsberg erecta and Cvi resulted in higher levels of chloroplastic Cu/Zn-SOD activity in Arabidopsis Cvi, and the relative transcript levels were higher in Cvi under normal growth conditions. In addition, Arabidopsis Cvi showed a higher tolerance to paraquat treatments (Abarca et al., 2001). The mechanisms of oxidative stress tolerance in salt cress are similar to those of Arabidopsis Cvi. Expression of the Arabidopsis Fe-SOD gene protected both the plasmalemma and PSII against the superoxide generated by scavenging radicals during leaf discs impregnated with metal viologen (Van Camp et al., 1996). Besides the oxidative stress tolerance, it has been reported that transgenic alfalfa overexpressing Mn-SOD exhibited higher tolerance to freezing and water-deficit stresses. Thus, the extreme tolerance to salt stress of salt cress may be supported by the high level expression of the Fe-SOD gene in cooperation with P5CS.
With the acquisition of salt tolerance, there is no subsequent Na+ accumulation in the cytoplasm. Theoretically, there are three mechanisms: (1) reduction of Na+ influx; (2) vascuolar compartmentalization of Na+; and (3) excretion of Na+ via plasma membrane Na+/H+ antiporters. Overexpression of the vacuolar Na+/H+ antiporter NHX1 has been shown to confer salt tolerance in Arabidopsis and tomato, suggesting the utility of this vascuolar compartmentation of Na+ (Apse et al., 1999; Zhang and Blumwald, 2001). In addition, overexpression of a plasma membrane Na+/H+ antiporter gene in a freshwater cyanobacterium showed extreme salt tolerance and allowed growth in seawater, indicating the effectiveness of excretion of Na+ via plasma membrane Na+/H+ antiporters for salt tolerance (Waditee et al., 2002). We found that less NaCl is accumulated in salt cress plants than in Arabidopsis after 10 or 24 h of treatment (Fig. 2B), and the expression of the SOS1 gene in salt cress under normal conditions is higher than that of Arabidopsis (Table III). Recently, overexpression of SOS1 was shown to confer salt tolerance by retrieving Na+ from the xylem of transgenic plants (Shi et al., 2003). These results suggest that the higher expression of SOS1 may support excretion of Na+ from the xylem in salt cress, thereby limiting Na+ accumulation in the shoot in salt cress during high salt stress. The SOS1 transcript in the overexpressed transgenic lines is present at only a slightly higher level than in the wild type under normal growth conditions, whereas compared with the wild type the level is much higher under salt stress. This suggests that SOS1 mRNA is unstable in the absence of salt stress in most or all cells and that salt stress causes a posttranscriptional stabilization of the transcript (Shi et al., 2003). However, in salt cress, the SOS1 gene is expressed at high levels even in the absence of stress conditions. In salt cress, a number of abiotic or biotic stress-inducible genes were expressed under normal growth conditions (Table III). Therefore, the machinery for degrading the SOS1 transcripts in salt cress may be down-regulated in normal growth conditions.
Interestingly, in salt cress, not only abiotic but also biotic stress-inducible genes were overexpressed under normal growth conditions (Table III). Pathogenesis-related protein PDF1.2, used as a general marker gene for pathogenesis, chitinase, P-protein, and β-glucosidase were expressed at higher levels in salt cress. Furthermore, the comparison of the genes expressed at higher levels in salt cress with our previous microarray analyses of various biotic or biotic-related treatments (Narusaka et al., 2003) revealed a number of genes induced by biotic stress or oxidative stress treatments (Fig. 4). Plant defense responses to microbial attack are regulated through a complex network of signaling pathways that involve three signaling molecules: SA, JA, and ET. The SA and JA signaling pathways are mutually antagonistic (Kunkel and Brooks, 2002). PDF1.2, chitinase, and β-glucosidase function in the JA and ET signaling pathways, and their expressions are induced by JA and ET but not SA (Penninckx et al., 1998; Hara-Nishimura and Matsushima, 2003). Incidentally, the P-protein gene was also expressed at higher levels in salt cress. Very recently, Chandok et al. (2003) demonstrated that the pathogen-induced NO-synthesizing enzyme is a variant form of P-protein of Gly decarboxylase. After an attack by a pathogen, NO induces the expression of PR-1 through a SA signaling pathway (Wendehenne et al., 2001). However, the P-protein is not a SA-inducible gene, and PR genes are not expressed at a high level in salt cress. In addition, in the cluster analysis shown in Figure 4, the branch of salt cress is close to the branch of JA and ET rather than to that of SA. This suggests that JA and ET signaling might be up-regulated in salt cress under normal growth condition.
In conclusion, we showed that salt cress had not only salt stress tolerance but also oxidative stress tolerance. Based on expression profiling using the full-length Arabidopsis cDNA microarray, we showed that various genes induced by abiotic and biotic stresses in Arabidopsis are overexpressed in unstressed conditions in salt cress. This suggests that stress-inducible signaling pathways are constitutive and active in salt cress even under normal growth conditions without stress.
MATERIALS AND METHODS
Plant Material and Salt Stress Treatments for Soil-Grown Plants
Salt cress (Thellungiella halophila) and Arabidopsis L. Heynh., ecotype Columbia, were sown on MS plates containing 1.2% (w/v) agar and 3% Suc. The seeds were stratified at 4°C for 7 d and then transferred to 22°C under continuous light for germination and growth. Two or 3 weeks after germination, seedlings of Arabidopsis and salt cress were transferred onto separate 9-cm plastic pots filled with a 1:1 perlite:vermiculite and watered with 1,000-fold diluted Hyponex (Hyponex, Osaka). One week after transfer, the seedlings were transferred into a 1,000-fold diluted Hyponex solution containing 500 mm NaCl for the salinity stress treatment.
Salinity Treatments in Hydroponic System
In addition to the above method, two- or 3-week-old seedlings of Arabidopsis and salt cress were transferred onto tea strainers filled with glass beads with a hydroponic culture (1,000-fold diluted Hyponex). One week after transfer, the tea strainers containing the seedlings were transferred into 1,000-fold diluted Hyponex containing 250 mm NaCl for salinity stress.
Measurement of Na+ Content
Three-week-old Arabidopsis and 4-week-old salt cress plants grown in the hydroponic system were exposed to 250 mm NaCl solution for 2, 5, 10, and 24 h. We sampled only rosette leaves because this portion was immersed in the hydroponic culture. Na+ content was determined by converting the result of electrolyte leakage from leaves into the Na+. A total of 0.1 g leaves from each treatment group were immersed in 5 mL of sterile distilled water. The solution was measured for conductivity with a conductivity meter (DS-15; Horiba, Kyoto) after the sample was boiled for 14 min.
Measurement of Pro Content
Three-week-old Arabidopsis and 4-week-old salt cress plants grown in the hydroponic system were ground in distilled water. The homogenate was boiled for 6 min and then centrifuged at 10,000g for 15 min at 4°C. The supernatant was precipitated with 10% TCA for 5 h and centrifuged at 10,000g for 20 min. The supernatant after TCA precipitation was derivatized, and the contents of free Pro were determined by HPLC (model LC Module I plus; Waters, Milford, MA).
Measurement of ABA Content
Three-week-old Arabidopsis and 4-week-old salt cress plants grown in the hydroponic system were used as samples. Endogenous ABA was measured using exactly the same procedure as previously described (Iuchi et al., 2000).
RNA Isolation and RNA Gel-Blot Analysis
Total RNA was prepared using TRIZOL reagent (Life Technologies, Rockville, MD), and mRNA was prepared using an mRNA isolation kit (Miltenyi Biotec, Auburn, CA). Isolated total RNA was also used for RNA gel-blot analysis. Total RNA was fractionated in a 1% agarose gel containing formaldehyde and was blotted onto a nylon filter. The filters were hybridized with 32P-labeled fragments at 57°C, washed twice with 0.1× SSC, 0.1% SDS, at 60°C for 15 min, and autoradiographed.
Full-Length cDNA Microarray Analysis
Full-length cDNA microarray analysis was carried as reported previously (Seki et al., 2001, 2002b). In this study, we used a cDNA microarray containing approximately 7,000 Arabidopsis genes (Seki et al., 2002b). mRNAs from salt cress plants and Arabidopsis plants were used for preparation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed in equal amounts and hybridized with the cDNA microarray.
Data Analysis
Image analysis and signal quantification were performed with QuantArray version 2.0 (GSI Lumonics, Oxnard, CA). Background fluorescence was calculated on the basis of the fluorescence signal of the negative control genes, the mouse nicotinic acetylcholine receptor epsilon-subunit (nAChRE) gene and the mouse glucocorticoid receptor homolog gene. Intensity-dependent normalization (Yang et al., 2002) was used to equalize hybridization signals generated from different samples. Gene-clustering analysis was performed with Cluster/TreeView (Eisen et al., 1998). Average linkage clustering with uncentered correlation was used.
Acknowledgments
We thank Dr. Tokihiko Nanjo for discussion, Setsuko Kawamura for her excellent technical assistance, and Emily Smith-Katai for critical reading of the manuscript.
This work was supported in part by the Genome Research Project in RIKEN (to K.S.) and by the Special Postdoctoral Researchers' Program from RIKEN (to T.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039909.
References
- Abarca D, Roldan M, Martin M, Sabater B (2001) Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J Exp Bot 52: 1417–1425 [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK (2001) Learning from the Arabidopsis experience. Plant Physiol 127: 1354–1360 [PMC free article] [PubMed] [Google Scholar]
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 [DOI] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Hellmann H, Daschner K, Binder S, Frommer WB (2001) A nuclear gene encoding mitochondrial delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J 27: 345–356 [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD (2000) Genome relationships: the grass model in current research. Plant Cell 12: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsushima R (2003) A wound-inducible organelle derived from endoplasmic reticulum: a plant strategy against environmental stresses? Curr Opin Plant Biol 6: 583–588 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB (2000) Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol 122: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg N, Bulow L (1998) Improving stress tolerance in plants by gene transfer. Trends Plant Sci 3: 61–66 [Google Scholar]
- Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ (1996) Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J 9: 537–548 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi-Kishor PBK, Hong Z, Miao G-H, Hu C-AA, Verma DPS (1995) Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108: 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Culotta VC (1995) The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA 92: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S, Van De Cotte B, Van Montagu M, Verbruggen N (2002) Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol 128: 73–83 [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Fujita M, Seki M, Kato T, Tabata S, Shinozaki K (2003) Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol 44: 541–548 [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999. a) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461: 205–210 [DOI] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999. b) Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J 18: 185–193 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Ishida J, Nakashima M, Kamiya A, Enju A, Sakurai T, Satoh M, Kobayashi M, et al (2003) The cDNA microarray analysis using an Arabidopsis pad3 mutant reveals the expression profiles and classification of genes induced by Alternaria brassicicola attack. Plant Cell Physiol 44: 377–387 [DOI] [PubMed] [Google Scholar]
- Negishi T, Nakanishi H, Yazaki J, Kishimoto N, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, Kikuchi S, et al (2002) cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J 30: 83–94 [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycooke JC, Jones ML, Stushnoff C (2003) Down-regulating α-galactosidase enhances freezing tolerance in transgenic petunia. Plant Physiol 133: 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. a) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. b) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tarczynski MC, Jensen RG, Bohnert HJ (1993) Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259: 508–510 [DOI] [PubMed] [Google Scholar]
- Thomas JC, Sepahi M, Arendall B, Bohnert HJ (1995) Enhancement of seed germination in high salinity by engineering mannitol expression in Arabidopsis thaliana. Plant Cell Environ 18: 801–806 [Google Scholar]
- Van Camp W, Capiau K, Van Montagu M, Inze D, Slooten L (1996) Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol 112: 1703–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waditee R, Hibino T, Nakamura T, Incharoensakdi A, Takabe T (2002) Overexpression of a Na+/H+ antiporter confers salt tolerance on a freshwater cyanobacterium, making it capable of growth in sea water. Proc Natl Acad Sci USA 99: 4109–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Hirschi KD, Sze H (2003) Plants pass the salt. Trends Plant Sci 8: 200–201 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6: 177–183 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]