Abstract
Suc-phosphate synthase (SPS) is a key regulatory enzyme in the pathway of Suc biosynthesis and has been linked to quantitative trait loci controlling plant growth and yield. In dicotyledonous plants there are three SPS gene families: A, B, and C. Here we report the finding of five families of SPS genes in wheat (Triticum aestivum) and other monocotyledonous plants from the family Poaceae (grasses). Three of these form separate subfamilies within the previously described A, B, and C gene families, but the other two form a novel and distinctive D family, which on present evidence is only found in the Poaceae. The D-type SPS proteins lack the phosphorylation sites associated with 14-3-3 protein binding and osmotic stress activation, and the linker region between the N-terminal catalytic glucosyltransferase domain and the C-terminal Suc-phosphatase-like domain is 80 to 90 amino acid residues shorter than in the A, B, or C types. The D family appears to have arisen after the divergence of mono- and dicotyledonous plants, with a later duplication event resulting in the two D-type subfamilies. Each of the SPS gene families in wheat showed different, but overlapping, spatial and temporal expression patterns, and in most organs at least two different SPS genes are expressed. Analysis of expressed sequence tags indicated similar expression patterns to wheat for each SPS gene family in barley (Hordeum vulgare) but not in more distantly related grasses. We identified an expressed sequence tag from rice (Oryza sativa) that appears to be derived from an endogenous antisense SPS gene, and this might account for the apparently low level of expression of the related OsSPS11 sense gene, adding to the already extensive list of mechanisms for regulating the activity of SPS in plants.
Suc occupies a central position in the metabolism of all plants and has many roles: transport sugar, storage reserve, compatible solute, and signal compound. It is synthesized by the enzymes Suc-phosphate synthase (SPS; EC 2.4.1.14) and Suc-phosphatase (SPP; EC 3.1.3.24) via the intermediate Suc-6F-phosphate (Leloir and Cardini, 1955). Several studies on agronomically important species within the Poaceae (grasses) have linked plant growth and productivity with SPS activity. In a comparison of various maize (Zea mays) genotypes, SPS activity was shown to correlate with growth rate, whereas the activities of key enzymes of photosynthesis (Rubisco) and starch synthesis (ADPGlc pyrophosphorylase) did not (Rocher et al., 1989). SPS has also been linked to relative growth rate in young maize plants (Causse et al., 1995a). The same group found evidence that maize contains at least two SPS genes, that the enzyme shows heterotic activity in hybrids of inbred lines, and that its activity is correlated with dry matter yield (Causse et al., 1995b; Prioul et al., 1999). SPS activity was also correlated with yield in an independent study of different maize lines (Sarquís et al., 1998) and linked to quantitative trait loci for grain yield (Bertin and Gallais, 2001). In rice (Oryza sativa), SPS activity correlates with leaf expansion rate (Seneweera et al., 1995), and recently a quantitative trait locus for plant height in rice was shown to coincide with the OsSPS1 gene, while transgenic rice plants with increased SPS activity grew taller than control plants (Ishimaru et al., 2003). In sugarcane (Saccharum officinarum), the source of 70% of the world's sugar supply, Suc accumulation in the stems is dependent on SPS activity (Zhu et al., 1997). Many of these studies are correlative and do not prove that SPS is a controlling factor in the growth and yield of these important crop plants. Nevertheless, it seems clear that SPS can be a valuable biochemical marker for these complex traits; therefore, it would be highly desirable to have a better understanding of the genetic control of the enzyme in the Poaceae.
The first SPS genes to be cloned came from maize (Worrell et al., 1991) and spinach (Spinacea oleracea; Klein et al., 1993; Sonnewald et al., 1993). Subsequently, SPS genes have been cloned from over 20 other plants, mostly dicotyledonous species (dicots), as well as cyanobacteria (Lunn and MacRae, 2003). As SPS genes from more species were described, a trend emerged for the sequences to cluster into two groups, with one group from dicots, e.g. spinach and potato (Solanum tuberosum), and the other from monocotyledonous plants (monocots), e.g. maize and rice. Based on Southern-hybridization analysis, it was once thought that each species contained only a single SPS gene (Worrell et al., 1991; Valdez-Alarcón et al., 1996). However, Sugiharto et al. (1997) cloned two different SPS cDNAs from sugarcane, one of which closely resembles the maize gene, whereas the other encodes a smaller protein with some similarity to the dicot SPSs known at that time. Two SPS cDNAs were also cloned from a dicot species of resurrection plant (Craterostigma plantaginea), one of which is more like the previously cloned genes from monocots (Ingram et al., 1997). These results showed that at least some plants contain multiple SPS genes and argued against a phylogeny based simply on a monocot-dicot divide.
Three different SPS genes are expressed in the leaves of Citrus unshiu (Komatsu et al., 1996), and four SPS genes were found when the Arabidopsis genome was sequenced (Langenkämper et al., 2002). Langenkämper et al. (2002) carried out a rigorous phylogenetic analysis of all the known SPS sequences and discovered that they cluster into three distinct families, which they designated A, B, and C. Most of the available sequences from dicots belong to the A family, including one of those from C. unshiu and two from Arabidopsis. However, these two species also have one gene each from the B and C families. B family genes have also been identified from two species of kiwifruit (Actinidia deliciosa and Actinidia chinensis; Langenkämper et al., 2002; Fung et al., 2003). Most of the available SPS sequences from monocots belong to the B family, except for a partial banana (Musa acuminata) SPS sequence in the A family. The smaller SPS isoform from sugarcane and a closely related partial sequence from barley (Hordeum vulgare) also seemed to be allied with the A family but were somewhat more divergent than the rest. None of the known monocot SPS genes belongs to the C family, but the relative scarcity of SPS sequences from monocots made it difficult to assess the true diversity of SPS genes in these plants.
A timely opportunity to address this has been presented by the sequencing of the rice genome, with publicly available sequences from both the japonica cv Nipponbare (http://rgp.dna.affrc.go.jp/IRGSP/) and the indica cv 93 to 11 (Yu et al., 2001). In addition, the International Triticeae EST Cooperative (ITEC; http://wheat.pw.usda.gov/genome/) has generated over half a million expressed sequence tags (ESTs) from wheat and more than 348,000 from barley, with smaller numbers from other members of the Triticeae tribe, providing a powerful resource for gene discovery in these species. There are also large publicly available collections of ESTs from maize, rice, sugarcane, and sorghum (Sorghum bicolor), as well as shotgun fragments of maize genomic sequence (Chandler and Brendel, 2002). From these sequence data we discovered five distinct families of SPS genes in the grasses, two of which encode unusual isoforms unique to these plants. The evolution, function, and regulation of these five SPS gene families and the enzymes they encode are discussed.
RESULTS
Wheat SPS Genes
We identified five families of SPS genes in wheat (Triticum aestivum), which were assigned the Roman numerals I-V. Within each family at least two, usually three, highly similar but not identical variants were present (Supplemental Data [available at www.plantphysiol.org], Appendix D), and these are designated by the suffixes a-c. The longest contiguous sequence from each family and further details are provided in the Supplemental Data (Appendix A). The number of amino acid residues, molecular mass, and pI of representative SPS proteins from each family are shown in Table I.
Table I.
Properties of SPS isoforms from wheat, rice, and maize
| Isoform | No. of Amino Acid Residues | Molecular Mass | pI |
|---|---|---|---|
| kD | |||
| Family I (=Family C) | |||
| Wheat1 | 1,055 | 115 | 6.14 |
| Rice11 | 1,106 | 119 | 6.35 |
| Maize1 | 1,051 | 114 | 7.01 |
| Family II (=Family A) | |||
| Wheat2 | 1,076 | 119 | 6.30 |
| Rice8 | 1,066 | 119 | 6.41 |
| Maize2 | 1,059 | 118 | 6.55 |
| Family III (=Family DIII) | |||
| Wheat9 | 964 | 108 | 7.33 |
| Rice2 | 963 | 108 | 7.26 |
| Maize3 | 964 | 108 | 7.62 |
| Family IV (=Family DIV) | |||
| Wheat4 | (972) | (109) | (7.49) |
| Rice6 | 977 | 109 | 7.23 |
| Maize4 | – | – | – |
| Family V (=Family B) | |||
| Wheat6 | (1,075) | (118) | (6.66) |
| Rice1 | 1,084 | 119 | 6.70 |
| Maize5 | 1,068 | 119 | 6.64 |
Values in parentheses are estimated (see Supplemental Data, Appendix A, for details).
The two or three variants within each SPS gene family in bread wheat, and the allohexaploid (AABBDD genome) nature of this species, suggested that there are homeologous SPS genes from each family in the A, B, and D genomes. This was investigated further by Southern-hybridization analysis on genomic DNA from 1) the diploid D-genome progenitor species Aegilops tauschii (DD), (2) the tetraploid species durum wheat (Triticum turgidum subsp. durum; AABB), and (3) bread wheat, with family specific probes. RFLPs were detected between the three species with at least one of the restriction enzymes used (Fig. 1), and each probe gave a different hybridization pattern showing there was no significant cross-hybridization with genes from the other families.
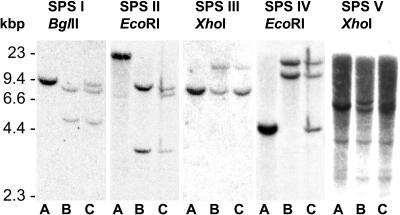
Figure 1.
Southern-hybridization analysis of SPS genes in diploid, tetraploid, and hexaploid species of wheat. Autoradiograms of (A) Aegilops tauschii (DD), (B) durum wheat (AABB), and (C) bread wheat (AABBDD) genomic DNA (20 μg) digested with BglII, EcoRI, or XhoI and hybridized with SPS gene family specific probes.
For SPSI and SPSIV the diploid, tetraploid, and hexaploid species contained one, two, and three separate hybridizing fragments, with bread wheat having the same sized fragments as both of the other two species. For SPSIII the D-genome fragment was the same size as the smaller of the two fragments from durum wheat and bread wheat. However, the lower band from bread wheat was more intense than the larger one, suggesting that the smaller hybridizing fragments were more abundant and is consistent with the lower band containing fragments from both the D genome and either the A or B genomes. The SPSII and SPSV RFLP patterns were more complex. A. tauschii contained two SPSII-hybridizing fragments, the larger of which was the most strongly hybridizing. There was no comparable fragment from bread wheat, but this species did have a fragment of the same size as the smaller, more weakly hybridizing fragment from A. tauschii, as well as fragments comparable to those from durum wheat. For SPSV, all three species contained three smaller invariant fragments in addition to the larger polymorphic fragments. Although the latter were very similar in size, it was possible to distinguish three separate bands in bread wheat comparable to each of those from A. tauschii and durum wheat.
Rice SPS Genes
There are 5 SPS genes in the rice genome, with one each on chromosomes 2, 6, 8, and 11, in addition to the previously described SPS gene on chromosome 1 (Sakamoto et al., 1995; Valdez-Alarcon et al., 1996). Further information on the sequences that are not annotated in the GenBank database is provided in the Supplemental Data (Appendix B). Each of the five genes (OsSPS1, OsSPS2, OsSPS6, OsSPS8, and OsSPS11) shows greater similarity to one of the five families of wheat SPS genes than to the other rice SPS genes (Table II), placing them in families V, III, IV, II, and I, respectively.
Table II.
Comparison of wheat, rice, and maize SPS nucleotide and protein sequences
| Wheat1 | Wheat2 | Wheat9 | Wheat4 | Wheat6 | Rice1 | Rice2 | Rice6 | Rice8 | Rice11 | Maize1 | Maize2 | Maize3 | Maize4ac | Maize4cdc | Maize4ec | Maize5a | Maize5ba | Maize5ca | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wheat1 | 52 | 50 | 50 | 54 | 55 | 50 | 49 | 51 | 83 | 78 | 52 | 51 | 49 | 57 | 42 | 56 | 53 | 55 | |
| Wheat2 | 54 | 64 | 64 | 54 | 55 | 65 | 65 | 88 | 52 | 52 | 85 | 65 | 74 | 69 | 54 | 56 | 52 | 56 | |
| Wheat9 | 54 | 70 | 82 | 52 | 53 | 91 | 85 | 65 | 51 | 49 | 65 | 89 | 96 | 83 | 77 | 54 | 54 | 52 | |
| Wheat4a | 51 | 69 | 81 | 56 | 54 | 84 | 87 | 65 | 51 | 50 | 64 | 83 | ND | 85 | 81 | 55 | ND | 53 | |
| Wheat6b | 62 | 58 | 55 | 58 | 86 | 53 | 52 | 55 | 56 | 53 | 55 | 54 | 52 | 64 | 42 | 84 | 81 | 86 | |
| Rice1 | 62 | 58 | 56 | 57 | 84 | 54 | 54 | 55 | 57 | 55 | 56 | 54 | 55 | 63 | 42 | 86 | 94 | 85 | |
| Rice2 | 53 | 70 | 89 | 82 | 57 | 57 | 87 | 67 | 51 | 50 | 66 | 93 | 96 | 85 | 80 | 54 | 58 | 53 | |
| Rice6 | 53 | 69 | 82 | 87 | 57 | 58 | 83 | 66 | 52 | 50 | 66 | 86 | 92 | 87 | 84 | 54 | 54 | 53 | |
| Rice8 | 53 | 88 | 71 | 69 | 61 | 58 | 72 | 70 | 52 | 52 | 88 | 67 | 77 | 70 | 56 | 57 | 51 | 57 | |
| Rice11 | 85 | 53 | 54 | 52 | 62 | 62 | 54 | 54 | 54 | 83 | 52 | 52 | 52 | 58 | 40 | 57 | 50 | 56 | |
| Maize1 | 82 | 55 | 53 | 52 | 62 | 61 | 54 | 53 | 53 | 84 | 52 | 49 | 48 | 57 | 39 | 55 | 54 | 53 | |
| Maize2 | 55 | 84 | 69 | 69 | 59 | 59 | 71 | 69 | 86 | 53 | 54 | 66 | 75 | 70 | 56 | 57 | 52 | 57 | |
| Maize3 | 53 | 67 | 86 | 82 | 57 | 58 | 88 | 83 | 70 | 53 | 53 | 69 | 96 | 86 | 77 | 55 | 58 | 54 | |
| Maize4ac | 64 | 83 | 92 | ND | 66 | 66 | 92 | 91 | 81 | 68 | 64 | 79 | 89 | ND | ND | 56 | 55 | ND | |
| Maize4cd | 55 | 72 | 82 | 86 | 63 | 63 | 83 | 86 | 72 | 56 | 57 | 71 | 83 | ND | ND | 66 | ND | 63 | |
| Maize4ea | 37 | 64 | 78 | 82 | 41 | 39 | 79 | 84 | 66 | 39 | 38 | 65 | 77 | ND | ND | 41 | ND | 40 | |
| Maize5a | 61 | 59 | 57 | 57 | 81 | 83 | 59 | 58 | 59 | 60 | 61 | 58 | 58 | 64 | 64 | 42 | 96 | 96 | |
| Maize5bc | 71 | 67 | 70 | ND | 83 | 86 | 69 | 66 | 65 | 69 | 68 | 65 | 67 | 67 | ND | ND | 93 | ND | |
| Maize5ca | 60 | 58 | 56 | 57 | 83 | 82 | 57 | 56 | 57 | 58 | 60 | 57 | 57 | ND | 63 | 41 | 94 | ND |
Sequence identity was determined using the GAP program of the Wisconsin GCG sequence analysis package. Values shown in bold are for the protein sequences. ND, Not determined for pairs of partial sequences with no overlapping regions of similarity.
Truncated at 5′-end/N terminus.
Sequence contains a gap with a predicted length of 180 bp (60 amino acids).
Truncated at 3′-end/C terminus.
Truncated at 5′-end/N terminus and 3′-end/C terminus.
The 3′-end of a 1,281-bp cDNA clone (AK069527), derived from chromosome 6 (AP005386 and AP003961), includes regions that have 84% to 95% identity with parts of exons 9 and 10 and intron 9 in the OsSPS11 gene but in the antisense orientation. The 5′-end of the cDNA and the first open reading frame reading from this end (encoding a 16-amino acid peptide) have no similarity with SPS sequences. The rice family I gene (OsSPS11) is poorly representated in the EST collection (Supplemental Data, Appendix D). This could simply reflect a low level of transcription; however, another possibility is suppression of OsSPS11 gene expression by a post-transcriptional mechanism involving the endogenous OsSPS11-like antisense transcript.
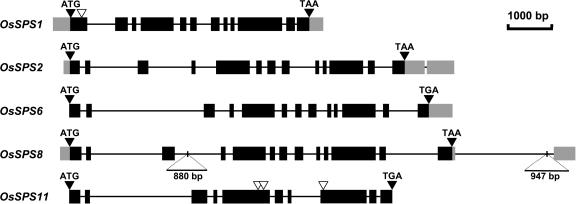
The five rice SPS genes have very similar exon-intron structures, with the OsSPS2, OsSPS6, and OsSPS8 genes containing 12 introns at equivalent positions in the coding regions (Fig. 2). Eight of the splice sites are also conserved in the other two genes, but OsSPS1 lacks the equivalent of intron 1, and OsSPS11 lacks introns 5, 6, and 10 (Fig. 2). OsSPS2 (family III) and OsSPS6 (family IV) are the most closely related of the rice SPS genes, with 83% identity in the coding regions (Table II). The chromosome regions containing these two genes show some colinearity, with genes encoding an H+-translocating pyrophosphatase and an Ile tRNA ligase located upstream of both OsSPS2 and OsSPS6.
Figure 2.
Schematic alignment of rice SPS genes. The gene number indicates on which chromosome the gene lies. Protein-coding regions are shown in black, and 5′- and 3′-UTRs in gray. Black triangles indicate the positions of the translation start (ATG) and stop (TAA, TGA) codons. The white triangles mark the positions where introns are absent from one gene in relation to the others. The size and position of two insertions in introns 3 and 13 of the OsSPS8 gene in japonica rice cv Nipponbare are indicated by the bars.
Maize SPS Genes
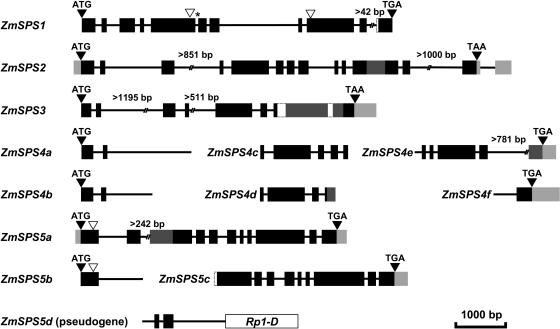
There are at least seven SPS genes in maize, including the original SPS (ZmSPS5a) sequence described by Worrell et al. (1991; Fig. 3). Sequencing of the maize genome to date has been targeted to gene-rich regions with low copy number or low methylation (Chandler and Brendel, 2002). The seven SPS genes we identified account for all of the SPS sequences in the public maize EST collection, which is derived from a wide range of tissues and developmental stages; therefore, it seems unlikely that additional SPS genes will be discovered as sequencing of the maize genome is completed. The maize genes fall into five groups that are allied to the five SPS gene families in wheat and rice (Table II) and were assigned numbers on the basis of their closest similarity to one of the wheat families. The ZmSPS1, ZmSPS2, and ZmSPS3 genes were the only maize genes found in their respective families, whereas there are at least two genes from family IV and two from family V in maize (Fig. 3). The two family IV sequences are unlikely to represent allelic forms of the same gene as they were both found in the genomic sequence from the inbred line B73 and are represented by ESTs from another inbred line W23 (Appendix D). Similarly, both the family V genes were present in the genomic sequence from cv B73 and represented by ESTs from two other inbred lines, F2 and F334. The maize SPS gene structures are very similar to each other and to the related genes from rice (Figs. 2 and 3). Further information on the maize SPS sequences is provided in the Supplemental Data (Appendix C).
Figure 3.
Schematic alignment of maize SPS and SPS-like genes. The gene number indicates the SPS gene family. Protein-coding exons within genomic sequences are shown in black with those regions derived only from maize cDNA (EST) sequences shown in dark gray. The 5′- and 3′-UTRs are shown in light gray. Gaps in the maize exon sequence are indicated by dashed lines, and gaps in the introns are indicated by two angled lines with the minimum size of the intron indicated above. Black triangles indicate the positions of the translation start (ATG) and stop (TAA, TGA) codons. The white triangles mark the positions where introns are absent from one gene in relation to the others, and the asterisk shows the position of an intron in the maize ZmSPS1 gene that is absent in the orthologous rice gene (OsSPS11).
SPS Gene Families in Plants
The compilation of sequences from wheat, rice, and maize described above showed that all three species have five SPS genes or groups of genes in common, with orthologous genes from the different species generally showing greater similarity to each other than they do to the other SPS genes from the same species (Table II). ESTs from each of the five SPS families were also found in three other species in the Poaceae: barley, sorghum, and sugarcane (Supplemental Data, Appendix D).
To investigate the relationships between the different genes, phylogenetic bootstrap analyses were carried out on the deduced protein sequences, along with all other available higher plant SPS sequences, as described by Langenkämper et al. (2002). Higher plant SPSs, in addition to the catalytic N-terminal glucosyltransferase domain, contain a C-terminal domain that resembles SPP (Lunn et al., 2000). Analyses using (1) the full-length protein, (2) the glucosyltransferase domain only, (3) the SPP-like domain only, or (4) a region around the active site gave essentially the same results, and a representative phylogenetic tree is shown in Figure 4. The sequences form four distinct clusters. The family II sequences from the grasses lie together within a larger cluster corresponding to the previously described A family. Likewise, the family V and family I sequences from the grasses constitute subfamilies within the broader B and C families, respectively. In addition to the A, B, and C families, there is a fourth cluster of SPS sequences containing both the family III and IV sequences from the grasses. This cluster represents a novel and distinctive family, designated family D, and is well supported by the bootstrap values in every analysis undertaken (Fig. 4; other data not shown).
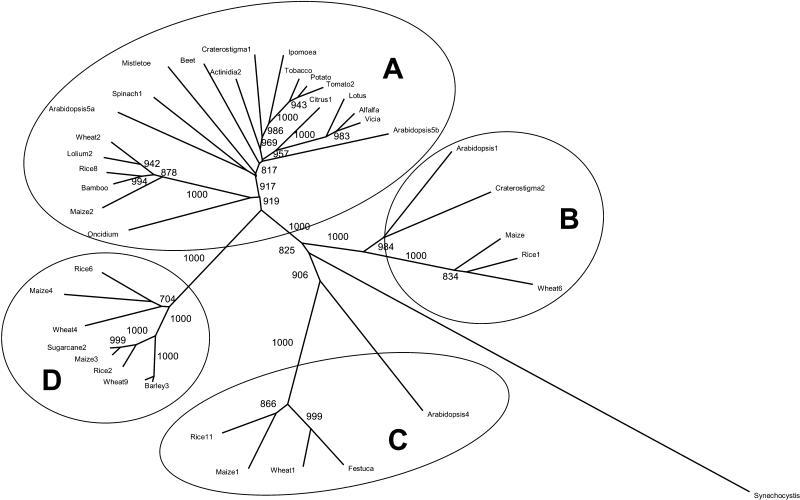
Figure 4.
Phylogenetic tree of full-length SPS protein sequences from plants and the cyanobacterium Synechocystis sp. PCC 6803. Unrooted neighbor-joining tree (no gaps) showing bootstrap values >700 (1,000 replicates). The GenBank accession numbers of the sequences are as described in Lunn and MacRae (2003), with the addition of alfalfa (Medicago sativa) AF322116, bamboo (Bambusa oldhamii) AY445835, and mistletoe (Viscum album subsp. album) AY331261. Lolium perenne and Festuca arundinacea sequences are from Demmer et al. (2003).
The D family only contains sequences from species in the Poaceae. Two sequences from nongraminaceous monocots, the orchid Oncidium sp. cv Goldiana (AY135211) and banana (U59489), belong to the A family and lie near the node of the grass branch in some analyses but in between the dicots and grasses in others (Fig. 4; other data not shown). Searches of the EST database failed to find any D family sequences from monocots other than members of the Poaceae or from dicots. The cyanobacterial SPS from Synechocystis sp. PCC 6803 (Curatti et al., 1998; Hagemann and Marin, 1999; Lunn et al., 1999) was also included in some analyses, and this consistently lies distantly from all the higher plant SPS sequences, with a branch point closest to the B and C families (Fig. 4; other data not shown).
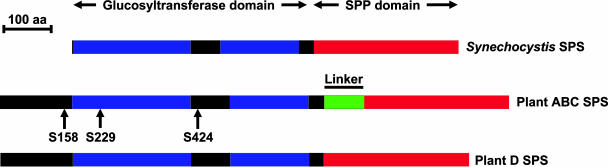
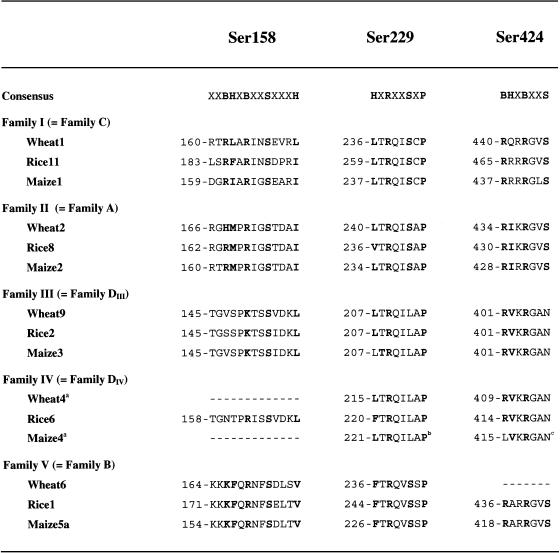
The D family SPS proteins are smaller than those from the other families, with molecular masses of 108 to 109 kD rather than 114 to 119 kD, and have higher pIs (Table I). Alignment of the SPS sequences showed that the linker region between the glucosyltransferase and SPP-like domains is about 80 to 90 amino acids shorter in the D family proteins (Fig. 5). SPS is modulated by multisite protein phosphorylation in response to light, osmotic stress, nitrogen supply, and temperature and also binds 14-3-3 proteins (Quy and Champigny, 1992; Reimholz et al., 1994; Huber and Huber, 1996). In addition to their smaller size, the D family SPSs are characterized by the lack of the two Ser residues involved in 14-3-3 protein binding (Ser-229) and osmotic stress activation (Ser-424), although some elements of the consensus phosphorylation site motifs (Huber and Huber, 1996) do appear to be present (Fig. 6). Although the D family proteins contain Ser residues equivalent to Ser-158 that is involved in light-dark regulation of spinach leaf SPS, the critical basic and hydrophobic residues at the −6 and −5 positions in the consensus motif (Huber and Huber, 1996) are missing (Fig. 6).
Figure 5.
Schematic alignment of SPS proteins from plants and the cyanobacterium Synechocystis sp. PCC 6803. The glucosyltransferase domain is shown in blue, the SPP-like domain in red, and the linker region in green. The positions of the phosphorylation sites involved in light-dark regulation (Ser-158), 14-3-3 protein binding (Ser-229), and osmotic stress activation (Ser-424) of the plant SPS are indicated by the arrows.
Figure 6.
Comparison of wheat, rice, and maize SPS protein sequences in the regions of the regulatory phosphorylation sites. Sequences were aligned using the PILEUP program of the GCG Wisconsin Sequence Analysis Package. Residues matching the consensus sequences (Huber and Huber, 1996) are shown in bold (B, basic; H, hydrophobic). Footnotes: a, Residue number estimated; b, Maize SPS4c (LTRQIIAP in Maize SPS4d); and c, Maize SPS4c and SPS4d.
SPS Gene Expression and Function
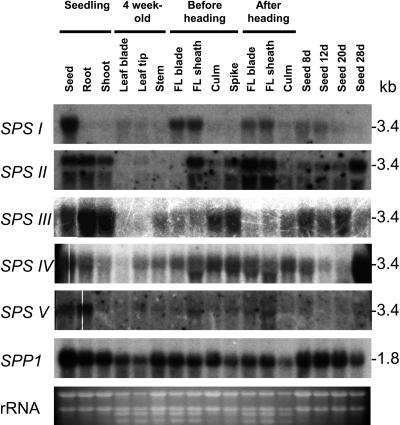
The spatial and temporal expression patterns of the different SPS gene families in wheat were investigated by northern-hybridization analysis on RNA from a wide range of tissues at different stages of development (Fig. 7). This is the first such comprehensive expression survey of all the SPS gene families in a single plant species. Given the very high similarity (>93% identity) between the homeologous wheat sequences, it was expected that the probes would hybridize with all of the mRNAs from the same family. Each probe hybridized specifically with RNAs in the size range expected for full-length SPS transcripts (3.3–3.6 kb), except for TaSPSII, which also hybridized with smaller RNA species (Fig. 7). There was a clear correlation between the signal intensities of the two TaSPSII-hybridizing bands from each sample, suggesting that the smaller bands did not arise by nonspecific cross-hybridization and might be accounted for by partial degradation of the TaSPSII transcripts.
Figure 7.
Northern-hybridization analysis of SPS and SPP1 gene expression in bread wheat. RNA was extracted from various tissues harvested from plants at different stages of development: germinating seedlings, 4-week-old vegetative plants, and older plants just before heading and after heading. FL, Flag leaf.
TaSPSI (family C) genes were most strongly expressed in the seeds of germinating seedlings and in the flag leaf blade and sheath (Fig. 7). Low level expression was also detected in expanding leaves from young vegetative plants. TaSPSII (family A) genes were expressed in all parts of germinating seedlings, in the flag leaf blade and sheath, particularly after heading, in the immature spike and in developing seeds, especially later in development. TaSPSIII (family D) genes were most highly expressed in germinating seedlings and in the culm and spike before heading, with some expression also in developing seeds during the earlier stages of development. In contrast, TaSPSIV (family D) genes were most strongly expressed in seeds during the latest stage of development, with some expression also found in germinating seedlings, especially in the seed and to a lesser extent in the young leaf tip, flag leaf, culm, and immature spike. In all experiments, longer exposure times than for the other probes were needed to detect signals from TaSPSV (family B) transcripts, suggesting that these are of low abundance, with the strongest signals coming from seeds and roots of germinating seedlings. The major SPP gene family in wheat (Lunn, 2003) appears to be constitutively expressed in both source (leaves and germinating seeds) and sink (developing seeds and roots) tissues.
The expression patterns of the different wheat SPS genes shown by the northern analysis are broadly consistent with the origins of the wheat SPS ESTs (Appendix D). Almost all of the wheat SPS ESTs derived from leaves belong to the C family. A particularly interesting finding was that all but one of the 23 B-family wheat ESTs came from immature or flowering spikes or from anthers. Strikingly similar patterns were also found for the barley ESTs, with all seven of the B-family ESTs coming from anthers, whereas over 40% of the barley C-family ESTs came from leaves (Appendix D). In contrast to wheat and barley, the C-family genes were very poorly represented in the EST collections from rice, sugarcane, and sorghum and not at all from maize, whereas maize B family ESTs were particularly abundant from leaves and shoots (Appendix D). Many of the rice OsSPS6 (D family, subfamily IV) and OsSPS8 (A family) ESTs came from leaves inoculated with the rice blast fungus (Magnaporthe grisea) but not from uninfected control leaves, indicating that infection with this pathogen induced their expression.
DISCUSSION
Evolution of SPS Gene Families in the Grasses
We discovered an unexpected diversity of SPS genes in grasses, with five separate gene families in each of the six species examined. Three of these families—II, V, and I—constitute subfamilies within the broader A, B, and C families, according to the classification of Langenkämper et al. (2002). This is the first report of C-family sequences from any monocot species. It seems clear that these three gene families must have arisen before the separation of the monocots and dicots, which is thought to have occurred about 200 million years ago (Mitchell-Olds and Clauss, 2002). The SPS genes in higher plants probably evolved from a cyanobacterial-like enzyme inherited during the endosymbiosis of the ancestor of chloroplasts (Lunn, 2002), and the conservation of many of the exon-intron splice sites within the A, B, and C family genes in Arabidopsis (Langenkämper et al., 2002), rice, and maize is consistent with all three families having a common origin. The cyanobacterial SPS from Synechocystis sp. PCC 6803 consistently joins the higher plant SPS family tree between the branches leading on the one hand to the C family and on the other to the A and B families (Fig. 4), suggesting that the ancestral plant SPS first diverged to form the C family and a proto-AB family.
In addition to the A, B, and C types, the grasses also possess two unusual forms of SPS, encoded by two closely related groups of genes (i.e. families III and IV) within the newly described D family. Although the D-type SPS was only found in grass species, nongraminaceous monocots are rather poorly represented in the public databases, so it is too early to judge whether the D-type is truly restricted to the grasses or is also found in other monocots. The D-family genes show some affinity with the A-family, e.g. in gene structure and sequence similarity (Table II), and the phylogenetic tree (Fig. 4) shows they share a more recent common ancestry than they do with the B and C families. The simplest explanation for the present-day distribution of the D family is that it diverged from the A family after the division of the monocots and dicots 200 million years ago but before the divergence of the grass lineage, which is estimated to have occurred 77 million years ago (Gaut, 2002). The similarity between subfamilies III and IV within the D family, and the colinearity of the OsSPS2 and OsSPS6 genes with at least two other genes on chromosomes 2 and 6 in rice, suggests that these subfamilies arose by duplication of a chromosomal region containing the ancestral D-family gene that had already acquired the distinctive D-type features. The presence of both D-type subfamilies in species from the Bambusoideae-Ehrhartoideae-Pooideae (wheat, barley, and rice) and Panicoideae-Arundinoideae-Centothecoideae-Chloridoideae (maize, sorghum, and sugarcane) clades implies that the duplication event occurred before these two clades diverged about 50 million years ago (Gaut, 2002).
One of the characteristic features of the unique D-type SPSs is that they have a much shorter linker region between the glucosyltransferase and SPP-like domains (Fig. 5). There is circumstantial evidence that proteolytic cleavage of SPS in response to sugar starvation could occur within this linker region, leading to loss of the SPP-like domain but not catalytic activity (Cotelle et al., 2000), and this processing has been postulated to prevent the enzyme forming a complex with SPP (Lunn and MacRae, 2003). Presumably the D-type SPSs might not be subject to such regulation. The conservation of regulatory phosphorylation sites in the A, B, and C families, with some remnants of the motifs found even in the D-type proteins (Fig. 6), implies that these features were originally present in the D-type but then lost. Curiously, the loss of the linker region and regulatory phosphorylation sites means that the D-type SPSs resemble the cyanobacterial (Synechocystis) enzyme (Fig. 5) and seems to be a reversal of the trend for the plant enzyme to become more complex than the presumed ancestral form.
The basic complement of five SPS gene families in the grasses has been retained in each of the three genomes of hexaploid wheat. However, in maize, which is thought to be an ancient tetraploid (White and Doebley, 1998), only the B-family and D-family (subfamily IV) genes appear to be duplicated. Presumably two copies of the A, C, and D (subfamily III) family genes would also have been present after the original genome duplication event, but one copy of each has since been lost. The presence of two B-family genes and the B-family-like ZmSPS5d pseudogene is consistent with previous reports of three loci, on chromosomes 3, 6, and 8, that cross-hybridize with a ZmSPS5a-derived probe (Causse et al., 1995a, 1995b; Prioul et al., 1999). Previously published data and the EST survey indicate that the B-family genes, ZmSPS5a and ZmSPS5c, encode the major SPS isoforms in maize leaves (Worrell et al., 1991; Appendix D). Maize, like most C4 plants, has two photosynthetic cell types in leaves, and there have been conflicting reports on the presence of SPS in bundle sheath cells (for discussion, see Lunn and Furbank, 1997). However, little or no SPS activity was found in the only report of direct measurements on isolated maize bundle sheath cells (Lunn and Furbank, 1997), suggesting that both maize B-family genes are expressed in mesophyll cells. Perhaps the high rates of photosynthetic Suc synthesis in maize required the retention of both copies of the duplicated B-family gene in order to express adequate amounts of the SPS protein in leaves.
Function of the SPS Gene Families in Grasses
The expression patterns of the various SPS gene families were investigated as a first step toward understanding their respective functions. Although most of the SPS ESTs found from wheat leaves belonged to the C-family, the northern analysis only showed weak expression of this, or any other, SPS gene family in the youngest, fully expanded leaves from vegetative plants, although D-family SPSIV transcripts were detected in the leaf tips (Fig. 7). The surprisingly low level of expression of SPS genes could indicate that there is little turnover of SPS proteins in source leaves. However, another possibility is that SPS gene expression follows a diurnal rhythm in leaves, like many other photosynthesis-related genes, and transcript levels happened to be low at the time of sampling. The C-family genes were more highly expressed in the flag leaf, along with D-family SPSIV and A-family SPSII genes (Fig. 7). Thus it appears that more than one isoform of SPS contributes to the SPS activity involved in photosynthetic Suc synthesis in leaves. Finer resolution of the gene expression patterns, e.g. by in situ hybridization, will be needed to say whether SPS genes from different families are expressed together in the same cells. Although the native SPS enzyme is known to exist as a dimer or tetramer in plants, we do not know if it can form hetero-oligomeric complexes or what the functional significance of such complexes might be. SPS is light activated in wheat leaves (Trevanion et al., 2004), and phosphorylation site motifs likely to be associated with this regulation are present in the A- and C-family isoforms expressed in leaves (Ser-158; Fig. 6).
SPS and SPP are also required for synthesis of Suc from starch reserves in germinating seeds, and transcripts from all five SPS gene families and from the SPP1 gene family were found in germinating wheat seeds. Northern signals from different probes are not directly comparable unless differences in labeling and hybridization efficiencies can be quantified, and transcript levels do not necessarily reflect the amount of the corresponding protein present in the tissue. Therefore, although we know which genes are expressed, we cannot say from these results alone which are the dominant isoforms in germinating wheat seeds.
Wheat, like many grasses, synthesizes complex carbohydrates (fructan in wheat and barley, starch in maize and rice) in its stem (culm) during vegetative growth. These reserves are later remobilized to supplement the supply of assimilates from current photosynthesis during grain filling, and the SPS activity in wheat stems increases at this time (Wardlaw and Willenbrink, 1994). The D-family genes SPSIII and SPSIV and, rather weakly, the SPSI and SPSII genes are expressed in wheat culms (Fig. 7), suggesting that multiple isoforms of SPS also contribute to Suc synthesis during fructan remobilization. SPS might also be required for remobilization of starch or other complex carbohydrates in the anther walls to supply the developing pollen grains with Suc (Datta et al., 2002), accounting for the expression of the B-family SPSV genes in anthers (Supplemental Data, Appendix D).
A rather unexpected finding was that SPSI, SPSII, SPSIII, and SPSIV genes, as well as SPP1 genes, are all expressed in developing seeds, which are net Suc-utilizing organs. SPS and SPP activities are readily detectable in developing seeds (J.E. Lunn, unpublished data), but their function in these sink organs is poorly understood. Developing wheat seeds can also store small amounts of fructan temporarily (Schnyder, 1993), and the enzymes could be involved in its remobilization. Futile cycles of Suc breakdown and resynthesis, requiring SPS and SPP, occur in many sink tissues, and fluxes through these cycles can sometimes be considerable (Rontein et al., 2002). Such futile cycles have been proposed to allow more flexible and sensitive control over carbohydrate metabolism and partitioning (Nguyen-Quoc and Foyer, 2001; Rontein et al., 2002) and might promote Suc unloading from the phloem into the developing seed by establishing concentration gradients within the sink tissue. SPS activity is also present in embryos from dormant mature wheat seeds; indeed the enzyme was first discovered in extracts from wheatgerm (Leloir and Cardini, 1955). Perhaps having the enzyme present in the mature seed avoids any lag in Suc synthesis when starch breakdown begins during germination. The induction of SPSII and SPSIV genes during late seed development (Fig. 7) could account for the activity in mature seeds.
The expression patterns of the SPS gene families in barley closely resemble those in wheat, but considering all six species together, we found no consistent trends that could suggest a specific or conserved function for any of the isoforms. Expression of the B-family genes in wheat and barley appears to be almost restricted to the anthers, whereas in maize and rice the B-family genes are widely expressed and probably encode the major SPS isoform in leaves (Worrell et al., 1991; Sakamoto et al., 1995; Valdez-Alarcon et al., 1996). The B-family SoSPS1 gene in sugarcane is also expressed in leaves (Sugiharto et al., 1997). In contrast, the C-family genes are widely expressed in wheat and barley and probably represent one of the major isoforms in leaves, whereas the C-family genes in rice, maize, sorghum, and sugarcane appear to be expressed very little, if at all (Supplemental Data, Appendix D). We know too little about the kinetic properties of the different SPS isoforms to understand what the physiological significance of these preferences might be, although it is interesting to note that the cool climate species, wheat and barley, differ from the warm climate species. It is also possible that the expression patterns of the SPS gene families have been modified during the domestication of these crop plants. The strong links between SPS and plant productivity suggest that the enzyme has indeed been under selective pressure, and investigation of the SPS gene families in wild species, particularly wild relatives of crop species, could give us a valuable insight into the process of domestication.
CONCLUSION
The grasses contain five families of SPS genes, corresponding to the previously described A, B, and C families in dicots plus a novel D family containing two subfamilies of genes. The D family appears to be restricted to the grasses and shows unique structural and regulatory features. There was overlap in the expression patterns of the different gene families in wheat, indicating that most Suc-synthesizing organs are likely to possess multiple isoforms of SPS. Although a complex picture emerges, we suggest that this analysis of SPS gene families provides the basis for future studies of the genetic and biochemical control of SPS in wheat and other grasses, along with the important agronomic traits that have been linked to this enzyme.
MATERIALS AND METHODS
Plant Material
Aegilops tauschii, durum wheat (Triticum turgidum subsp. durum cv Fransawi), and bread wheat (Triticum aestivum cvs Hartog and Cadenza) plants were grown in 20-cm plastic pots containing a mixture of soil, perlite, sand, and peat moss (50:25:15:10 by volume) and Osmocote slow release fertilizer, in a naturally illuminated glasshouse with 18°C day and 13°C night temperatures.
Identification of Wheat and Barley SPS Sequences
The maize SPS (GenBank accession no. M97550) was used to search the wheat sequences in the GenBank EST database using the TBLASTN program (Altschul et al., 1990). Ten of the corresponding cDNA clones, representing the diversity of sequences found, were obtained and fully sequenced. The sequences have been submitted to the GenBank database with accession numbers BE418274, AF347064, AF347065, AF347066, AF347067, AF347068, AF347069, AF354298, AF534907, and AY425710.
Further sequences were identified by reverse transcription (RT)-PCR using primers based on regions highly conserved in other SPS sequences. RNA was extracted from leaves of 4-week-old wheat (Triticum aestivum cv Cadenza) plants using Trizol reagent (Invitrogen, Paisley, UK). RT-PCR was carried out using SUPERSCRIPT II RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA) with the following primer pairs: (1) RT-PCR1R (first strand synthesis) and RT-PCR1F; (2) RT-PCR2R (first strand synthesis) and RT-PCR2F (Table III). The contiguous sequence of TaSPSIVa was extended by 5′-RACE using the 5′-RACE 1 primer (Table III) for first strand synthesis. After removal of the mRNA template by digestion with avian myeloblastosis virus ribonuclease H, the single stranded cDNA was tailed at the 3′ end by incubation with dATP and terminal transferase. Second strand cDNA synthesis and subsequent PCR were carried out using an oligo(dT) anchored forward primer and a nested reverse primer (5′-RACE 2, Table III).
Table III.
Primers used for cloning of the wheat SPSIVa sequence by RT-PCR and 5′-RACE and for synthesis of wheat SPS and SPP probe
| Purpose | Primer Sequence (5′ → 3′) |
|---|---|
| Wheat SPSIVa Cloning | |
| RT-PCR1(F) | CMWGARTTTGTYGAYGGWGC |
| RT-PCR1(R) | CGRCAYTCWCCAAAIGCTTTNAC |
| RT-PCR2(F) | GGTCATRCTTGCAAGGAAGC |
| RT-PCR2(R) | GAAGYTTATASAGTGCATCTGC |
| 5′-RACE1 | GTGAACGTTAAGTGCTCCAG |
| 5′-RACE2 | GCAGCAACTCCTGCACTGCC |
| Hybridization Probes | |
| SPS I (F) | TACTGCGACCACCCGTCG |
| SPS I (R) | GTCATGCCGGTCGAGAGC |
| SPS II (F) | TACGCTCAAGTCTAGACATCC |
| SPS II (R) | CAAAGGATGTATCTCTGATGC |
| SPS III (F) | TTGACTCTTGGCATGAGATCTCC |
| SPS III (R) | CAAAACCAGCAGGATGCATGC |
| SPS IV (F) | TTAACTCTTAGCCCAAGATACC |
| SPS IV (R) | CTGTAGGAGCCATGCCTGC |
| SPS V (F) | GGAATTCCAGGACCTATCG |
| SPS V (R) | GACAATTGGGTGTCAGACCGGACCG |
| SPP 1 (F) | AACTTATGGTGAAGCCATGG |
| SPP 1 (R) | CTTCGCATTTTCTGCATGCC |
F, forward; R, reverse.
Two barley SPS sequences, HvSPS2 and HvSPS3, were compiled from cDNA and EST sequences and submitted to the GenBank third party annotation database with accession numbers BK001785 and BK001784.
DNA Sequencing
Sequencing was carried out on both strands by the dideoxy chain termination method using Big Dye chain terminator chemistry (Applied Biosystems, Foster City, CA).
Maize Genomic Sequence Assembly
Maize SPS gene fragments were identified by searching the GenBank genome survey sequence database with the maize SPS (M97550), sugarcane SPS2 (AB001338), and rice SPS sequences, using the BLASTN and TBLASTN programs (Altschul et al., 1990). Maize genomic sequences with >97% identity were aligned, and consensus sequences were extracted to search the genome survey sequence database for overlapping gene fragments, which were assembled into contiguous sequences. This process was repeated in an iterative manner until no further overlapping sequences were found. Discontinuous assemblies of genomic sequence were linked by maize cDNA sequences, or by orthologous cDNA (EST) sequences from sorghum or sugarcane.
Phylogenetic Analysis
All full-length and partial amino acid sequences were aligned using ClustalX (1.8; Thompson et al., 1997). The aligned file was then edited manually using the GeneDoc program (Nicholas and Nicholas, 1997) into the following groups: (1) full-length sequences, (2) full-length monocot species, (3) glucosyltransferase domain (from Met1 to spinach Ile672 or maize Val678), (4) Suc-phosphatase domain (reading from spinach Arg770 or maize Arg777 to the stop codon), or (5) a 280-amino acid region from spinach Gly199 to Cys425 or maize Gly201 to Cys425 around the active site of the enzyme. This active site region is slightly larger than that used in Langenkämper et al. (2002). Minor corrections in alignment of sequences were also made manually. The alignment was reimported into ClustalX and neighbor joining tree algorithms, with and without gaps, with 1,000 bootstrap replicates carried out. This analysis did not allow outgroups or rooting of trees. Alignments were also saved as Phylip files and a subset (active site and full-length protein) analyzed using the neighbor joining algorithm or protein parsimony algorithm as implemented in PHYLIP 3.57 (http://evolution.genetics.washington.edu/phylip/doc/main; Felsenstein, 1996). Neighbor-joining trees were analyzed, with and without gaps, with 1,000 bootstrap replicates (using seqboot) and the consensus (majority rule) tree exported. Protein parsimony trees were analyzed after creating a protein distance matrix (without an evolutionary clock, using the Dayhoff PAM model of amino acid substitution) and the consensus tree exported. Trees were viewed in TreeView (Page, 1996). Clusters that were formed in more than 70% of the bootstrap samplings were regarded as significant, and the value of the node is indicated on the phylogram. Analyses of full-length sequences using the protein parsimony method were bootstrapped 500 times. Phylip-based trees were developed either with Synechocystis as an outgroup or without an outgroup or were rooted with Synechocystis. All trees were compared for fidelity of sequence association.
Southern-Hybridization Analysis
Southern-hybridization analysis was carried out as described by Aoki et al. (2002) using genomic DNA extracted from 1 g of young wheat tissue as described in Shure et al. (1983). Membranes were hybridized with radiolabeled DNA probes at 65°C for 16 h. DNA probes were prepared by PCR using the oligonucleotide primer pairs shown in Table III, with appropriate wheat SPS (TaSPS1, TaSPS2, TaSPS9, TaSPS7, and TaSPS5) and SPP (TaSPP1; Lunn, 2003) cDNA clones as templates. After hybridization, membranes were washed in 2× SSC (1× SSC: 150 mm NaCl and 15 mm trisodium citrate), 0.1% (w/v) SDS for 30 min at 65°C prior to exposure to x-ray film (Kodak Biomax, Coburg, Australia).
Northern-Hybridization Analysis
Northern-hybridization analysis was carried out as described by Aoki et al. (2002), using RNA extracted from 0.5 g of fresh wheat tissue as described in Wadsworth et al. (1988). Membranes were hybridized with radiolabeled DNA probes, prepared as described above, at 60°C for 16 h. After hybridization, membranes were washed in 0.2× SSC, 0.1% SDS for 30 min at 60°C and exposed to x-ray film.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers BE418274, AF347064, AF347065, AF347066, AF347067, AF347068, AF347069, AF354298, AF534907, AY425710, BK001785, and BK001784.
Supplementary Material
Acknowledgments
We thank Professors Sylvie Cloutier, Olin Anderson, and Yasunari Ogihara and their colleagues for the generous gifts of the wheat EST cDNA clones, and the members of the International Triticeae EST Cooperative, the International Rice Genome Sequencing Project, and the Maize Genome Sequencing Project for providing public access to unpublished sequence data. C.K.C. was supported by a CASE PhD studentship from the BBSRC.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042457.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aoki N, Whitfeld P, Hoeren F, Scofield G, Newell K, Patrick J, Offler C, Clarke B, Rahman S, Furbank RT (2002) Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol Biol 50: 453–462 [DOI] [PubMed] [Google Scholar]
- Bertin P, Gallais A (2001) Genetic variation for nitrogen use efficiency in a set of recombinant inbred lines. II – QTL detection and coincidence. Maydica 46: 53–68 [Google Scholar]
- Causse M, Rocher JP, Henry AM, Charcosset A, Prioul JL, de Vienne D (1995. a) Genetic dissection of the relationship between carbon metabolism and early growth in maize, with emphasis on key-enzyme loci. Mol Breed 1: 259–272 [Google Scholar]
- Causse M, Rocher J-P, Pelleschi S, Barrière Y, de Vienne D, Prioul J-L (1995. b) Sucrose-phosphate synthase: an enzyme with heterotic activity correlated with maize growth. Crop Sci 35: 995–1001 [Google Scholar]
- Chandler VL, Brendel V (2002) The maize genome sequencing project. Plant Physiol 130: 1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotelle V, Meek SEM, Provan F, Milne FC, Morrice N, MacKintosh C (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J 19: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L, Folco E, Desplats P, Abratti G, Limones V, Herrera-Estrella L, Salerno GL (1998) Sucrose-phosphate synthase from Synechocystis sp. PCC 6803: identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J Bacteriol 189: 6776–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R, Chamusco KC, Chourey PS (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmer J, Forster RL, Gibson JB, Shenk MA, Norriss MG, Glenn M, Saulsbury KM, Hall C (May 15, 2003) Compositions from the grasses Lolium perenne and Festuca arundinacea. Patent WO 03/040306 A2, pp 1–166
- Felsenstein J (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266: 418–427 [DOI] [PubMed] [Google Scholar]
- Fung RWM, Langenkämper G, Gardner RC, MacRae E (2003) Differential expression within an SPS gene family. Plant Sci 164: 459–470 [Google Scholar]
- Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154: 15–28 [Google Scholar]
- Hagemann M, Marin K (1999) Salt-induced sucrose accumulation is mediated by sucrose-phosphate synthase in cyanobacteria. J Plant Physiol 155: 424–430 [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 431–444 [DOI] [PubMed] [Google Scholar]
- Ingram J, Chandler JW, Gallagher L, Salamini F, Bartels D (1997) Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiol 115: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru K, Ono K, Kashiwagi T (2003) Identification of a new gene controlling plant height in rice using the candidate-gene approach. Planta 218: 388–395 [DOI] [PubMed] [Google Scholar]
- Klein RR, Crafts-Brandner SJ, Salvucci ME (1993) Cloning and developmental expression of the sucrose-phosphate synthase gene from spinach. Planta 190: 498–510 [DOI] [PubMed] [Google Scholar]
- Komatsu A, Takanokura Y, Omura M, Akihama T (1996) Cloning and molecular analysis of cDNAs encoding three sucrose phosphate synthase isoforms from a citrus fruit (Citrus unshiu Marc.). Mol Gen Genet 252: 346–351 [DOI] [PubMed] [Google Scholar]
- Langenkämper G, Fung RWM, Newcomb RD, Atkinson RG, Gardner RC, MacRae EA (2002) Sucrose phosphate synthase genes in plants belong to three different families. J Mol Evol 54: 322–332 [DOI] [PubMed] [Google Scholar]
- Leloir LF, Cardini CE (1955) The biosynthesis of sucrose phosphate. J Biol Chem 214: 157–165 [PubMed] [Google Scholar]
- Lunn JE (2002) Evolution of sucrose synthesis. Plant Physiol 128: 1490–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE (2003) Sucrose-phosphatase gene families in plants. Gene 303: 187–196 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Ashton AR, Hatch MD, Heldt HW (2000) Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase in plants. Proc Natl Acad Sci USA 97: 12914–12919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Furbank RT (1997) Localisation of sucrose-phosphate synthase and starch in leaves of C4 plants. Planta 202: 106–111 [DOI] [PubMed] [Google Scholar]
- Lunn JE, MacRae EA (2003) New complexities in the synthesis of sucrose. Curr Opin Plant Biol 6: 208–214 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Price GD, Furbank RT (1999) Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 40: 297–305 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Clauss MJ (2002) Plant evolutionary genomics. Curr Opin Plant Biol 5: 74–79 [DOI] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH (2001) A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52: 881–889 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr. (1997) GeneDoc. A tool for editing and annotating multiple sequence alignments: Multiple sequence alignment editor and shading utility version 2.6.002. http://www.psc.edu/biomed/genedoc
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Prioul J-L, Pelleschi S, Séne M, Thévenot C, Causse M, de Vienne D, Leonardi A (1999) From QTLs for enzyme activity to candidate genes in maize. J Exp Bot 50: 1281–1288 [Google Scholar]
- Quy LV, Champigny ML (1992) NO3− enhances the kinase activity for phosphorylation of phosphoenolpyruvate carboxylase and sucrose phosphate synthase proteins in wheat leaves; evidence from the effects of mannose and okadaic acid. Plant Physiol 99: 344–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimholz R, Geigenberger P, Stitt M (1994) Sucrose-phosphate synthase is regulated via metabolites and protein phosphorylation in potato tubers, in a manner analogous to the enzyme in leaves. Planta 192: 480–488 [Google Scholar]
- Rocher JP, Prioul JL, Lecharny A, Reyss A, Joussaume M (1989) Genetic variability in carbon fixation, sucrose-P-synthase and ADP glucose pyrophosphorylase in maize plants of differing growth rate. Plant Physiol 89: 416–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein D, Dieuaide-Noubhani M, Dufourc EJ, Raymond P, Rolin D (2002) The metabolic architecture of plant cells. Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 277: 43948–43960 [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Satozawa T, Kishimoto N, Higo K, Shimada H, Fujimura T (1995) Structure and RFLP mapping of a rice sucrose-phosphate synthase (SPS) gene that is specifically expressed in the source organ. Plant Sci 112: 207–217 [Google Scholar]
- Sarquís JI, Gonzalez H, Sánchez de Jimenez E, Dunlap JR (1998) Physiological traits associated with mass selection for improved yield in a maize population. Field Crops Res 56: 239–246 [Google Scholar]
- Schnyder H (1993) The role of carbohydrate storage and redistribution in the source-sink relations of wheat and barley during grain filling: a review. New Phytol 123: 233–245 [Google Scholar]
- Seneweera SP, Basra AS, Barlow EW, Conroy JP (1995) Diurnal regulation of leaf blade elongation in rice by CO2. Plant Physiol 108: 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M, Wessler S, Federoff N (1983) Molecular identification and isolation of the Waxy locus in maize. Cell 35: 225–233 [DOI] [PubMed] [Google Scholar]
- Sonnewald U, Quick WP, MacRae E, Krause KP, Stitt M (1993) Purification, cloning and expression of spinach leaf sucrose-phosphate synthase in Escherichia coli. Planta 189: 174–181 [DOI] [PubMed] [Google Scholar]
- Sugiharto B, Sakakibara H, Sumadi, Sugiyama T (1997) Differential expression of two genes for sucrose-phosphate synthase in sugarcane: molecular cloning of the cDNAs and comparative analysis of gene expression. Plant Cell Physiol 38: 961–965 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevanion SJ, Castleden CK, Foyer CH, Furbank RT, Quick WP, Lunn JE (2004) Regulation of sucrose-phosphate synthase in wheat (Triticum aestivum) leaves. Funct Plant Biol (in press) [DOI] [PubMed]
- Valdez-Alarcon JJ, Ferrando M, Salerno G, Jimenez-Moraila B, Herrera-Estrella L (1996) Characterization of a rice sucrose-phosphate synthase encoding gene. Gene 170: 217–222 [DOI] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG (1988) A procedure for the small scale isolation of plant RNA suitable for RNA suitable for RNA blot analysis. Anal Biochem 172: 279–283 [DOI] [PubMed] [Google Scholar]
- Wardlaw IF, Willenbrink J (1994) Carbohydrate storage and mobilisation by the culm of wheat between heading and grain maturity: the relation to sucrose synthase and sucrose-phosphate synthase. Aust J Plant Physiol 21: 255–271 [Google Scholar]
- White S, Doebley J (1998) Of genes and genomes and the origin of maize. Trends Genet 14: 327–332 [DOI] [PubMed] [Google Scholar]
- Worrell AC, Bruneau JM, Summerfelt K, Boersig M, Voelker TA (1991) Expression of maize sucrose phosphate synthase in tomato alters leaf carbohydrate partitioning. Plant Cell 3: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu SN, Wang J, Li SG, Wong KSG, Liu B, Deng Y, Dai L, Zhou Y, Zhang XQ, et al (2001) A draft sequence of the rice (Oryza sativa sp indica) genome. Chin Sci Bull 46: 1937–1942 [Google Scholar]
- Zhu YJ, Komor E, Moore PH (1997) Sucrose accumulation in the sugarcane stem is regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase. Plant Physiol 115: 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.