Abstract
Many cold-regulated genes of Arabidopsis are inducible by abscisic acid (ABA) as well as by cold. This has been thought to occur via two separate signaling pathways, with ABA acting via ABA-responsive promoter elements and low temperature activating the C-repeat element (CRT; dehydration-responsive) promoter element via CBF (DREB1) transcription factors. We show here that ABA is also capable of activating the CRT promoter element. Although the more recently discovered ABA-inducible CBF4 transcription factor might have accounted for this, we show here that CBF1-3 transcript levels also increase in response to elevated ABA levels. This increase in CBF1-3 transcript levels appears to be at least in part due to increased activity of the CBF promoters in response to ABA. A total of 125 bp of the CBF2 promoter, which has previously been shown to be sufficient for cold-, mechanical-, and cycloheximide-induced expression, was also sufficient for ABA-induced expression. However, the ABA-responsive promoter element-like motif within this region is not needed for ABA-induced expression. An observed increase in CBF protein levels after ABA treatment, together with previous data showing that increased CBF levels are sufficient for cold-regulated gene induction, suggests that ABA-induced increases in CBF1-3 transcript levels do have the potential to activate the CRT. Our data indicate therefore that activation of the CRT may also occur via a novel ABA-inducible signaling pathway using the normally cold-inducible CBFs.
The phytohormone abscisic acid (ABA) is involved in mediating responses to a number of environmental stresses including drought (Leung and Giraudat, 1998). Cellular ABA accumulates in response to such stresses, and increases in its concentration can lead to a number of physiological adaptations, including stomatal closure and growth inhibition, as well as up-regulation of specific genes. Many of the genes that are inducible by ABA are also expressed in response to cold and/or drought conditions; for example, the Arabidopsis cold-regulated (COR) genes RAB18, LTI78, and KIN2 (Kurkela and Franck, 1990; Lång and Palva, 1992; Nordin et al., 1993; Mäntylä et al., 1995). Several signaling pathways leading to COR gene expression have been described, including both ABA-dependent and ABA-independent pathways (Shinozaki and Yamaguchi-Shinozaki, 2000). The gene LTI78 (also known as COR78 or RD29A) has been studied as a paradigm system for a gene whose expression can be controlled by osmotic stress, low temperature, and ABA (Yamaguchi-Shinozaki and Shinozaki, 1994; Ishitani et al., 1997; Liu et al., 1998) via either ABA-dependent or ABA-independent routes (Shinozaki and Yamaguchi-Shinozaki, 2000).
Two cis-acting elements have been identified in the promoter of LTI78 and other COR genes that control expression under different stress conditions. The dehydration-responsive (DRE) element (Yamaguchi-Shinozaki and Shinozaki, 1994) has been shown to mediate both cold- and osmotic stress-inducible ABA-independent gene expression (Liu et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000). The same element is also referred to as the C-repeat element (CRT; Stockinger et al., 1997). The CRT/DRE element, an 8-bp motif, with a 5-nucleotide core element of CCGAC, may be activated by members of either the CBF or DREB2 families of transcription factors (TFs). The CBF family (also known as DREB1; Gilmour et al., 1998; Shinwari et al., 1998; Medina et al., 1999) cause COR gene activation under low temperature conditions (Stockinger et al., 1997; Liu et al., 1998), whereas the DREB2 family are involved in drought/osmotic stress induction through the same element (Liu et al., 1998). Constitutive overexpression of CBF TFs causes COR gene expression in the absence of a cold stimulus (Jaglo-Ottosen et al., 1998; Liu et al., 1998); however, overexpression of DREB2 is insufficient for induction of COR gene expression (Liu et al., 1998). These data indicate that in the case of DREB2 TFs, some posttranscriptional modification is required before activation of COR gene expression can proceed. Since the original identification of three very closely related members of the CBF gene family (CBF1-3 or DREB1A-C), three other CBF-related genes have been identified (Haake et al., 2002; Sakuma et al., 2002). Sakuma et al. (2002) have termed these DREB1D, E, and F. None of these three genes are inducible by cold although we have demonstrated that DREB1D (which we have described as CBF4) is inducible by ABA (Haake et al., 2002). For the sake of clarity, from hereon we use the term CBF to describe the cold-inducible CBF1-3 TF genes collectively and the term CBF4 (also known as DREB1D) to describe specifically the CBF1-like gene that is not cold inducible. We refer to the cis-element as the CRT/DRE.
Two ABA-inducible pathways leading to gene expression have been identified: one using a bZIP family of TFs (ABRE-binding factors or AREBs), which bind to the ABA-responsive elements (ABREs; Uno et al., 2000), and the MYC and MYB proteins, which interact with MYCR/MYBR elements (Abe et al., 1997, 2003). MYC and MYB recognition sequences do not occur in the LTI78 promoter but are found in other gene promoters, such as the gene RD22 (Yamaguchi-Shinozaki and Shinozaki, 1993; Abe et al., 2003).
The accumulation of ABA in response to osmotic stresses is thought to account for ABA-dependent drought-inducible expression of COR genes, via these ABA-response elements. However, although ABA levels rise transiently in response to low temperature, ABA does not continue to accumulate in response to cold. This observation has contributed to the belief that ABA does not play a role in low temperature signaling (Shinozaki and Yamaguchi-Shinozaki, 2000). The CBF signaling pathway leading to CRT/DRE activation has been described as an ABA-independent route leading to COR gene expression (Liu et al., 1998). By isolating expression via the CRT/DRE, we have now shown that this pathway can also be activated by ABA.
RESULTS
ABA Induces Expression via the CRT/DRE Promoter Element
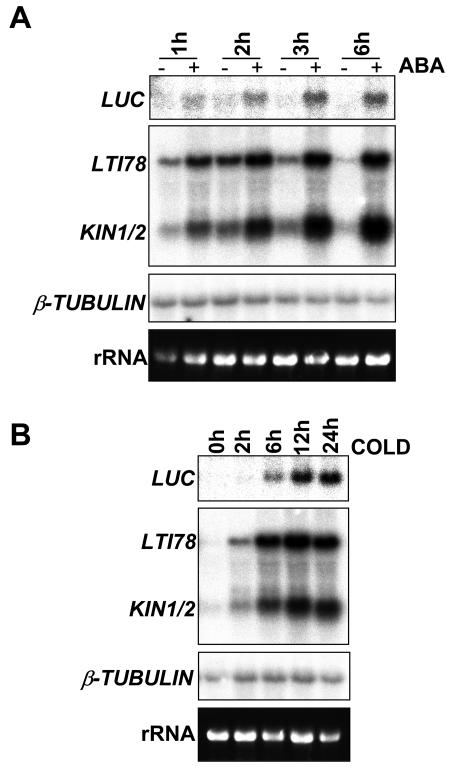
We measured the possible activation of COR gene expression via the CRT/DRE element by ABA, using Arabidopsis plants expressing an artificial promoter construct (Boyce et al., 2003) in which four copies of the CRT/DRE are linked to a minimal CaMV promoter driving expression of a firefly luciferase (LUC) gene. We have shown previously that this construct reports both cold-induced and osmotic-induced activation of expression via this element (Boyce et al., 2003). Seven-day-old seedlings were treated with 100 μm ABA or 0.1% ethanol (control) for 1, 2, 3, or 6 h or cooled to 5°C for 2, 6, 12, or 24 h. Northern analysis of LUC transcripts revealed that ABA caused an elevation of LUC transcripts compared with control samples in less than 1 h, and these levels reached a maximum at around 3 h of treatment (Fig. 1A). As we have observed previously, LUC expression continued to increase throughout a 24-h period in the cold (Fig. 1B). Cold- and ABA-induction of LUC was measured in samples loaded on and probed on the same membrane, so that quantitative comparisons between transcript levels could be made between treatments. When compared with the intensity of bands corresponding to cold-induced LUC transcript levels on the same northern blot, the maximal level of induction achieved with 100 μm ABA was greater than that achieved after 6 h cold induction. In this and subsequent northern analyses, rRNA stained with ethidium bromide was used to control for differences in the amounts of RNA loaded per lane. The transcript levels of the constitutively expressed gene β-TUBULIN gene (Knight et al., 1999) were also monitored on this blot to confirm that loading was equal between lanes in this gel.
Figure 1.
Induction of CRT::LUC and COR gene expression by ABA or low temperature treatment. Plants were treated for (A) 1, 2, 3, or 6 h with 100 μm ABA (+) or 0.1% ethanol control (−) or for (B) 0, 2, 6, 12, or 24 h at 5°C before harvesting tissue for northern analysis. Samples were loaded on the same gel and transferred to a filter, which was probed with a LUC probe and reprobed to detect levels of the COR genes LTI78 and KIN1/2. rRNA stained with ethidium bromide was used to compare loading between lanes and the blot was also reprobed with a β-TUBULIN probe to confirm equal loading between lanes.
We also examined in the same RNA samples, expression of LTI78 and KIN1/KIN2, three genes known to be inducible by cold and ABA (Kurkela and Borg Franck, 1992; Nordin et al., 1993). LTI78 transcript levels increased after 1 h in ABA and slightly thereafter up to 6 h (Fig. 1A), while in response to low temperature, transcript accumulated to similar levels after 6 h as seen after 6 h ABA, but continued to rise in response to further cold treatment, reaching maximum levels at around 12 h cold (Fig. 1B). KIN1/KIN2 was expressed to greater levels in response to ABA than in response to cold, and transcript levels continued to rise up to 6 h after the onset of ABA treatment.
ABA Induces Expression of CBF1-3
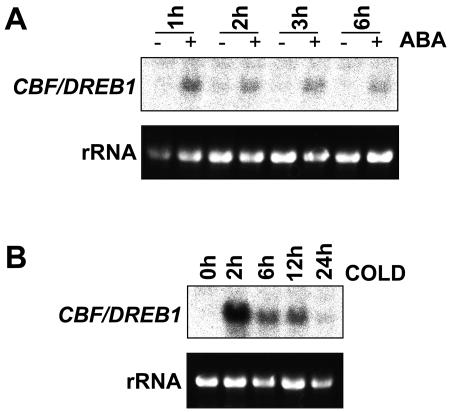
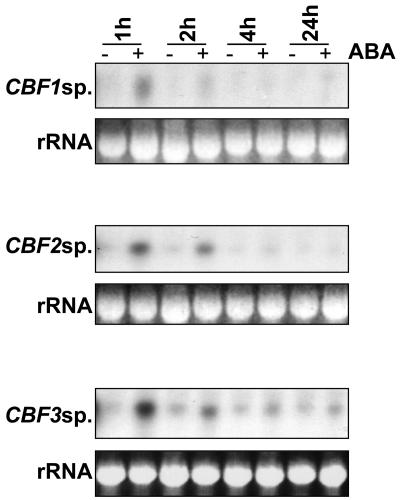
We made the observation that ABA increases CRT/DRE activation before we identified CBF4, a gene encoding an ABA-inducible TF with homology to CBF1-3 (Haake et al., 2002). Therefore, to account for our observation we examined expression of CBF1-3 in response to ABA (Figs. 2 and 3). We probed a replicate northern filter corresponding to the same cold- and ABA-treated samples as described above, using a generic probe to detect CBF1-3 transcripts. This probe corresponded to the entire CBF1 coding region and does not hybridize to the CBF4 transcript. Similarly to previous observations by ourselves and others (Gilmour et al., 1998; Shinwari et al., 1998), we observed that cold induction of CBF peaks early after the onset of cooling, with a large peak in transcript level occurring around 2 h (Fig. 2B). CBF transcript levels increased after 1 h treatment with ABA, declining after this time; however, at all time-points transcript levels were higher in ABA-treated samples than in the corresponding untreated controls (Fig. 2A). We also observed an increase in CBF transcripts in response to osmotic stress (data not shown), which could be as a result of endogenous ABA accumulation under these conditions. We next checked whether the increase in CBF expression was due to increased transcripts of only one, two, or all of the three CBF genes. Using gene specific probes (as described previously [Gilmour et al., 1998]), we measured levels of each of the three CBF transcripts (Fig. 3). All three transcripts were increased by ABA and peaked after 1 h ABA treatment and were clearly detectable after 2 h. Only CBF3 was still obviously elevated after 4 h.
Figure 2.
Induction of CBF expression by ABA or low temperature treatment. The filter used for Figure 1 was reprobed with a probe for CBF. A, Samples from plants treated for 1, 2, 3, or 6 h with 100 μm ABA (+) or 0.1% ethanol control (−). B, Samples from plants treated for 0, 2, 6, 12, or 24 h at 5°C before harvesting tissue for northern analysis. rRNA stained with ethidium bromide was used to compare loading between lanes.
Figure 3.
Analysis of ABA-inducibility of individual CBF genes. Samples were extracted from plants treated for 1, 2, 4, or 24 h with 100 μm ABA (+) or 0.1% ethanol control (−) and transcript levels of CBF1, CBF2, and CBF3 detected by northern blot using gene-specific probes. rRNA stained with ethidium bromide was used to compare loading between lanes.
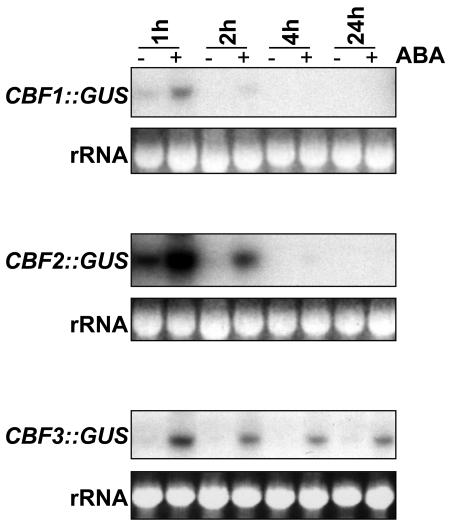
We decided to investigate further the increase in CBF expression caused by ABA treatment to ascertain whether the effect of ABA was simply to stabilize existing transcripts or to activate the CBF gene promoters and thus increase transcription itself. We measured β-glucuronidase (GUS) expression on a northern blot in response to ABA, using plants expressing a CBF promoter fusion with GUS for each of the three CBF genes in which the GUS coding region was fused to the full CBF promoter (Zarka et al., 2003). CBF-dependent GUS transcript levels for all three genes increased after 1 h in ABA, indicating that the addition of ABA did cause increased levels of CBF transcription (Fig. 4; the data shown in Figs. 3 and 4 correspond to the same RNA samples). As with the CBF3 native transcript, expression was prolonged up to 24 h in the case of CBF3::GUS. Similar results were obtained using a CBF3::LUC promoter fusion (data not shown), indicating that the results were independent of the reporter chosen.
Figure 4.
Activation of CBF1, 2, and 3 promoters in response to ABA. Samples were extracted from plants expressing CBF promoter GUS fusions and treated for 1, 2, 4, or 24 h with 100 μm ABA (+) or 0.1% ethanol control (−) and GUS transcript levels detected by northern blot. rRNA stained with ethidium bromide was used to compare loading between lanes.
Which Region of the CBF2 Promoter Confers ABA Inducibility?
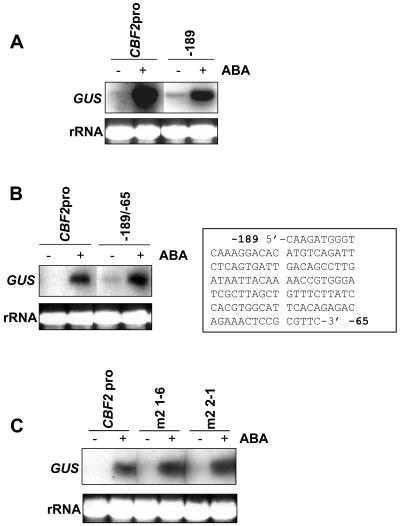
We previously identified a 125-bp region (−189 to −65) of the CBF2 promoter that is sufficient to impart cold-, mechanical-, and cycloheximide-induced gene expression (Zarka et al., 2003). These sequences are present within the promoter fragment designated −189 (Zarka et al., 2003); it contains the CBF2 promoter with a 5′ deletion to a point 189 bp upstream from the start of transcription fused to the GUS reporter gene. As shown in Figure 5A, the −189 promoter deletion was also responsive to ABA. In addition, the sequence between −189 and −65 could impart ABA induction when present as a dimer fused to a minimal CaMV 35S promoter::GUS reporter gene (Fig. 5B).
Figure 5.
Activation of the wild type and mutated −189 deleted CBF2 promoter in response to ABA. Samples were extracted from plants expressing the GUS gene fused to various full-length and deletion versions of the CBF2 promoter, in response to a 1.5-h treatment with with 100 μm (+) or 0.1% ethanol control (−) and GUS transcript levels detected by northern blot. rRNA stained with ethidium bromide was used to compare loading between lanes. A, Comparison of GUS expression driven by either the full-length CBF2 promoter (CBF2pro) or by a 5′ deletion of this promoter to −189 bp upstream of the start codon. B, Comparison of GUS expression driven by either the full-length promoter (CBF2pro) or the −189/−65 sequence from the CBF2 promoter, as a dimer fused to the CaMV minimal promoter (Zarka et al., 2003). The −189/−65 sequence is shown in the inset. C, Comparison of GUS expression driven by either the full-length CBF2 promoter (CBF2pro) or the mutated version of the −189 deletion construct (m2) in which a putative ABA-responsive element/bZIP binding site sequence CACGTG has been changed to TCTAGA. Two independent transgenic lines (m2 1-6 and m2 2-1) are shown.
Within the −189 region at position −139, is a potential ABA-responsive element/bZIP binding site (Busk and Pagès, 1998; Massari and Murre, 2000) with the sequence CACGTG (Zarka et al., 2003). To test whether ABA induction by the −189 deletion fragment required this sequence, we tested plants (independent lines m2 1-6 and 2-1) expressing a mutated version of the −189 promoter fragment in which the putative ABRE/bZIP element had been changed to TCTAGA (Zarka et al., 2003). The −189 promoter fragment with this mutation was still capable of imparting ABA induction at a level similar to that imparted by the wild-type −189 promoter fragment and the whole CBF2 promoter (Fig. 5C).
Increases in CBF Protein Levels in Response to Cold and ABA
Our data show a relatively small increase in steady-state CBF transcript levels in response to ABA. We therefore sought to discover whether ABA could actually have sufficient effect to alter the amount of CBF TF available to activate the CRT/DRE. To do this, we tested CBF protein accumulation.
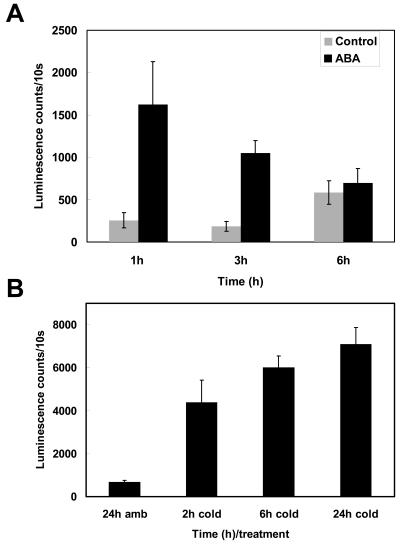
To date, we have been unable to raise antibodies to the CBF proteins of a sufficient titer for in vivo studies. This has also been the experience of other workers in this field (J. Salinas, K. Shinozaki, personal communication); therefore, we chose an alternative approach to determining CBF protein accumulation in Arabidopsis. We produced Columbia wild-type plants expressing a full-length CBF coding sequence driven by its native promoter and fused to an aequorin coding sequence in the C-terminal position as a tag for protein expression. This system has the advantage of being particularly sensitive to very small amounts of protein. We tested the potential of plants to accumulate this fusion protein, as a measure of their potential to accumulate native CBF protein in response to ABA. Figure 6 shows that CBF-AEQUORIN accumulated to significantly higher levels than controls after 1 h ABA treatment. After 3 h ABA treatment, although decreasing, levels were still significantly higher in ABA-treated plants (Fig. 6A). After 6 h exposure to ABA, the levels of CBF-AEQUORIN were not significantly greater than those in control treated plants (Fig. 6A). CBF-AEQUORIN levels were elevated in control treatments after 6 h, probably the result of prolonged incubation in liquid medium. We also demonstrated that as expected, CBF-AEQUORIN accumulated in response to cold treatment (Fig. 6B). The data shown are typical of those recorded for three independent transgenic lines expressing CBF-AEQUORIN.
Figure 6.
Accumulation of CBF-AEQUORIN fusion protein in response to cold and ABA. Seven-day-old plants were used, expressing a CBF-AEQUORIN fusion protein. Aequorin activity was measured by an in vitro assay. The values shown are means of three replicates from 5 plants/sample, after the subtraction of background luminescence. Five seedlings were harvested per replicate, and results are typical of those obtained with three different transgenic lines. Error bars represent SEM. Plants were treated for 1, 3, or 6 h with 100 μm ABA or 0.1% ethanol (control, A), or for 0, 2, 6, or 24 h at 5°C (B).
DISCUSSION
The phytohormone ABA is produced by plants under cold and drought stress conditions and plays an important role in mediating stress tolerance. ABA, cold, and osmotic stresses are known to induce expression of many of the same genes in Arabidopsis (Xiong et al., 2002), presumably because the encoded proteins that aid in combating cellular dehydration are necessary under both drought and freezing conditions. Many COR genes contain the CRT/DRE promoter element, which is cold- and osmotic stress-inducible (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997; Liu et al., 1998). Induction via this element involves the action of the CBF (DREB1; cold) and DREB2 (drought) families of TFs (Stockinger et al., 1997; Liu et al., 1998) and their homologs (Haake et al., 2002; Sakuma et al., 2002). Some COR genes also contain ABREs activated by ABRE-binding factors specific to ABA (Choi et al., 2000; Kang et al., 2002). The ABA-dependent and ABA-independent pathways leading to COR gene activation have been considered to exist entirely separately (Shinozaki and Yamaguchi-Shinozaki, 2000) although there has been some recent speculation that the CBF and DREB2 families of TF might in some way be activated by ABA (Xiong et al., 2002). We sought to test whether ABA induction of COR genes can occur via the CRT/DRE.
Under the conditions we used, ABA caused greater induction of KIN1/KIN2 and LTI78 than did cold treatment, even after 24 h (Fig. 1). It is well known that these COR genes are readily inducible by ABA; however, this has until now been assumed to occur solely through ABREs in the COR gene promoters. Monitoring expression of the CRT::LUC construct allowed us to examine the possible contribution to this level of COR gene expression attributable to the activation of the CRT/DRE specifically. Examination of the effect of ABA on the accumulation of LUC transcripts revealed that at least some of the ABA inducibility of the COR genes could occur via the CRT/DRE element. This is the first direct demonstration of ABA-dependent signaling through the CRT/DRE in Arabidopsis. However, in maize it is interesting to note that two DRE-like elements (DRE1 and DRE2), which are inducible by ABA, have been identified in the RAB17 gene promoter (Busk et al., 1997). ABA also up-regulates the transcript levels of DBF1 and DBF2, trans-acting factors that bind to DRE2 (Kizis and Pages, 2002).
We have previously shown that the sfr6 mutant of Arabidopsis, which is deficient in cold acclimation and COR gene expression, is unable to properly induce the CRT/DRE element in response to cold and osmotic stress (Knight et al., 1999; Boyce et al., 2003). Interestingly, ABA regulation of CRT/DRE-containing genes is also affected in sfr6 (Knight et al., 1999; Boyce et al., 2003), consistent with CRT/DRE being ABA-sensitive. Indeed, ABA induction of CRT::LUC expression is reduced in sfr6 (H. Knight and M.R. Knight, unpublished data).
In addition to CBF1-3, Sakuma et al. (2002) have identified three CBF-related genes encoding proteins that bind to the CRT/DRE. Although they found no evidence that any of these genes was ABA-inducible, we have shown that one of them, CBF4 (DREB1D), can be expressed in response to ABA (Haake et al., 2002). Our data in this paper suggested that ABA could act via one of the TFs that usually activate the CRT/DRE element. Therefore, we elected to test whether or not ABA could increase expression of the low temperature-inducible CBF1-3 TF family.
Our data showed that ABA does increase CBF transcript levels (Fig. 2), and increases in the levels of all three of the CBF transcripts monitored (CBF1-3) contribute to this elevation (Fig. 3). The increases in expression seen with each of the three promoter GUS fusions (Fig. 4) indicates that at least part of the increase in native transcript levels (Figs. 2 and 3) is due to an increase in transcription itself. The precise regulatory elements involved in this ABA-responsiveness remain to be determined. However, the 125-bp region between −189 and −65 of the CBF2 promoter, which was previously shown to be responsive to low temperature, mechanical agitation, and cycloheximide (Zarka et al., 2003), was found here to also impart ABA induction (Fig. 5). This region of the CBF2 promoter includes a potential ABRE/ZIP binding site (Busk and Pagès, 1998; Massari and Murre, 2000) at position −139, but mutational analysis indicated this sequence did not contribute significantly to the ability of the −189 deletion of the CBF2 promoter to impart ABA induction. However, initial analysis of ICEr1 and ICEr2 mutations in the 125-bp region are consistent with these sequences (already shown to be cold-inducible) also playing a role in ABA induction (D.G. Zarka and M.F. Thomashow, unpublished data).
The effect of ABA upon DREB1A (CBF3) and DREB2A has been tested previously, but no significant increase in expression was reported (Liu et al., 1998; Medina et al., 1999). The increase in CBF1-3 transcript levels we report may have been more easily observed in the system we used, as plants were smaller and possibly more accessible and/or sensitive to the ABA. The fact that two different groups report differently the responsiveness of CBF4 (DREB1D) to ABA (Haake et al., 2002; Sakuma et al., 2002) might suggest that the induction of these genes is dependent upon context and that the age and condition of plants influence flux through different branches of this signaling network. Indeed, we have found this to be true in the case of plants that have experienced one form of stress before being exposed to another (Knight et al., 1998). When considering the response to ABA, water relations during the growing period are likely to be very significant, and whether or not the plants have been gown on soil or in tissue culture may greatly influence their responsiveness to ABA.
Contrary to our results, Narusaka et al. (2003) showed that artificial reporter constructs in which all cis elements apart from the DRE (CRT) elements were eliminated, fail to be activated by ABA treatment, indicating that this element cannot, in isolation, respond to ABA (Narusaka et al., 2003). However, this lack of response to ABA is consistent with data from the same laboratory that indicated CBF1-4 are not ABA-responsive. As we have argued above, this indicates that ABA treatments and/or the physiological state of the plants used were different in the studies conducted by this research group.
The relatively small increases in the levels of steady-state CBF transcripts in response to ABA would only be of any significance if they are converted into changes in CBF TF protein levels, which can then activate the CRT/DRE. We therefore tested whether the relative levels of translated CBF protein were significantly elevated in response to ABA. We observed that CBF protein, as measured by detection of a CBF-AEQUORIN fusion protein, accumulated rapidly in response to ABA (Fig. 6). These significant increases in CBF protein accumulation in response to ABA could account for (part of) the activation of the CRT/DRE. The kinetics of CBF-AEQUORIN accumulation were similar to those of the transcript accumulation, with large increases seen after only 1 h of ABA treatment. This might indicate that in response to ABA, translation of CBF TFs follows rapidly after transcription.
It is interesting to speculate as to under which conditions in nature ABA might accumulate in the manner required for activation of this pathway. Although ABA accumulates in response to low temperature, the increases are transient rather than the cumulative increases seen in response to drought stress (Thomashow, 1999). It has therefore been argued that endogenous ABA accumulating during low temperature does not reach levels sufficient to induce signaling pathways (Shinozaki and Yamaguchi-Shinozaki, 2000), and it is possible that levels would not be high enough to activate the CRT/DRE signaling pathway. It may be possible, however, that in being responsive to ABA as well as cold, the CBF pathway can integrate additional information relating to changes in the plant's environment. ABA may be able to potentiate cold-induced CBF signaling even if the ABA and cold stimuli do not occur concurrently. If this is the case, the CBF1-3 TFs may have a unique role, distinct from that of CBF4, which cannot respond to low temperature but only ABA and drought (Haake et al., 2002). Alternatively, the activation of CBF1-3 by ABA may be more likely to operate under drought conditions, during which ABA accumulation will occur to more significant levels. If this was the case, it would mean that cross-talk is occurring between what have been considered two separate pathways involving two distinct sets of TFs and that under certain circumstances the drought pathway is recruiting CBF.
In this study we have focused on CBF and not DREB2, as DREB2 requires posttranscriptional modification before it is able to influence CRT/DRE-driven expression. Measurement of increased DREB2 protein levels, therefore, cannot be assumed to signify increased CRT/DRE activation, whereas increasing the level of CBF protein is known to be sufficient to activate the CRT/DRE (Stockinger et al., 1997; Liu et al., 1998). Overexpression of CBF4 causes ectopic expression of the COR genes in the absence of stress (Haake et al., 2002), as does overexpression of CBF1-3 (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Gilmour et al., 2000). Thus, our data suggest that both the cold-inducible CBF TFs and the non-cold-inducible CBF4 could be involved in activation of the CRT/DRE by ABA. We cannot, however, discount the possibility that a similar mechanism also exists that employs the DREB2 TFs, and this remains worthy of further study once the nature of the posttranslational modification is understood.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Col-0 expressing CRT::LUC and Ws-2 expressing the promoter::GUS fusions) were grown on solid Murashige and Skoog or Gamborg's B5 medium, as previously described (Knight et al., 1999; Zarka et al., 2003) and experiments performed when they were 7 to 10 d old.
DNA Constructs and Plants Transformation
The CRT::LUC construct and plants used were as described previously (Boyce et al., 2003). The CBF promoter GUS fusions were constructed using approximately 1 kb of promoter sequence fused to the GUS reporter gene and have been described previously, as have the wild-type and mutated m2 versions of the −189 deleted CBF2 promoter (Zarka et al., 2003). The CBF-AEQUORIN construct consisted of the full coding sequence of CBF1 and 1,019 bp of the 5′ sequence upstream of the start codon, fused to a coding sequence for aequorin. The CBF promoter and coding sequence fragment was amplified by PCR, using the primers 5′-GGTCTAGATGTTTTCTTCACCTTACCA-3′ (forward) and 5′-CCGTCGACCGTAACTCCAAAGCGACACGTCA-3′ (reverse) to engineer a 5′ XbaI restriction site and a 3′ SalI site. The engineered XbaI-SalI CBF fragment was cloned into a previously produced version of the pDH51 vector (Pietrzak et al., 1986) that contained the aequorin sequence as a SalI-PstI fragment (Knight et al., 1996) to produce an in-frame fusion with the aequorin coding sequence. The whole chimeric construct (minus the 35S promoter sequence from pDH51) was then transferred to the Agrobacterium binary vector, pBIN19 (Bevan, 1984), as an XbaI-EcoRI insert. The resultant binary vector plasmid was purified from Escherichia coli and then transferred to Agrobacterium strain C58C1 by a freeze-thaw method (Holsters et al., 1978). Col-0 wild-type plants were transformed with Agrobacterium by the floral dip method (Clough and Bent, 1998) and primary transformants selected on kanamycin (50 μg/mL) timentin (200 μg/mL) selection plates. Lines expressing the aequorin construct were identified using in vitro assays of aequorin activity as described previously (Knight et al., 1996) and in detail below.
Plant Treatment with Cold or ABA
Cold treatments were performed by floating seedlings on water contained in plastic tubes, which were then cooled to 5°C in a cooled water-bath or maintained at ambient temperature. (±) cis, trans-ABA (Sigma, Poole, UK and St. Louis) was dissolved in ethanol as a stock solution of 100 mm and diluted 1:1,000 to give a working concentration of 100 μm. Ethanol at 0.1% was used as a control treatment. Seedlings were floated on ABA solution in transparent perspex multi-well plates (Greiner, Stonehouse, UK). In all cases, tissue was harvested and transferred to microcentrifuge tubes, frozen in liquid nitrogen, and stored at −80°C. Approximately 50 to 100 seedlings were used per RNA sample, and 5 seedlings/sample for in vitro determination of LUC or aequorin levels.
Measurement of Gene Expression Using RNA Gel-Blot Hybridization
Total RNA was prepared from seedling tissue using RNEASY plant RNA minipreps (Qiagen, Dorking, UK), and 10 μg/sample was electrophoresed through 1.0% agarose (Life Technologies, Paisley, UK) formaldehyde gels (Sambrook et al., 1989). RNA was transferred to nylon membranes (Böehringer Mannheim, Mannheim, Germany) by capillary action. Blots were prehybridized and hybridized as described previously (Hajela et al., 1990; Stockinger et al., 1997; Knight et al., 1999). Probes for KIN1/KIN2, LTI78, and β-TUBULIN were prepared from the products of PCR using specific primers as described previously (Knight et al., 1996, 1997, 1999). The generic CBF probe was prepared by PCR using specific primers (5′-ccttatccagtttcttgaaacagag-3′ [forward] and 5′-cgaatattagtaactccaaagcgac-3′ [reverse]). The product, (approximately 700 bp) was cloned into a pBluescript vector (Stratagene, La Jolla, CA) and the fragment released with restriction enzymes and used as probe. Gene-specific CBF probes and GUS probe were prepared as described previously (Baker et al., 1994; Gilmour et al., 1998). [CRT]4::LUC transcript levels were measured by using a probe for approximately 1,200 bp of the coding region of firefly LUC released by BglII/NcoI restriction digest of the vector pLUK07 (Mankin et al., 1997). Probes were labeled using 32P-CTP by random priming.
Measurement of CBF-AEQUORIN Fusion Protein Levels
In vitro reconstitution of aequorin was performed as described previously (Knight et al., 1996) by homogenizing 5 7-d-old seedlings in 0.5 mL reconstitution buffer (0.5 m sodium chloride, 10 mm Tris-HCl, pH 7.4, 5 mm EDTA, 5 mm β-mercaptoethanol, 0.1% gelatin), spinning the tubes for 10 min in a microcentrifuge, and adding coelenterazine to 100 μL of supernatant to a final concentration of 1 μm coelenterazine. Reconstitution was allowed to occur over 2 h in darkness, after which 50 μL of the reconstituted aequorin mixture was added to 0.5 mL 200 mm Tris, 5 μm EDTA, pH 7.4 and discharged in the luminometer by addition of an equal volume of 50 mm CaCl2. Three replicate samples were taken for each treatment, and the experiment was performed with three independent transgenic lines expressing the CBF-AEQUORIN fusion.
Acknowledgments
We thank Vakkas Tekin and Pauline White for excellent technical assistance.
This work was supported by the Biotechnology and Biological Sciences Research Council (research grant to M.R.K.), by the U.S. Department of Agriculture National Research Initiative program (grant no. 98–35100–6999 to M.F.T.), by the U.S. Department of Energy (DEFB0291ER20021 to M.F.T.), and by the Michigan Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043562.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Bevan M (1984) Binary agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR (2003) The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J 34: 395–406 [DOI] [PubMed] [Google Scholar]
- Busk PK, Jensen AB, Pages M (1997) Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J 11: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–443 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF (1990) Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol 93: 1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J (1978) Transfection and transformations of A. tumefaciens. Mol Gen Genet 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30: 679–689 [DOI] [PubMed] [Google Scholar]
- Knight H, Brandt S, Knight MR (1998) A history of stress alters drought calcium signalling pathways in Arabidopsis. Plant J 16: 681–687 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1997) Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Knight H, Veale E, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg Franck M (1992) Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol 19: 689–692 [DOI] [PubMed] [Google Scholar]
- Kurkela S, Franck M (1990) Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol 15: 137–144 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199–222 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki Y, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin SL, Allen GC, Thompson WF (1997) Introduction of a plant intron into the luciferase gene of Photinus pyralis. Plant Mol Biol Rep 15: 186–196 [Google Scholar]
- Mäntylä E, Lång V, Palva ET (1995) Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol 107: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET (1993) Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21: 641–653 [DOI] [PubMed] [Google Scholar]
- Pietrzak M, Shillito RD, Hohn T, Potrykus I (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14: 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, NY
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14: S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238: 17–25 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]