Abstract
The life cycle of flowering plants alternates between a diploid sporophytic and a haploid gametophytic generation. After fertilization of each the egg and central cells by one male gamete, the development of both fertilization products occurs coordinated with the maternally derived seed coat and carpel tissues forming the fruit. The reproduction program is likely to involve the concerted activity of many genes. To identify genes with specific functions during reproduction, we have analyzed the expression profile of more than 22,000 genes present on the Arabidopsis ATH1 microarray during three stages of flower and fruit development. We found 1,886 genes regulated during reproductive development and 1,043 genes that were specifically expressed during reproduction. When compared to cells from an Arabidopsis suspension culture, S-phase genes were underrepresented and G2 and M-phase genes were strongly enriched in the set of specific genes, indicating that important functions during reproduction are exerted in the G2 and M phases of the cell cycle. Many potential signaling components, such as receptor-like protein kinases, phosphatases, and transcription factors, were present in both groups of genes. Members of the YABBY, MADS box, and Myb transcription factor families were significantly overrepresented in the group of specific genes, revealing an important role of these families during reproduction. Furthermore, we found a significant enrichment of predicted secreted proteins smaller than 15 kD that could function directly as signaling molecules or as precursors for peptide hormones. Our study provides a basis for targeted reverse-genetic approaches aimed to identify key genes of reproductive development in plants.
During evolution, both animals and plants developed distinct pathways of sexual reproduction. The haploid life cycle starts with the formation of the meiotic products, for instance arrested primary oocytes and spermatocytes in mammals or micro- and megaspores in plants. The haploid phase ends when a sperm cell fuses with an egg cell to form the diploid zygote, which initiates embryo development. In animals, gametes are derived from a population of germ cells committed early during development to their reproductive fate and which usually rest until the onset of sexual maturity. By contrast, stem cells in shoot apical meristems of flowering plants divide continuously during postembryonic development. Only after formation of vegetative organs, the meristem produces specialized reproductive organs in which meiosis occurs—the stamens and carpel-borne ovules of the flower (for review, see Bowman and Eshed, 2000; Carles and Fletcher, 2003). As early as 1790, Goethe suggested that floral organs are derived from leaves and adapted to their specialized tasks (Goethe, 1790).
Animal gametes differentiate directly from the products of meiosis, whereas in plants the meiotic products (spores) typically undergo two or three mitotic divisions to give rise to the male or female gametophytes, respectively (Drews et al., 1998; Grossniklaus and Schneitz, 1998). In several plant species, including Arabidopsis, the female gametophyte consists of seven cells: three antipodal cells, two synergid cells, one egg cell, and one central cell that contains two polar nuclei (Drews et al., 1998; Grossniklaus and Schneitz, 1998). The mature male gametophyte comprises two sperm cells contained within a vegetative cell (McCormick, 1993; da Costa-Nunes and Grossniklaus, 2003).
During fertilization one sperm cell nucleus fuses with the egg cell nucleus, giving rise to the diploid embryo, whereas the other sperm cell fuses with the homodiploid central cell nucleus, generating the triploid endosperm (Drews and Yadegari, 2002). Subsequent seed and fruit development are highly coordinated processes between the embryo and endosperm, as well as the maternally derived testa (seed coat) and carpels. Because of the significant economical importance of flowers, seeds, and fruits, the complex processes involved in plant reproduction are not only of academic interest but also of high economic relevance.
Many genes have been identified genetically that have a function in plant reproduction. The establishment of floral organ identity and initiation of seed development have been studied extensively, and many regulators of these processes have been identified (McElver et al., 2001; Zik and Irish, 2003; for review, see Chaudhury et al., 2001). High-throughput RNA profiling technologies can now complement the genetic and molecular approaches to provide new insights into plant reproduction. In particular, oligonucleotide-based microarrays can produce reliable, high-quality data (Lockhart et al., 1996; Hennig et al., 2003) to establish new biological knowledge on the transcriptional programs that are active during developmental processes (Spellman et al., 1998; Harmer et al., 2000; Becker et al., 2003; Honys and Twell, 2003). Here, we report a comprehensive Affymetrix GeneChip analysis of the Arabidopsis transcriptome at key steps of reproductive development. We identified 1,043 genes that were specifically expressed during reproduction. Among those genes were many potential signaling components, such as receptor-like protein kinases, phosphatases, and transcription factors. In addition, we found a significant enrichment of predicted secreted proteins smaller than 15 kD that could function directly as signaling molecules or as precursors for peptide hormones. Our study provides the basis for targeted reverse-genetic approaches aimed to identify important regulators of reproductive development in plants.
RESULTS AND DISCUSSION
Identification of Genes Involved in Plant Reproductive Development
Arabidopsis wild-type plants were grown under standard growth conditions for 6 weeks, and closed flower buds shortly before pollination (stage I), open pollinated flowers (stage II), and siliques 2 d after pollination (stage III) were harvested (Fig. 1). A fraction of each sample was cleared and analyzed by microscopy to ensure that homogenous developmental stages were used. We focused on the developmental stage of the female gametophyte before fertilization and the embryo and endosperm after fertilization. Stage I samples contained flowers with fully developed female gametophytes. Stage II samples contained developing seeds with only a few endosperm nuclei and either a zygote or a one-celled embryo proper, while stage III samples contained seeds with an embryo proper of four to eight cells.
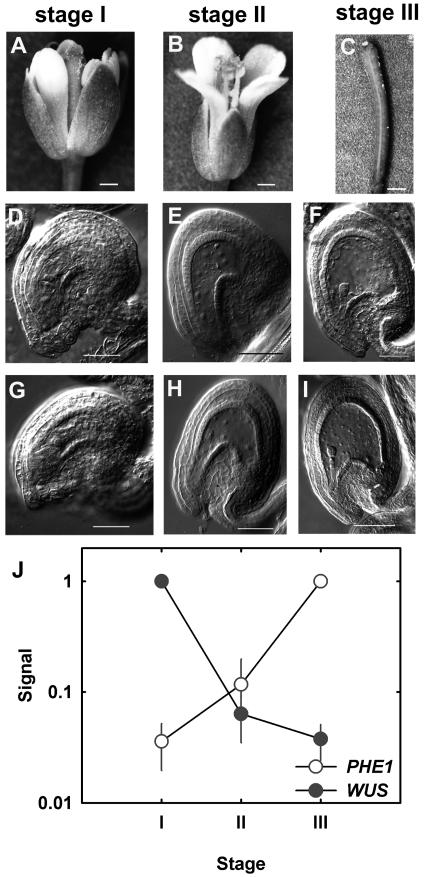
Figure 1.
Developmental stages used for this study. The developmental stages of flowers (A and B) and siliques (C) shown in sections A to C correspond to the pictures of cleared gametophytes and seeds shown below in sections D to I. A, Closed flower (flower was manually opened to show development of gynoecium and anthers). B, Open flower after fertilization. C, Open flower after fertilization. D, Cleared female gametophytes before fusion of central-cell nuclei. E, Cleared seeds containing only endosperm nuclei. F, Cleared seeds containing four-cell embryos. G, Cleared female gametophytes after fusion of central-cell nuclei. H, Cleared seeds containing only endosperm nuclei and one-celled embryo proper. I, Cleared seeds containing eight-cell embryos. Bars: A, 290 μm; B, 360 μm; C, 600 μm; D to I, 50 μm. J, Expression profiles of PHERES and WUSCHEL.
The experiment was performed in duplicate and labeled RNA samples were hybridized individually to Affymetrix ATH1 microarrays. First, we wanted to estimate the reliability of our data, in particular the similarity of the replicates. The coefficient of variation (cv) is commonly used as a quantitative measure of data quality (Piper et al., 2002; Jiao et al., 2003). The replicate measurements had cv values of 0.15 to 0.17 (Table I). These numbers are similar to previous experiments (Köhler et al., 2003) and considerably smaller than those reported in a recent study on reproducibility of yeast (Saccharomyces cerevisiae) transcriptome measurements (Piper et al., 2002). Expression of several regulatory genes during reproductive development is well documented. Such genes can serve as an independent quality test for the microarray data. Both WUSCHEL, a gene required for ovule development (Gross-Hardt et al., 2002), and PHERES1, a gene involved in early processes of seed development (Köhler et al., 2003), showed the expected expression pattern (Fig. 1J). Similarly, expression profiles of MEDEA, AGAMOUS, and SUPERMAN resembled previously published data (Drews et al., 1991; Grossniklaus et al., 1998; Sakai et al., 2000) demonstrating the validity of the microarray approach (data not shown).
Table I.
Reproducibility of microarray data
| Stage I | Stage II | Stage III | Seedlingsa | |
|---|---|---|---|---|
| cv | 0.17 | 0.15 | 0.14 | 0.19 |
Coefficients of variation are shown for different hybridizations as described in the text.
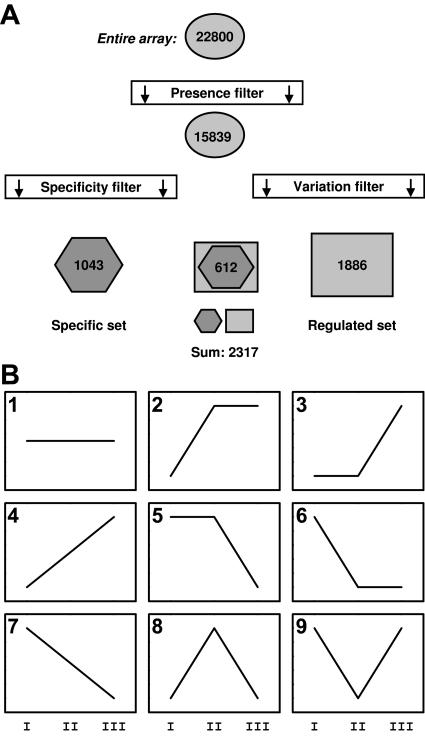
The entire data set of the transcriptional profiling experiment is available at the authors' homepage and in the supplemental online material, which can be viewed at www.plantphysiol.org. Transcripts of 15,454 genes were detected in flowers (P < 0.04 in both measurements of either stage I or stage II). Similarly, 13,367 genes were expressed significantly in seedlings or leaves. Most genes were expressed in both leaves and flowers, but transcripts from 13% of the flower genes were never detected in leaves or seedlings. By contrast, only 4% of the transcripts found in leaf and seedling samples were not detected in flowers. Interestingly, flowers expressed mainly the same genes as leaves, and there were many more flower-specific than leaf- or seedling-specific genes, which is in agreement with the hypothesis that flower organs represent modified leaves as was proposed by Goethe more than 200 years ago (Goethe, 1790). Based on all our samples (stages I, II, and III), transcripts of 15,839 genes were detected (P < 0.04), and these genes were called present genes (Table SI). To eliminate unchanged house-keeping genes, we selected from the set of present genes only those genes for which transcripts in both replicates were either decreased (P > 0.975, signal log ratio < −1) or increased (P < 0.025, signal log ratio > 1) between any two stages (stage I versus II, stage I versus III, stage II versus III; Fig. 2A). This variation filter revealed 1,886 genes changed during reproductive development (regulated genes). To identify genes that are predominantly expressed in reproductive organs and less important in the vegetative phase, we compared data from this profiling experiment to data from other experiments of our laboratory and data obtained from Nottingham Arabidopsis Stock Centre (Craigon et al., 2004). The data set includes two replicates each of 14-d-old seedlings and rosette leaves of 25-d-old plants, and four root samples (in total eight microarrays). We filtered genes that were included in the set of present genes in our samples and had no detectable transcripts (P ≥ 0.04) in all leaf, seedling, and root samples. For a gene to be included in our list, we also required that its minimal expression value (signal) in flowers should be at least twice as high as the maximal signal in any other sample. This filtering (specificity filter) identified 1,043 genes that we consider reproduction related because their expression is highest in reproductive organs (Supplemental Table II, specific genes).
Figure 2.
Summary of data analysis strategies. A, Summary of filtering strategies (for details see text). B, Models of dynamic gene expression patterns.
Functional Classification of Genes Involved in Reproductive Development
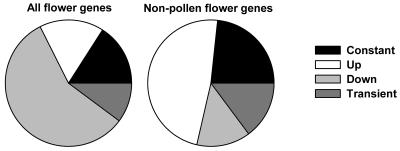
Because our set of specific genes also includes genes with constant transcript amounts during the experiment, we used a regression approach to identify dynamic expression patterns. We expected that any gene in the specific set should fall into one of nine different models (Fig. 2B). Accordingly, each gene was assigned to the model class it fit best. From the 1,043 specific genes, expression of 166 did not change significantly over the time course assayed in the experiment, 172 were up-regulated, and 599 were down-regulated during flower and silique development. A total of 106 genes were transiently induced or repressed (Fig. 3). If genes that are expressed in anthers and pollen (Honys and Twell, 2003; Craigon et al., 2004) are excluded from the set of specific genes, 189 specific nonpollen genes remained. Thus, genes expressed in pollen contribute most to the set of floral-specific genes. Because only 14% of the nonpollen genes are down-regulated and half of them are up-regulated, mainly following kinetics of model 3, genes involved in early stages of seed development dominate the set of 189 remaining specific genes. Of the 1,043 specific genes, 244 have no close homolog (E < −30), and 58 of the 189 specific nonpollen genes have no close homolog, suggesting a specialized function and qualifying them as good candidates for targeted reverse-genetic approaches. Functional categories were assigned to genes from the different model classes of specific genes. The results in Table II show that transcriptional reprogramming during reproductive development involves genes from all major functional classes. Significant enrichment was detected for genes encoding proteins involved in metabolism, transcription, and cellular organization.
Figure 3.
Distribution of reproductive genes in categories. Genes found to be specific for reproduction were grouped according to their expression profile. Shown are the relative group sizes for all such genes or only for genes not detected in anthers and pollen.
Table II.
Distribution of reproductive genes in categories
| Category | All | Constant | Up | Down | Transient |
|---|---|---|---|---|---|
| %a | |||||
| Cell cycle | 4.0 | 6.0 (22) | 2.9 (11) | 2.0 (28) | 1.9 (5) |
| Cell fate | 3.3 | 4.7 (17) | 2.9 (11) | 2.7 (37) | 4.2 (11) |
| Cell defense | 6.5 | 4.9 (18) | 7.2 (27) | 5.8 (79) | 9.9 (26) |
| Cellular communication | 5.6 | 4.9 (18) | 2.9 (11) | 6.1 (83) | 2.3 (6) |
| Cellular transport | 4.2 | 5.2 (19) | 2.4 (9) | 4.6 (63) | 3.1 (8) |
| Cellular organization | 3.9 | 3.6 (13) | 5.9 (22) | 6.7 (92) | 5.0 (13) |
| Energy | 2.6 | 1.1 (4) | 2.1 (8) | 1.9 (26) | 2.3 (6) |
| Metabolism | 9.5 | 6.6 (24) | 13.3 (50) | 14.3 (195) | 15.6 (41) |
| Protein activity regulation | 0.1 | 0.0 (0) | 0.0 (0) | 0.1 (1) | 0.0 (0) |
| Protein fate | 5.2 | 4.9 (18) | 7.2 (27) | 4.0 (55) | 3.8 (10) |
| Protein synthesis | 2.1 | 2.2 (8) | 1.1 (4) | 0.7 (9) | 1.5 (4) |
| Interaction with cellular environment | 1.5 | 1.1 (4) | 0.8 (3) | 1.8 (25) | 0.0 (0) |
| Storage protein | 0.1 | 0.0 (0) | 0.3 (1) | 0.3 (4) | 0.0 (0) |
| Systemic interaction with environment | 0.8 | 1.1 (4) | 0.8 (3) | 1.0 (13) | 1.5 (4) |
| Transcription | 7.6 | 12.1 (44) | 7.4 (28) | 5.0 (69) | 4.6 (12) |
| Transport facilitation | 2.5 | 1.6 (6) | 2.4 (9) | 3.4 (47) | 1.5 (4) |
| Transposable elements | 1.3 | 0.5 (2) | 0.3 (1) | 0.6 (8) | 0.0 (0) |
| Unclassified proteins | 39.2 | 39.5 (144) | 40.2 (151) | 39.0 (534) | 42.7 (112) |
Frequency of annotated genes having defined automatically derived functional categories is shown. Total number of genes is given in parentheses. Note that one gene can have more than one annotated function. Numbers in bold indicate significant enrichment (P < 0.05 after step-down Bonferoni correction for multiple testing).
Metabolic Regulation during Reproductive Development
Many of the genes regulated during reproductive development encode enzymes for metabolic functions. During later stages of seed development, specific pathways are activated for seed filling (Girke et al., 2000; Ruuska et al., 2002; Sreenivasulu et al., 2004). To detect underlying patterns among the various transcriptional changes of genes encoding metabolic enzymes, we used visualization tools to map gene expression data to the metabolic network (AraCyc, Mueller et al., 2003; PathwayMap, M. Hirsch-Hoffman, O. Laule, A. Fürholz, and W. Gruissem, unpublished data). Visual inspection of the data revealed dominating effects in a few metabolic pathways. Most obviously, expression of genes for the central cellular catabolism, in particular glycolysis and citrate cycle, including pyruvate kinase, phosphoglycerate mutase, citrate synthase, and malate dehydrogenase, was strongly reduced over the time course analyzed in the experiment. Similar to observations in developing seeds (Ruuska et al., 2002), not all glycolytic enzymes were regulated similarly. In addition, many genes for enzymes in anabolic pathways were down-regulated. In this case, however, usually only one gene was affected per pathway. Reductions were observed in pathways for biosynthesis of various amino acids and fatty acid biosynthesis. In part, these observations could reflect the exceptional high metabolic activity of developing pollen that dominated in stage I. Surprisingly, chlorophyll biosynthesis was consistently induced throughout the time series. In addition to most enzymes from the porphyrin pathway itself, also enzymes supplying building blocks for this pathway were induced. This included, for instance, enzymes from the nonmevalonate isoprenoid pathway, cystathione gamma synthetase, which is involved in S-adenosylmethionine generation and Asp aminotransferase, which serves to recycle generated ammonium. Oxygen and ATP are limiting in developing embryos, and photosynthesis in embryos is important for oxygen supply (Eastmond et al., 1996; Ruuska et al., 2002; Borisjuk et al., 2003; Rolletschek et al., 2003). Although expression of genes for chlorophyll biosynthesis was increased, no significant changes were found for photosynthesis genes (data not shown). Other authors reported an expression peak of genes for photosynthesis only 4 to 8 d after fertilization (Sreenivasulu et al., 2004). Therefore, active chlorophyll biosynthesis is established before the core photosynthetic genes become activated. Moreover, it is likely that after pollination, not only the embryo but also the maternal tissues in testa and silique walls strongly increase their photosynthetic capacity and contribute actively to seed development. This hypothesis is supported by measurements of Brassica napus silique wall metabolism (King et al., 1997).
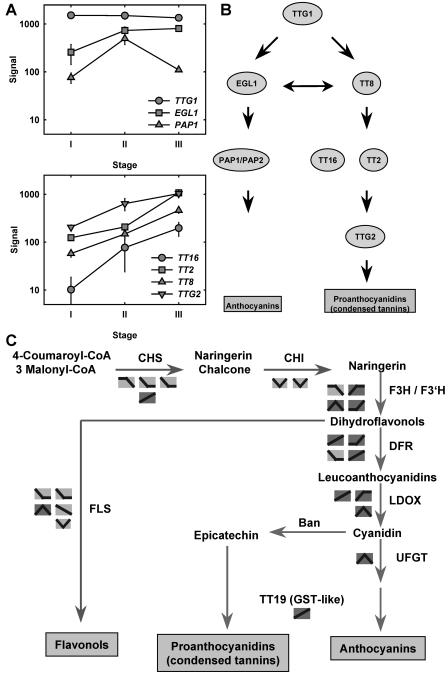
A second group of metabolic pathways found to be strongly affected were flavonoid and anthocyanin biosynthesis. These pathways are controlled by a hierarchy of several transcription factors (Marles et al., 2003; Fig. 4). TRANSPARENT TESTA GLABROUS 1 (TTG1) forms the top of this hierarchy and is also required for other functions (Zhang et al., 2003). The TTG1 gene is strongly expressed in the sampled tissue, but no significant regulation was observed (Fig. 4A). PAP1, encoding a specific regulator of the anthocyanin-generating branch, is transiently induced. EGL1, which is also involved in activating the branch leading to anthocyanins, is transiently increased from stage I to stage II and then remains constant. By contrast, expression of TT8, TT2, TT16, and TTG2, which are all required to activate genes involved in the proanthocyanin synthesis pathway, are continuously up-regulated. This pattern of expression is mirrored by the expression of structural genes: the flavonoid branch is predominantly down-regulated, the anthocyanin branch is transiently induced, and the proanthocyanin branch is continuously activated (Fig. 4C). These observations provide a molecular and genomic basis for previous physiological observations regarding the importance of flavonoid biosynthesis during flower development (Shirley, 1996). Flavonoids are synthesized in several floral organs and are required for pollen function, anthocyanins are transiently expressed in Arabidopsis pistils after pollination, and proanthocyanins are used in the developing testa to form condensed tannins (Xie et al., 2003).
Figure 4.
Flavonoid metabolism during reproduction. A, Expression profiles of transcription factors regulating flavonoid biosynthetic genes during reproduction. TTG1/2, TRANSPARENT TESTA GLABROUS 1/2; TT2/8/16, TRANSPARENT TESTA 2/8/16; EGL1, ENHANCER OF GLABRA 1; PAP1/2, PRODUCTION OF ANTHOCYANIN PIGMENT 1/2. B, Scheme of the hierarchy of transcription factors regulating flavonoid biosynthetic genes (modified after Zhang et al., 2003). C, Scheme of flavonoid biosynthesis and observed changes of gene expression during reproduction. CHS, CHALCONE SYNTHASE; CHI, CHALCONE ISOMERASE; F3H, FLAVANONE 3-β-HYDROXYLASE; F3′H, FLAVONOID 3′-HYDROXYLASE; DFR, DIHYDROFLAVONOL REDUCTASE; LDOX, LEUCOANTHOCYANIDIN OXIDASE; BAN, BANYULS; UFGT, UDP-FLAVONOID GLYCOSYL TRANSFERASE; TT19, TRANSPARENT TESTA 19.
Cell Cycle Activity during Development
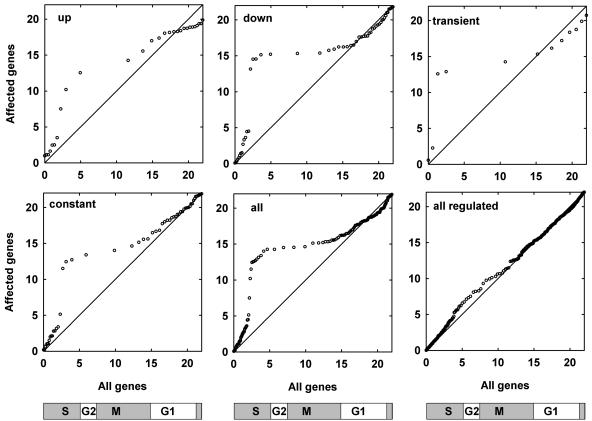
Based on the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000), 61 genes were described belonging to a core set of cell cycle regulators (Vandepoele et al., 2002). Fifty-five of them are present on the ATH1 microarray, and 51 were in the set of present genes. However, not a single core cell cycle gene belonged to the regulated genes. The proteins encoded by the genes in the core set fulfill basic cellular functions and are therefore not strongly regulated during the developmental time course tested. Because flower and fruit development involve high cell proliferation activity, we asked in which cell cycle phase the selected genes are preferentially expressed. Based on a published data set (Menges et al., 2003), we constructed quantile-quantile plots for specific genes with particular expression profiles and for the set of all genes with a reliably changing expression in the synchronized cell suspension culture (Fig. 5). The expression maxima of the genes from the regulated set did not deviate substantially from the maxima distribution of all genes expressed in suspension cells. However, the set of specific genes, as well as the subsets for all particular expression models, contained significantly less S-phase genes than the reference set. This underrepresentation of S-phase genes was accompanied by a strong enrichment for G2 and M-phase genes. This suggests that S-phase during reproductive development relies on proteins that are important in other tissues as well. By contrast, G2 and M phase involve many specific proteins. Thus, flower-specific functions are particularly important during G2 and M phase of the cell cycle. Such functions could, for instance, involve control of the orientation of the division plane, which is essential for plant morphogenesis.
Figure 5.
Cell cycle regulation of reproductive genes. Quantile-quantile plots were constructed based on various subsets of reproductive genes that were expressed also in a cell-suspension culture. For each gene of a subset, times of peak expression during the 22-h cell cycle were used and plotted versus all cell cycle regulated genes in the suspension culture. Data points along the diagonal indicate a similar distribution of expression peaks of the selected subset and the entire population of cell cycle regulated genes. A slope ≫1 results if fewer genes than expected from the subset show maximal expression within a certain time window. Conversely, a slope close to 0 shows that more genes than expected from the subset show maximal expression within this time window. The schemes below the plots indicate cell cycle phases.
Protein Kinases, Transcription Factors, and Other Putative Signaling Proteins
The set of genes regulated during reproductive development contained many potential signal transduction components. Among them were 14 phosphatases, 61 protein kinases, and 118 transcription factors. Similarly, among the specific reproductive genes were 5 phosphatases, 42 protein kinases, and 64 transcription factors. Most of the protein kinases preferentially expressed in flowers are receptor-like kinases, suggesting an important role of receptor-like kinase signaling during reproductive development. This phase of plant development involves many processes depending on cell-cell communication (e.g. pollen tube growth, guidance, and reception; double fertilization; development of embryo and endosperm; development and growth of testa and carpel walls; and floral organ abscission) that are likely to involve many receptor-like kinases.
Genes predicted to encode transcription factors were enriched in some subfamilies of specific genes. We wondered whether particular transcription factor families might be more important for reproductive development than others. The Arabidopsis Gene Regulatory Information Server database of Arabidopsis transcription factors (Davuluri et al., 2003) lists 1,467 genes encoding transcription factors from 35 families. To identify families that are particularly important in reproductive development, we determined how many members of a given family belong to the set of regulated genes (Table III). We selected overrepresented families based on a combination of absolute and relative filter criteria: If less than two genes from a given family were present in this set, the family was ignored; if two to 10 genes were present and this number equaled at least 30% of the entire family size, the family was selected. Similarly, the family was selected if more than 10 genes were present and they represented at least 5% of the family size. Based on these filters, we found seven families particularly important during reproductive development (selected from the set of regulated genes; Table III). These are the C2C2-YABBY family, MADS family, NAC family, CCAAT-HAP3 family, MYB family, AP2-EREBP family, and the bHLH family. By contrast, transcripts for members of several other TF families were abundant in the tested samples, but none of them varied during development, demonstrating a more basic function in cellular metabolism and proliferation. The E2F/DP family, for instance, has eight members in Arabidopsis; six of them are present on the ATH1 microarray, and transcripts for all six were detected in the samples. However, none of them was regulated. Similarly, none of the 21 GRAS family members, the 21 DOF family members, or the 11 HSF family members changed abundance during the time course investigated.
Table III.
Regulated transcription factors
| Family Name | Total | On Chip | Detected | Percent | Regulated | Percent | P Value |
|---|---|---|---|---|---|---|---|
| C2C2-YABBY | 5 | 4 | 4 | 100.0 | 3 | 75.0 | 0.005 |
| MADS | 100 | 75 | 38 | 50.7 | 14 | 18.7 | 0.005 |
| NAC | 90 | 83 | 44 | 53.0 | 13 | 15.7 | 0.023 |
| CCAAT-HAP3 | 10 | 8 | 6 | 75.0 | 3 | 37.5 | 0.030 |
| MYB | 137 | 129 | 68 | 52.7 | 17 | 13.2 | 0.044 |
| AP2-EREBP | 120 | 108 | 46 | 42.6 | 12 | 11.1 | 0.190 |
| bHLH | 146 | 107 | 76 | 71.0 | 10 | 9.3 | 0.392 |
| C2C2-Gata | 28 | 20 | 16 | 80.0 | 3 | 15.0 | 0.231 |
| C2H2 | 98 | 85 | 57 | 67.1 | 9 | 10.6 | 0.274 |
| WRKY | 69 | 58 | 34 | 58.6 | 8 | 13.8 | 1.000 |
| GRF | 9 | 8 | 7 | 87.5 | 2 | 25.0 | 0.143 |
| SBP | 16 | 16 | 16 | 100.0 | 2 | 12.5 | 0.381 |
| TUB | 10 | 11 | 10 | 90.9 | 1 | 9.1 | 0.113 |
| HB | 64 | 63 | 42 | 66.7 | 5 | 7.9 | 0.596 |
| Trihelix | 29 | 27 | 22 | 81.5 | 2 | 7.4 | 0.597 |
| bZIP | 70 | 64 | 47 | 73.4 | 4 | 6.3 | 0.774 |
| ABI3VP1 | 18 | 17 | 11 | 64.7 | 1 | 5.9 | 0.755 |
| C3H | 163 | 124 | 85 | 68.5 | 7 | 5.6 | 0.886 |
| C2C2-CO-like | 30 | 24 | 16 | 66.7 | 1 | 4.2 | 0.863 |
| G2-like | 44 | 36 | 24 | 66.7 | 1 | 2.8 | 0.949 |
| E2F-DP | 8 | 6 | 6 | 100.0 | 0 | 0.0 | 1.000 |
| CCAAT-DR1 | 2 | 2 | 2 | 100.0 | 0 | 0.0 | 1.000 |
| Alfin-like | 7 | 7 | 7 | 100.0 | 0 | 0.0 | 1.000 |
| ARF | 22 | 18 | 17 | 94.4 | 0 | 0.0 | 1.000 |
| TCP | 26 | 20 | 18 | 90.0 | 0 | 0.0 | 0.653 |
| GRAS | 25 | 25 | 21 | 84.0 | 0 | 0.0 | 1.000 |
| CPP | 8 | 6 | 5 | 83.3 | 0 | 0.0 | 1.000 |
| MYB-related | 9 | 8 | 6 | 75.0 | 0 | 0.0 | 1.000 |
| CCAAT-HAP5 | 13 | 10 | 7 | 70.0 | 0 | 0.0 | 1.000 |
| Orphan | 3 | 3 | 2 | 66.7 | 0 | 0.0 | 1.000 |
| C2C2-Dof | 36 | 32 | 21 | 65.6 | 0 | 0.0 | 1.000 |
| ARR-B | 15 | 12 | 7 | 58.3 | 0 | 0.0 | 1.000 |
| HSF | 21 | 21 | 11 | 52.4 | 0 | 0.0 | 1.000 |
| EIL | 6 | 6 | 3 | 50.0 | 0 | 0.0 | 1.000 |
| CCAAT-HAP2 | 10 | 10 | 5 | 50.0 | 0 | 0.0 | 1.000 |
The same analysis was carried out for the genes found to be specifically expressed during reproductive development (Table IV). In this case, three families of transcription factors were found to be particularly important for plant reproduction: YABBY transcription factors (2 out of 4), MADS box genes (15 out of 75), and MYB transcription factors (11 out of 129) belong to this set. Heat shock factors, response regulators, E2F/DP-like proteins, and 19 other families of transcription factors had no or only one member in the set of specific genes. Genetic studies have shown that YABBY and MADS box transcription factors play important roles during floral organ development. YABBY transcription factors specify abaxial cell fate in floral organs and ovules, whereas MADS box genes specify flower organ identity and interact genetically with YABBY genes (Golz and Hudson, 1999; Bowman, 2000; Theissen et al., 2000; Ng and Yanofsky, 2001). This important role of MADS box and YABBY genes is reflected at the transcriptional level. Interestingly, among the specifically expressed MADS box genes are several genes belonging to the type I class (AGL30, AGL33, AGL35, PHE1, AGL66, and AGL104; De Bodt et al., 2003; Parenicova et al., 2003). Even though there are more type I (68) than type II MADS box genes (MIKC class, 39), only one type I gene (PHE1) has been functionally characterized to date (Köhler et al., 2003). However, the evolutionary history of type I genes led to the conclusion that this subclass of MADS box genes might have important roles in Arabidopsis that are currently not well understood (Alvarez-Buylla et al., 2000; Martinez-Castilla and Alvarez-Buylla, 2003; Nam et al., 2004). Our data suggest that some of these genes function in reproductive development, and alterations in their sequences would likely impact reproductive output, i.e. plant fitness. This interpretation could explain why the signature of positive Darwinian selection was frequently found among type I MADS box genes (Martinez-Castilla and Alvarez-Buylla, 2003).
Table IV.
Specific transcription factors
| Family Name | Total | On Chip | Detected | Percent | Flower Genes | Percent | P Value |
|---|---|---|---|---|---|---|---|
| MADS | 100 | 75 | 38 | 50.7 | 15 | 20.0 | 0.000 |
| MYB | 137 | 129 | 68 | 52.7 | 11 | 8.5 | 0.038 |
| C2C2-YABBY | 5 | 4 | 4 | 100.0 | 2 | 50.0 | 0.015 |
| Trihelix | 29 | 27 | 22 | 81.5 | 4 | 14.8 | 0.037 |
| SBP | 16 | 16 | 16 | 100.0 | 2 | 12.5 | 0.167 |
| ABI3VP1 | 18 | 17 | 11 | 64.7 | 2 | 11.8 | 0.183 |
| GRF | 9 | 8 | 7 | 87.5 | 1 | 12.5 | 0.306 |
| CCAAT-HAP3 | 10 | 8 | 6 | 75.0 | 1 | 12.5 | 0.306 |
| TUB | 10 | 11 | 10 | 90.9 | 1 | 9.1 | 0.395 |
| C2H2 | 98 | 85 | 57 | 67.1 | 7 | 8.2 | 0.099 |
| bHLH | 146 | 107 | 76 | 71.0 | 7 | 6.5 | 0.222 |
| NAC | 90 | 83 | 44 | 53.0 | 4 | 4.8 | 0.526 |
| WRKY | 69 | 58 | 34 | 58.6 | 2 | 3.4 | 0.743 |
| HB | 64 | 63 | 42 | 66.7 | 2 | 3.2 | 0.783 |
| bZIP | 70 | 64 | 47 | 73.4 | 2 | 3.1 | 0.790 |
| G2-like | 44 | 36 | 24 | 66.7 | 1 | 2.8 | 0.807 |
| AP2-EREBP | 120 | 108 | 46 | 42.6 | 3 | 2.8 | 0.871 |
| C3H | 163 | 124 | 85 | 68.5 | 1 | 0.8 | 0.997 |
| E2F-DP | 8 | 6 | 6 | 100.0 | 0 | 0.0 | 1.000 |
| CCAAT-DR1 | 2 | 2 | 2 | 100.0 | 0 | 0.0 | 1.000 |
| Alfin-like | 7 | 7 | 7 | 100.0 | 0 | 0.0 | 1.000 |
| ARF | 22 | 18 | 17 | 94.4 | 0 | 0.0 | 1.000 |
| TCP | 26 | 20 | 18 | 90.0 | 0 | 0.0 | 1.000 |
| GRAS | 25 | 25 | 21 | 84.0 | 0 | 0.0 | 1.000 |
| CPP | 8 | 6 | 5 | 83.3 | 0 | 0.0 | 1.000 |
| C2C2-Gata | 28 | 20 | 16 | 80.0 | 0 | 0.0 | 1.000 |
| MYB-related | 9 | 8 | 6 | 75.0 | 0 | 0.0 | 1.000 |
| CCAAT-HAP5 | 13 | 10 | 7 | 70.0 | 0 | 0.0 | 1.000 |
| C2C2-CO-like | 30 | 24 | 16 | 66.7 | 0 | 0.0 | 1.000 |
| Orphan | 3 | 3 | 2 | 66.7 | 0 | 0.0 | 1.000 |
| C2C2-Dof | 36 | 32 | 21 | 65.6 | 0 | 0.0 | 1.000 |
| ARR-B | 15 | 12 | 7 | 58.3 | 0 | 0.0 | 1.000 |
| HSF | 21 | 21 | 11 | 52.4 | 0 | 0.0 | 1.000 |
| EIL | 6 | 6 | 3 | 50.0 | 0 | 0.0 | 1.000 |
| CCAAT-HAP2 | 10 | 10 | 5 | 50.0 | 0 | 0.0 | 1.000 |
Intracellular Targeting of Floral Proteins
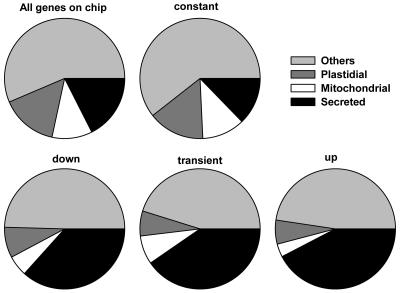
Many signal sequences required for targeting proteins to particular cellular compartments are well characterized. We used TargetP (Emanuelsson et al., 2000) to predict the subcellular localization of proteins encoded by genes of the specific set (Fig. 6). Genes belonging to the constant model subclass had a distribution of protein targeting similar to that of the entire set of genes represented on the ATH1 microarray: About 16% were predicted plastidial, 12% predicted mitochondrial, and 13% predicted secreted. By contrast, the up- and down-regulated subsets, as well as the transiently changed subsets, were strongly enriched for secreted proteins (37%–42%). This marked enrichment suggests important functions for secreted proteins during reproductive development. In plants, growth and cellular differentiation involves considerable reorganization of the cell wall. Among the secreted proteins were many potential enzymes modifying cell wall structure, e.g. expansins, pectinesterases, and galactosidases. Interestingly, 18% (60 out of the 333) of the predicted secreted proteins in model subclasses 2 to 8 had a molecular mass of less than 15 kD, while only 9% of all genes present on the ATH1 microarray encode proteins of less than 15 kD. Because only a few enzymes are smaller than 15 kD, many of the predicted secreted proteins could function directly as signaling molecules or as precursors for peptide hormones. Several systems mediating cell-cell signaling are based on small, secreted proteins or peptides, e.g. the WUSCHEL-CLAVATA1 (CLV1)-CLV3 system or sporophytic self-incompatibility in the Brassicaceae (Kachroo et al., 2002; Gross-Hardt and Laux, 2003; Matsubayashi, 2003). Most of the 60 small, secreted proteins are unknown or hypothetical proteins, but At3g25905 encodes the CLV3-related CLE27 protein (Cock and McCormick, 2001). Using reverse transcription followed by semiquantitative PCR, Sharma and colleagues showed that CLE27 is preferentially expressed in flower buds (Sharma et al., 2003). Also, the expression of small, putatively secreted Cys-rich proteins similar to the S locus Cys-rich protein involved in self-incompatibility in the Brassicaceae (Kachroo et al., 2002) has been documented in cells of the female gametophyte of maize (Zea mays; Cordts et al., 2001). Together with the aforementioned identification of 42 protein kinase genes, the 60 small, secreted proteins encoded by genes from the specific set form a starting point for future analysis of cell-cell signaling during reproductive development.
Figure 6.
Distribution of intracellular localization of proteins encoded by reproductive genes. Predictions of targeting are based on TargetP.
CONCLUSION
Our analysis provides the first study, to our knowledge, of the transcriptional program during the early phases of plant reproductive development on a nearly full-genome scale. We identified more than 2,300 genes that were specifically regulated during these developmental transitions or expressed preferentially in the tested samples. Using genetic screens, several important regulators of seed development have been identified. However, with the complete sequence information of the Arabidopsis genome available, we become aware that the function of several of the predicted genes is still unknown. Possible reasons include functional redundancy among the identified genes or that mutants in those genes give rise to quantitative phenotypes, which are difficult to identify with traditional genetic screens. It will be the challenge of future research to assign a function to those genes. Our expression analysis data allows the formulation of testable hypotheses that will guide reverse-genetic approaches. The identification of genes involved in reproductive processes in Arabidopsis has an important economic impact, as their homologs in crop plants are potential targets for improving yield and seed quality by conventional breeding or biotechnological approaches.
MATERIALS AND METHODS
Array Hybridization and Evaluation
Experimental procedures are described according to the minimum information about a microarray experiment standards (Brazma et al., 2001).
Experimental Design
Plants (Arabidopsis L. Heynh., accession Landsberg erecta) were grown in growth chambers at 70% humidity and daily cycles of 16 h light at 21°C and 8 h darkness at 21°C. Plant material used for the experiments was pooled from 12 plants. Stage I and stage II samples contained complete flower buds (stage I) or flowers (stage II). For stage III samples only siliques without withering flower organs were harvested. About 10% of the tissues for each sample were cleared and analyzed by microscopy to ensure that homogenous developmental stages were harvested. The entire experiment was performed twice providing independent biological replicates.
Array Design
Affymetrix Arabidopsis ATH1 GeneChips were used throughout the experiment (Affymetrix, Santa Clara, CA). The exact list of probes present on the arrays can be obtained from the manufacturer's Web site (http://www.affymetrix.com). Analysis was based upon annotations compiled by The Arabidopsis Information Resource (http://www.arabidopsis.org).
Samples
Total RNA was prepared from frozen tissue using Trizol and purified with RNeasy columns (Qiagen, Hilden, Germany). Fifteen micrograms of total RNA were used to prepare cDNA with the Superscript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions using oligodT-T7 oligonucleotides (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG(dT)24). The cDNA was subjected to in vitro transcription in the presence of 2 mm each of biotin-11-CTP and biotin-16-UTP (ENZO Life Sciences, Farmingdale, NY) using the MegaScript high yield transcription kit (Ambion, Austin, TX). After purification of the cRNA on RNeasy columns (Qiagen), 15 μg of cRNA were fragmented in a volume of 40 μL as recommended by Affymetrix.
Hybridizations
The fragmented 15 μg of labeled cRNA were denatured for 5 min at 99°C and hybridized to the arrays for 16 h as recommended by Affymetrix. Washing and detection of labeled cRNA using streptavidin-phycoerythrin were performed according to manufacturer's instructions using the EukGE-WS2v3 protocol involving two streptavidin-phycoerythrin-labeling steps.
Measurements
The arrays were scanned using a confocal scanner Agilent GS 2500 (Palo Alto, CA).
Evaluation, Normalization, and Data Analysis
Raw data were processed with the statistical algorithm of Affymetrix Microarray Suite 5.0 as described (Liu et al., 2002). Subsequent data processing and display were performed using Microsoft Access 2000 (Redmond, WA), Sigmaplot 8.0 (SPSS, Chicago), and the statistic package R (version 1.6.1) that is freely available at http://www.r-project.org/ (Ihaka and Gentleman, 1996).
For detection of specific genes, data from the NASCArray microarray database (Craigon et al., 2004) were used (slides for roots: Urwin_A-1-Urwin-Con_SLD, Jones_A1-jones-WT1_SLD, Jones_A1-jones-WT2_SLD, and Yap_A2-AMF; slides for pollen and microspores: Honys_BCP1_SLD, Honys_BCP2_SLD, Honys_MPG1_SLD, Honys_TCP1_SLD, Honys_TCP2_SLD, Honys_UNM1_SLD, and Honys_UNM2_SL).
Supplementary Material
Acknowledgments
We thank Nicole Schönrock (ETH Zürich) for seedling and leaf data sets. We thank the anonymous reviewers for their helpful comments on the manuscript.
This work was supported by the Human Frontier Science Program (grants to C.K.), Deutsche Forschungsgemeinschaft (grants to L.H.), the Functional Genomics Centre Zurich, and the University of Zurich.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043182.
References
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martinez-Castilla L, Yanofsky MF (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H, Walenta S, Panitz R, Wobus U, Weber H (2003) Energy status and its control on embryogenesis of legumes: ATP distribution within Vicia faba embryos is developmentally regulated and correlated with photosynthetic capacity. Plant J 36: 318–329 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed I (2000) Formation and maintenance of the shoot apical meristem. Trends Plant Sci 5: 110–115 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC (2003) Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci 8: 394–401 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordts S, Bantin J, Wittich PE, Kranz E, Lörz H, Dresselhaus T (2001) ZmES genes encode peptides with structural homology to defensins and are specifically expressed in the female gametophyte of maize. Plant J 25: 103–114 [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 (Database Issue): D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes JA, Grossniklaus U (2003) Unveiling the gene expression profile of pollen. Genome Biol 5: 205.1–205.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Florquin K, Rombauts S, Rouze P, Theissen G, Van De Peer Y (2003) Genomewide Structural Annotation and Evolutionary Analysis of the Type I MADS-Box Genes in Plants. J Mol Evol 56: 573–586 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA (1998) Genetic analysis of female gametophyte development and function. Plant Cell 10: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Eastmond P, Kolacna L, Rawsthorne S (1996) Photosynthesis by developing embryos of oilseed rape. J Exp Bot 47: 1763–1769 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethe JWv (1790) Versuch die Metamorphose der Pflanzen zu Erklären. C.W. Ettinger, Gotha, Germany
- Golz JF, Hudson A (1999) Plant development: YABBYs claw to the fore. Curr Biol 9: R861–R863 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Laux T (2003) Stem cell regulation in the shoot meristem. J Cell Sci 116: 1659–1666 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9: 227–238 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hennig L, Menges M, Murray JAH, Gruissem W (2003) Arabidopsis transcript profiling on Affymetrix GeneChip arrays. Plant Mol Biol 53: 457–465 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5: 299–314 [Google Scholar]
- Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M, et al (2003) A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol 133: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Nasralah ME, Nasrallah JB (2002) Self-incompatibility in the Brassicaceae: receptor-ligand signalling and cell-to-cell communication. Plant Cell 14 (Suppl): S227–S238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SP, Lunn JE, Furbank RT (1997) Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol 114: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, et al (2002) Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18: 1593–1599 [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Marles MA, Ray H, Gruber MY (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64: 367–383 [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla LP, Alvarez-Buylla ER (2003) Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc Natl Acad Sci USA 100: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y (2003) Ligand-receptor pairs in plant peptide signaling. J Cell Sci 116: 3863–3870 [DOI] [PubMed] [Google Scholar]
- McCormick S (1993) Male gametophyte development. Plant Cell 5: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Kim J, Lee S, An G, Ma H, Nei M (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, Daran-Lapujade P, Bro C, Regenberg B, Knudsen S, Nielsen J, Pronk JT (2002) Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J Biol Chem 277: 37001–37008 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Weber H, Borisjuk L (2003) Energy status and its control on embryogenesis of legumes: embryo photosynthesis contributes to oxygen supply and is coupled to biosynthetic fluxes. Plant Physiol 132: 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Krizek BA, Jacobsen SE, Meyerowitz EM (2000) Regulation of SUP Expression Identifies Multiple Regulators Involved in Arabidopsis Floral Meristem Development. Plant Cell 12: 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC (2003) The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Shirley BW (1996) Flavonoid biosynthesis: new functions for an old pathway. Trends Plant Sci 1: 377–382 [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Altschmied L, Radchuk V, Gubatz S, Wobus U, Weschke W (2004) Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J 37: 539–553 [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter KU, Saedler H (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42: 115–149 [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003) Flower development: initiation, differentiation, and diversification. Annu Rev Cell Dev Biol 19: 119–140 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.