Abstract
The stem–loop binding protein (SLBP) binds the 3′ end of histone mRNA and is present both in nucleus, and in the cytoplasm on the polyribosomes. SLBP participates in the processing of the histone pre-mRNA and in translation of the mature message. Histone mRNAs are rapidly degraded when cells are treated with inhibitors of DNA replication and are stabilized by inhibitors of translation, resulting in an increase in histone mRNA levels. Here, we show that SLBP is a component of the histone messenger ribonucleoprotein particle (mRNP). Histone mRNA from polyribosomes is immunoprecipitated with anti-SLBP. Most of the SLBP in cycloheximide-treated cells is present on polyribosomes as a result of continued synthesis and transport of the histone mRNP to the cytoplasm. When cells are treated with inhibitors of DNA replication, histone mRNAs are rapidly degraded but SLBP levels remain constant and SLBP is relocalized to the nucleus. SLBP remains active both in RNA binding and histone pre-mRNA processing when DNA replication is inhibited.
INTRODUCTION
The replication-dependent histone mRNAs in metazoans are the only eukaryotic mRNAs that do not end in poly(A) tails. Instead the replication-dependent histone mRNA's end in a highly conserved 26 nt sequence that contains a 16 nt stem–loop (1,2). Replication-dependent histone genes also lack introns; consequently, the only processing reaction required for formation of a mature histone mRNA is cleavage after the stem–loop to form the 3′ end (2). Since histone mRNAs lack a poly(A) tail, it is likely that the 3′ end of histone mRNAs and associated factors are important for their translation, much as the poly(A) tail, together with the poly(A) binding protein (PABP), plays an important role in the translation of polyadenylated eukaryotic mRNAs (3–5). The 3′ end of histone mRNA is also important for its localization to polyribosomes (6) and mRNAs ending in the stem–loop are translated with efficiencies comparable to polyadenylated mRNAs (7). We have recently shown that the stem–loop binding protein (SLBP) stimulates translation of reporter mRNAs ending in the stem–loop both in vivo and in vitro (8).
The levels of histone mRNA are 35-fold higher during S phase than G1 phase (9) and are coupled to the rate of DNA replication (10,11). Inhibition of DNA replication results in the rapid degradation of the polyribosomal histone mRNA (12). The 3′ end of histone mRNA is the cis-acting element responsible for this regulatory response (13). A 3′ to 5′ exonuclease associated with polyribosomes has been implicated in the degradation of histone mRNA (14–17), consistent with a role for the 3′ end of histone mRNA in the regulation of histone mRNA half-life.
A trans-acting factor, SLBP (also known as the hairpin binding protein, HBP) that interacts with the stem–loop has been identified in mammalian cells (18,19). SLBP is required for efficient histone pre-mRNA processing (2,20), associating with the histone pre-mRNA in the nucleus, and likely accompanies the histone mRNA to the cytoplasm as part of the histone messenger ribonucleoprotein (mRNP) (21). SLBP is also cell cycle regulated (22). As cells enter the S phase the synthesis of SLBP is activated by increasing the translation rate of the SLBP mRNA. SLBP is rapidly degraded at the end of S phase as a result of its phosphorylation (22,23). Here, we demonstrate that some of the polyribosomal histone mRNAs are bound to SLBP. Histone mRNA is rapidly degraded when cells in S phase are treated with inhibitors of DNA replication, but SLBP is stable, and relocates to the nucleus.
MATERIALS AND METHODS
Cell culture
Mouse myeloma cells were cultured in DMEM with 10% horse serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Life Technologies). Typically, cells were grown to a density of 3 × 105 to 5 × 105 cells/ml prior to harvesting. For treatment with DNA and protein synthesis inhibitors, a single culture was divided and treated with the inhibitors of translation or DNA synthesis for the time described in each experiment. The following concentrations were used for drug treatments: 0.1 mM cycloheximide (CHX), 2 μM 5-fluorodeoxyuridine (FUdR), 5 mM hydroxyurea (HU) or 5 μg/ml aphidicolin (Apd).
Preparation of nuclear extract and polyribosomes
Cells were fractionated as described previously (24). Briefly, cells (from 1–6 l of suspension culture) were allowed to swell in hypotonic buffer, lysed with a Dounce homogenizer and nuclei pelleted by low-speed centrifugation (1000 g, 10 min). Nuclear extracts were prepared from mouse myeloma cells exactly as previously described (20,25). A synthetic histone H2a-614 pre-mRNA was used for assaying histone pre-mRNA processing as previously described (20).
Polyribosomes were prepared by first removing the mitochondria by centrifugation at 10 000 g for 10 min. The cytoplasm was then layered over a 1 M sucrose cushion and centrifuged at 100 000 g for 4 h to pellet the polyribosomes. Polyribosomes were suspended in 10 mM HEPES–KOH, pH 7.9, 1.5 mM MgCl2, 20 mM KCl, 0.5 mM DTT, 0.75 mM spermidine, 0.15 mM spermine by gentle homogenization and stored at −80°C.
Measurement of SLBP synthesis rate
SLBP synthesis rate was determined essentially as described previously (22). Cells were preincubated in DMEM, without methionine and cysteine supplemented with 10% dialyzed fetal bovine serum, for 1 h prior to labeling to deplete intracellular stores of methionine and cysteine. SLBP synthesis rate in Chinese Hamster Ovary (CHO) cells treated with inhibitors of DNA synthesis was measured by pulsing cells with 1 mCi of [35S]methionine–cysteine for 15 min intervals after inhibition of DNA synthesis with 5 μg/ml Apd (NEN Life Science Products, Boston, MA). Whole-cell extracts were incubated with anti-SLBP and the SLBP–antibody complexes recovered by binding to protein A agarose beads. The bound proteins were eluted in SDS loading buffer and resolved by gel electrophoresis and detected by autoradiography (22).
Detection of phosphorylated SLBP
An aliquot of 2.5 × 106 mouse myeloma cells were grown in 100 mM culture dishes with 5 ml of phosphate-free media supplemented with 10% dialyzed horse serum. Cells were grown in phosphate-free media for 1 h prior to labeling to deplete the intracellular stores of phosphate. Cells were then labeled for 3 h with 1 mCi of [32P]orthophosphate (ICN) at a concentration of 0.2 mCi/ml. Whole-cell lysates were prepared by incubating cells in NP-40 lysis buffer (0.1% NP-40, 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 50 mM NaF, 1 mM DTT) for 10 min. Insoluble material was removed by centrifugation at 13 000 g and the soluble supernatant used for subsequent immunoprecipitations.
SLBP RNA binding activity
Mobility shift assays to detect SLBP binding were performed using a 30 nt histone RNA stem–loop (25). The SLBP was detected by western blotting using the anti-SLBP antibody as described previously (18).
Antibody precipitation of polyribosomes
The SLBP antibody used for RNA precipitation was affinity purified against the antigenic C-terminal peptide coupled to a Sulfolink column (Pierce, Rockford, IL). Typically 10 μg of peptide was coupled to the Sulfolink column and the antibody purified from 2–5 ml of serum. Antibody was eluted from the column, neutralized to pH 7.0 and stored in sodium azide.
All operations were performed at 4°C. Polyribosomes (5–10 μg of polyribosomal RNA) were diluted into 100 μl of NP-40 buffer [0.1% NP-40, 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM DTT, 10 mM EDTA, 40 U RNAsin (Promega, Madison, WI)]. Lysates were pre-cleared by incubation with 10–20 μl of protein A beads for 30 min to remove any non-specific binding material. Beads were removed by sedimentation in a microcentrifuge at 4000 r.p.m. (1500 g) for 4 min. The supernatant was removed and incubated for 1 h with the affinity-purified anti-SLBP (0.1–0.2 μg) and then with 10 μl of protein A agarose beads for an additional 1 h. The beads were recovered by centrifugation at 2000 r.p.m. (400 g) for 2 min and the complexes washed three times with the NP-40 lysis buffer.
RNA was prepared from both the supernatants and the pellets. The supernatant from the precipitation (100 μl) was mixed with an equal volume of urea lysis buffer (ULB, 7 M urea, 2% SDS, 0.35 M NaCl,10 mM EDTA, 10 mM Tris–HCl, pH 7.5) and extracted with phenol–chloroform and the RNA recovered by precipitation with ethanol. To prepare RNA from the immunoprecipitates, the beads were suspended in 200 μl of ULB and processed as above.
Histone mRNA levels in mouse RNA samples were measured by S1 nuclease mapping using the following probes. The histone H3.2 mRNAs were mapped using the H3.2-614 gene labeled at the SalI site and the H3.3 mRNAs were mapped using the H3.3 gene labeled at the 5′ end of the BglII site (26). The H1b and H1c genes were mapped using the H1c gene labeled at the NotI site (27). Actin mRNA was analyzed using a ribonuclease protection assay.
Histone mRNA levels in HeLa cell mRNP precipitates were measured by RT–PCR. Four micrograms of RNA from each supernatant and an equivalent fraction of each precipitate (typically one-tenth of the total precipitate) were reverse transcribed into cDNA with Superscript II (Invitrogen Life Technologies) primed with random hexamers according to the manufacturer's protocols. Each cDNA synthesis reaction was purified using QIAquick PCR Purification Kit (Qiagen Sciences, MD) and eluted with water in a volume of 30 μl. Each PCR reaction was performed in a 25 μl reaction volume, 1.5 μl of purified cDNA and 5 pmol of each primer were incubated in 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 2.5 U Taq DNA polymerase (New England BioLabs, MA) for 28 cycles (92°C, 30 s; 60°C, 30 s; 72°C, 30 s). The primers for each gene were as follows: HIST1H1A (AF531299.1, forward GGAGAAGAACAACAGCCGCAT, reverse TTGAGCTTGAAGGAACCCGAG, 104 bp amplicon); HIST1H2AB (AY131983.1, forward ACAACAAGAAGACCCGCATCAT, reverse TATTAGGCAAAACGCCACCCT, 116 bp amplicon); HIST1H2BG (AF531290.1, forward ACAAGCGCTCGACCATTACCT, reverse TGGTGACAGCCTTGGTACCTTC, 107 bp amplicon); HIST1H4B (AY128655.1, forward GGATAACATCCAAGGCATCACC, reverse CGCCACGAGTCTCCTCATAAAT, 100 bp amplicon); H3F3B (NM_005324.3, forward TGGTTTTTCGCTCGTCGACT, reverse GGCCATTTTCTTTCACCCAAC, 102 bp amplicon); FOXM1 (NM_021953, forward GTGGCGATCTGCGAGATTTTG, reverse TCTTTCCCTGGTCCTGCAGAAG, 123 bp amplicon); CCNE1 (NM_001238.1, forward GGAAGAGGAAGGCAAACGTGA, reverse TTATTGTCCCAAGGCTGGCTC, 108 bp amplicon). PCR products were resolved on a 2% agarose gel in 0.5× TAE and visualized by staining with ethidium bromide.
Sucrose gradient fractionation of polyribosomes
Mouse myeloma cells were lysed in hypotonic buffer and the nuclei and mitochondria removed by centrifugation. The cytoplasm was adjusted to 0.1% NP-40, 0.1 M NaCl, 5 mM Mg(OAc)2 and layered on a 15–40% sucrose gradient containing 0.1 M NaCl, 5 mM Mg(OAc)2, 10 mM Tris, pH 7.5. Polyribosomes were fractionated by centrifugation at 39 000 r.p.m. for 2.5 h in an SW841 rotor. Aliquots of 0.5 ml were collected and RNA was prepared from 0.3 ml of each aliquot. RNA prepared from each fraction was analyzed by agarose gel electrophoresis to identify the monoribosome peak (fractions 6 and 7). Protein was prepared from each fraction and analyzed by SDS–gel electrophoresis followed by western blotting for SLBP. SLBP binding was analyzed with a radiolabeled stem–loop probe by native gel electrophoresis as described previously (28). Histone H2a mRNA levels were measured by S1 nuclease mapping as described previously (11,26).
Immunostaining
HeLa cells stably expressing an hemagglutinin(HA)-tagged SLBP (23) were plated onto coverslips at low density and synchronized by a double thymidine block as described in (22). S-phase cells (3 h after release from the thymidine block) were treated with either HU or CHX for 1 h. Cells were then fixed with 4% paraformaldehyde, washed with phosphate-buffered saline and permeabilized with 0.5% Triton X-100. Detection of the HA–SLBP was carried out using a monoclonal α-HA antibody kindly provided by Dr Yue Xiong at a dilution of 1:250 followed by incubation with a cy3 labeled anti-mouse antibody (1:500, Jackson Immunoresearch). Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml).
RESULTS
Histone mRNAs are rapidly (<5 min) processed and transported to the cytoplasm (29) where they are efficiently translated. The histone mRNA 3′ end binds a protein, SLBP, which associates with the histone pre-mRNA in the nucleus and is necessary for histone pre-mRNA processing (20). SLBP also stimulates translation of histone mRNA (8), suggesting that it is associated with histone mRNA during at least the initial round(s) of translation. Here, we show that histone mRNA is co-precipitated with SLBP from polyribosomes, and have determined the subcellular localization of SLBP and its fate after degradation of histone mRNA.
Histone mRNAs are precipitated with anti-SLBP
To directly determine whether SLBP is associated with histone mRNA on polyribosomes, we incubated isolated polyribosomes with anti-SLBP (in the presence of EDTA to dissociate the polyribosomes), and recovered the SLBP–antibody complex with protein A agarose beads. RNA was prepared from the bound and unbound fractions and analyzed for the presence of different mRNAs by nuclease protection assays. We performed the precipitation under a variety of conditions with various detergents and for different amounts of time to determine the conditions for optimal precipitation of histone mRNA with minimal background binding. Essentially similar results were obtained using a variety of non-ionic detergents. Using either 0.1% NP-40 or 0.1% Tween-20 we were able to precipitate 20–30% of the histone mRNA present on polyribosomes with the affinity-purified anti-SLBP antibody (Figure 1A, lanes 3 and 5).
Figure 1.
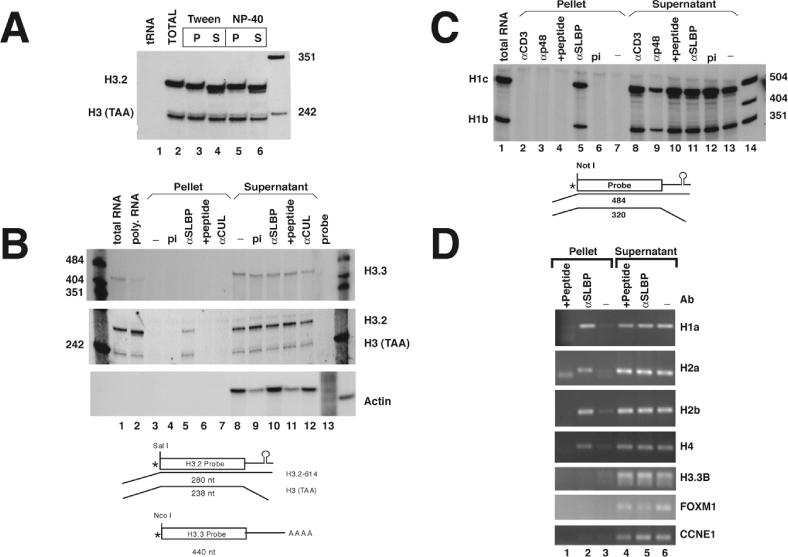
SLBP is bound to histone mRNAs in the cell. (A) Polyribosomes isolated from mouse myeloma cells were suspended in buffer containing 10 mM EDTA as described in Materials and Methods. α-SLBP was added in the presence of either 0.1% Tween-20 (lanes 3 and 4) or 0.1% NP-40 (lanes 5 and 6). RNA was prepared from the immunoprecipitates (lanes 3 and 5) and from the supernatant fraction (lanes 4 and 6) and analyzed by S1 nuclease mapping using the histone H3-614 gene as a probe. RNA prepared from polyribosomes was analyzed in lane 2 and yeast tRNA in lane 1. There is a small size differences (2–4 nt) observed between the total RNA (lane 2) and the immunoprecipitated mRNA compared with the RNA that did not immunoprecipitate. The final lane is pUC18 digested with HpaII. (B) Immunoprecipitations were performed from mouse myeloma polyribosomes with preimmune serum (pi) (lanes 4 and 9), α-SLBP (lanes 5 and 10), α-SLBP preincubated with 1 μg of antigenic peptide (lanes 6 and 11) or with an unrelated antibody, α-CUL (lanes 7 and 12). RNA was prepared from both supernatants and precipitates and analyzed for the polyadenylated H3.3 histone mRNA (upper panel) or the replication-dependent H3.2-614 mRNA (middle panel) by S1 nuclease mapping. The same RNA samples were analyzed for the mouse β-actin mRNA using a ribonuclease protection assay (lower panel). Analysis of total RNA or polyribomosal RNA are shown for H3.3 and H3.2 histone mRNAs but not for actin (lanes 1 and 2). A diagram of the S1 nuclease assay is shown below the figure. (C) Immunoprecipitations were performed with preimmune serum (pi) (lanes 6 and 12), α-SLBP (lanes 5 and 11), α-SLBP with antigenic peptide (lanes 4 and 10) or with the unrelated antibodies α-p48 and α-Cdk4 (lanes 2 and 3, and 8 and 9). RNA was prepared and analyzed by S1 nuclease mapping with the mouse H1c gene which maps both the H1c and H1b mRNAs (27). S1 nuclease mapping of total mRNA from the same cells is shown (lane 1). Lanes 7 and 13 shows polyribosomes incubated in buffer and then treated with protein A beads. Lane 14 is marker pUC18 digested with HpaII. A diagram of the S1 nuclease assay is shown below the figure. (D) Immunoprecipitations were performed from polyribosomes isolated from HeLa cells with either no antibody (lanes 3 and 6), α-SLBP (lanes 2 and 5) or α-SLPB with antigenic peptide (lanes 1 and 4). RNA was prepared and analyzed by RT–PCR with primers specific to H1a (HIST1H1A), H2a (HIST1H2AB), H2b (HIST1H2BG) and H4 (HIST1H4B). The same RNA samples were assayed for the presence of the polyadenylated histone H3.3 mRNA (H3F3B) and two additional cell-cycle-regulated mRNAs, FOXM1 and CCNE1 (62). PCR products were detected by ultraviolet (UV) illumination of ethidium bromide stained 2% agarose gels.
There was a detectable difference in the length of the histone mRNA in the precipitated and non-precipitated fractions (see Figure 1A, lanes 3–6). The histone mRNA precipitated with anti-SLBP was similar in length to the mRNA formed in the processing reaction. Most of the histone mRNA that was in the supernatant was several nucleotides shorter, suggesting that it had been trimmed by exonucleases associated with the polyribosomes (15,17). Since the histone mRNA in the total polyribosomal RNA preparation was largely full length (Figure 1A, lane 2), it is likely that the shortening of the RNA occurred during the incubation with the antibody, and/or subsequent washing procedures.
The precipitation of histone mRNA was specific for the anti-SLBP. There were substantial amounts of histone H3.2 mRNA present in the bound fraction only when anti-SLBP was used (Figure 1B, middle panel, lane 5) but not when the antibody was preincubated with the antigenic peptide (Figure 1B, lane 6) or when the precipitation was performed with an unrelated antibody (Figure 1B, lane 7), e.g. against Cullin-1 (30). Overall 20–30% of the histone mRNA was precipitated by the anti-SLBP antibody depending on the experiment.
In order to further determine the specificity of the precipitation, we measured the levels of two different mRNAs that we expected would not be precipitated by the anti-SLBP. There was no precipitation of the histone H3.3 mRNA, an mRNA for a replacement histone (Figure 1B, lane 5, upper panel), which does not contain a histone stem–loop but, instead, ends in a poly(A) tail (31), or of the abundant actin mRNA (Figure 1B, lane 5, lower panel).
We also analyzed the immunoprecipitates for two histone H1 mRNAs, H1c and H1b. These two mRNAs are both measured using the same probe in the S1 nuclease assay since their sequences are nearly identical for a large portion of the coding region (27). Both H1c and H1b mRNAs were precipitated by the anti-SLBP antibody (Figure 1C, lane 5) but not by the antibody preincubated with the antigenic peptide (Figure 1C, lane 4), with preimmune serum (lane 6), or with antibodies to the histone binding protein p48 or anti-CD3 antibodies (Figure 1C, lanes 2 and 3).
We also tested the ability of the anti-SLBP to precipitate the histone mRNAs from the HeLa cell polyribosomes. The precipitated RNAs were analyzed for histone mRNAs and several control mRNAs (histone H3.3, and two cell-cycle-regulated mRNAs FOXM1 and CCNE1) using RT–PCR. We designed primers to allow us to detect multiple members of the core histone mRNA family (32) and histone H1a mRNA. All the histone mRNAs were precipitated by SLBP and precipitation was competed by the peptide antigen (Figure 1D), while the other mRNAs were not precipitated by anti-SLBP. Since all of the replication-dependent histone mRNAs that we have tested were precipitated by the anti-SLBP it is likely that SLBP is bound to this class of messages on the polyribosomes, which comprise more than 60 different mRNAs (32).
CHX treatment alters the subcellular localization of SLBP
When cells are fractionated into nuclear, cytosolic and polyribosomal fractions, >90% of the histone mRNA is present in the polyribosomal fraction (6,33). We determined the distribution of SLBP by western blotting and immunostaining. When cells were fractionated biochemically, SLBP was found in all subcellular fractions with much of the protein in the polyribosomal fraction, as reported in (24) (Figure 2A, lanes 3 and 4). As expected from its role in histone pre-mRNA processing, significant amounts of SLBP were also found in the nuclear fraction (Figure 2A, lanes 1 and 2). There were also small amounts of SLBP found in the cytosolic fraction (Figure 2A, lanes 5 and 6).
Figure 2.
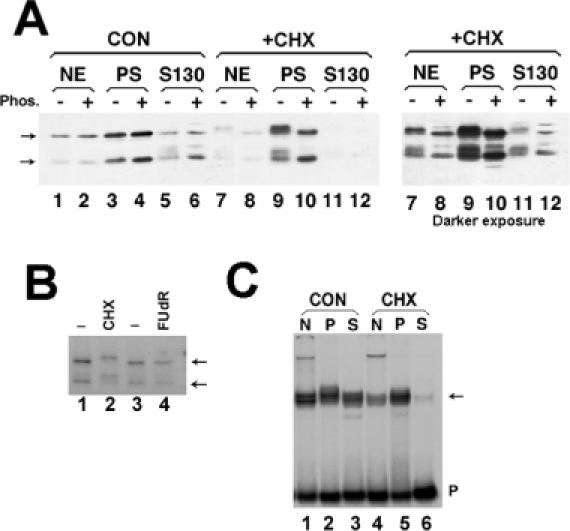
Effect of protein synthesis inhibitors on SLBP modification and localization (A) Mouse myeloma cells were grown normally (lanes 1–6) or treated with 0.1 mM CHX for 1 h (lanes 7–12). Both cultures were fractionated in parallel into nuclear extract (lanes 1, 2, 7, 8), polyribosomes (lanes 3, 4, 9, 10) or post-polyribosomal supernatant (S130, lanes 5, 6, 11, 12). The right hand panel is a longer exposure of the CHX fractionation (lanes 7–12). Extracts from equivalent amounts of cells were analyzed by gel electrophoresis and the SLBP detected by western blotting. There was some proteolysis of SLBP resulting in removal of ∼10 kDa from the N-terminus (18). Aliquots of the fractions in even-numbered lane were treated with protein phosphatase prior to electrophoresis. The position of unmodified SLBP and the proteolytic product are indicated by the arrows. (B) Mouse myeloma cells were labeled with H3[32PO4] and left untreated (lanes 1 and 3), treated with CHX for 2 h (lane 2) or treated with the DNA synthesis inhibitor FudR for 1 h (lane 4). Total cell lysates were prepared and the SLBP immunoprecipitated and resolved by SDS-PAGE. Phosphorylated SLBP was detected by autoradiography. The position of unmodified SLBP and the proteolytic product are indicated by the arrows. (C) The nuclear, polyribosomal and S130 extracts from equivalent amounts of mouse myeloma cells [same extracts as in panel (A)] were assayed by mobility shift assay using a radiolabeled stem–loop RNA as a probe (40). The stem–loop/RNA complex [(C) arrow] was resolved by native polyacrylamide gel electrophoresis.
Treatment of cells with inhibitors of protein synthesis results in an increase in histone mRNA concentrations (12,34). Blocking translation with CHX, which inhibits the translocation step in protein synthesis, results in stabilization of histone mRNA on polyribosomes and a 2- to 3-fold increase in the steady-state amount of histone mRNA (12,34,35). When cells are treated with CHX for 1 h and then fractionated, histone mRNA was associated with ribosomes, as was the great majority of the SLBP (Figure 2A, lanes 9 and 10). There was a decrease in both the amount of nuclear SLBP and cytosolic SLBP and a concomitant increase in the amount of SLBP associated with polyribosomes (Figure 2A, lanes 7–12). Since protein synthesis has been inhibited, the change in the subcellular distribution of SLBP must reflect a redistribution of pre-existing SLBP.
Histone gene transcription and processing continues when cells are treated with CHX (12), utilizing the SLBP in the nucleus for processing. Cytoplasmic histone mRNAs are stabilized, resulting in an increase in histone mRNA levels during CHX treatment. Ultimately, the majority of SLBP accumulates in the cytoplasm associated with polyribosomes in CHX-treated cells.
Although there was no significant change in the overall amount of SLBP, treatment with CHX results in a lower mobility of SLBP on SDS–polyacrylamide gels (Figure 2A, lanes 7–12). This is a result of phosphorylation, since treatment of the samples with calf intestinal phosphatase resulted in a shift in mobility back to the original mobility. There was no shift in mobility of the SLBP in the samples from control cells (Figure 2A, lanes 1–6).
To further confirm that SLBP is phosphorylated, we incubated cells with 32PO4 and isolated the SLBP by immunoprecipitation. Note that the SLBP in control cells is phosphorylated since the SLBP is also labeled with 32PO4 in control cells, but this phosphorylation does not alter the mobility of the protein (Figure 2A, lanes 1–6 and Figure 2B, lane 1). The mobility of the phosphorylated SLBP is lower in CHX-treated cells (Figure 2B, lane 2) but not in FUdR-treated cells (Figure 2B, lanes 3 and 4) suggesting that SLBP is hyperphosphorylated in CHX-treated cells. We assayed the RNA binding activity of SLBP in the subcellular fractions using a mobility shift assay. There was reduced binding activity of SLBP in the nuclear and cytosolic fractions of CHX-treated cells (Figure 2C), consistent with the results of western blotting.
We also determined the localization of SLBP by immunofluorescence in a cell line expressing HA-tagged SLBP. These cells express similar amounts of HA-tagged SLBP as endogenous SLBP and the HA–SLBP is regulated in parallel with the endogenous SLBP (23). The HA–SLBP was detected by immunostaining with the anti-HA antibody. We analyzed S-phase cells (3 h after release from a double thymidine block) all of which express SLBP and histone mRNA. SLBP is present in both the nucleus and cytoplasm of S-phase cells (Figure 3, left 2 columns). When cells are treated with CHX, SLBP redistributes to the cytoplasm (Figure 3, middle two columns), in agreement with the cell fractionation results.
Figure 3.
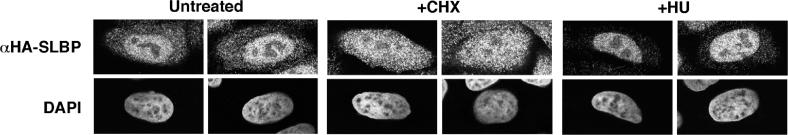
Localization of SLBP in cells treated with CHX or HU. HeLa cells expressing HA–SLBP were synchronized by double thymidine block. The cells were released into S phase for 3 h. Separate cultures were treated for 1 h with CHX (middle two panels) or HU (right two panels). Untreated cells in S phase are shown in the left two panels. Cells were fixed and the HA–SLBP detected by immunofluorescence (top panels) and DNA detected with DAPI (bottom panels) as described in Materials and Methods.
SLBP levels do not change when histone mRNA is degraded
Histone mRNAs are rapidly degraded when cells are treated with inhibitors of DNA replication (11,12) and at the end of the S phase (9,22). SLBP expression is tightly regulated during the cell cycle, accumulating at about the same time as histone mRNA and being rapidly degraded at the end of S phase at about the same time as the histone mRNA is degraded (22,23). Given the role of the 3′ end of histone mRNA in the coordinate regulation of histone mRNA levels during the cell cycle (9,36–38), and in regulating histone mRNA degradation (13), we asked whether there were changes in SLBP as a result of treatments that altered histone mRNA metabolism.
After treating CHO cells for 45 min with three different inhibitors of DNA replication (HU, Apd and FUdR), there was at least a 20-fold reduction in histone mRNA levels (Figure 4A, bottom). However, there was no decrease, but rather a slight increase (∼2-fold) in total cellular SLBP with all three inhibitors (Figure 4A, top). SLBP was resolved into two forms by gel electrophoresis, with the minor, slower migrating form being phosphorylated. There was no change in the ratio of phosphorylated and unphosphorylated forms of SLBP when DNA replication was inhibited, consistent with the results of 32PO4 labeling (Figure 2B). Thus SLBP is not degraded or detectably modified when histone mRNA is degraded. These results demonstrate that degradation of SLBP does not accompany histone mRNA degradation when DNA synthesis is inhibited.
Figure 4.
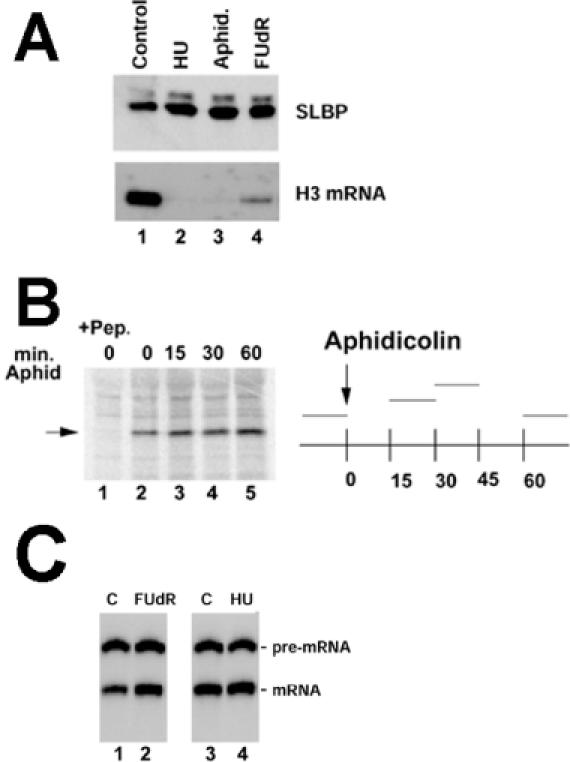
SLBP levels are not decreased when histone mRNA is degraded. (A) CHO cells were treated with HU, Apd or FUdR for 1 h. Lysates and RNA were prepared and analyzed for SLBP by western blotting (top) or histone H3 mRNA by S1 nuclease mapping (bottom). The slower migrating band represents a small amount of phosphorylated SLBP. (B) CHO cells were pulse labeled with [35S]methionine for 15 min, at the time periods indicated on the graph at the right. Apd was added to the cells at 0 min and then [35S]methionine added at 15, 30 or 60 min. Equal amounts of cell lysate were precipitated with anti-SLBP and the immunoprecipitates analyzed by gel electrophoresis (left); the position of SLBP is indicated by the arrow. In lane 1, the antibody was incubated with the antigenic peptide prior to immunoprecipitation. Above each lane the time in minutes after Apd treatment is indicated. The 0 h points were not treated with Apd (lanes 1 and 2). (C) Nuclear extracts were prepared from exponentially growing mouse myeloma cells (lanes 1 and 3) or mouse myeloma cells treated with FUdR (lane 2) or HU (lane 4) for 1 h. A synthetic radiolabeled 320 nt histone H2a pre-mRNA was incubated in the extract for 30 min, the RNA purified, analyzed by gel electrophoresis and the RNA products detected by autoradiography (20). The position of the substrate pre-mRNA and the 266 nt processed mRNA are indicated.
Since the translation of SLBP mRNA is regulated during the cell cycle (22), we also examined whether inhibition of DNA replication affected the rate of SLBP synthesis in CHO cells. The synthesis rate of the SLBP was determined at various times after treatment with Apd by pulse labeling with [35S]methionine for 15 min beginning at the indicated time followed by immunoprecipitation of SLBP. The 0 h time point represents cells that have not been treated with Apd. The synthesis rate of SLBP increases ∼2-fold when cells are treated with inhibitors of DNA replication (Figure 4B, lanes 3–5). Analysis of the SLBP mRNA levels shows a similar small increase (data not shown) when cells are treated with inhibitors of DNA replication. Thus, there is clearly not a decrease in the synthesis of SLBP or rapid degradation of SLBP when DNA synthesis is inhibited, in contrast to the rapid disappearance of SLBP at the end of S phase (22).
SLBP returns to the nucleus after inhibition of DNA replication
When cells are treated with inhibitors of DNA replication and histone mRNA is rapidly degraded, there is no decrease in the amount of SLBP. To determine whether there was a redistribution of SLBP when histone mRNAs were degraded as a result of inhibition of DNA replication, we treated the S-phase cells expressing the HA–SLBP with HU and then analyzed the distribution by immunofluoresence with the anti-HA antibody. When DNA replication was inhibited and histone mRNA degraded, we observed a relocalization of SLBP to the nucleus (Figure 3, right panels).
Extracts from cells treated with DNA synthesis inhibitors are active in histone pre-mRNA processing
Treatment of cells with inhibitors of DNA replication results in a rapid decrease in histone mRNA levels, with a much smaller decrease in the rate of histone gene transcription (11,12). To determine whether there was an effect on histone pre-mRNA processing in cells treated with inhibitors of DNA replication, nuclear extracts were prepared from control cells and cells treated with HU or FUdR. Both inhibitors result in a rapid reduction in histone mRNA levels (Figure 4A). However, extracts prepared from either FUdR- or HU-treated cells processed synthetic histone pre-mRNA at least as well as extracts prepared from control cells (Figure 4C). Thus there is no difference in histone pre-mRNA processing in nuclear extracts prepared from growing cells or cells treated with inhibitors of DNA replication.
SLBP sediments with histone mRNA on small polyribosomes
To obtain additional evidence that SLBP was associated with histone mRNA on polyribosomes, we prepared cytosolic fractions from exponentially growing mouse myeloma cells and fractionated them on 10–40% sucrose gradients to resolve the polyribosomes. We analyzed polyribosome gradients from control cells (Figure 5), cells treated with CHX (Figure 6), and cells treated with HU (not shown, Supplementary Figure A). After fractionation, the presence of histone mRNA was determined by an S1 nuclease protection assay, SLBP levels by western blotting, SLBP binding activity by mobility shift assay and the ribosomal RNA in each fraction by ethidium bromide staining on an agarose gel.
Figure 5.
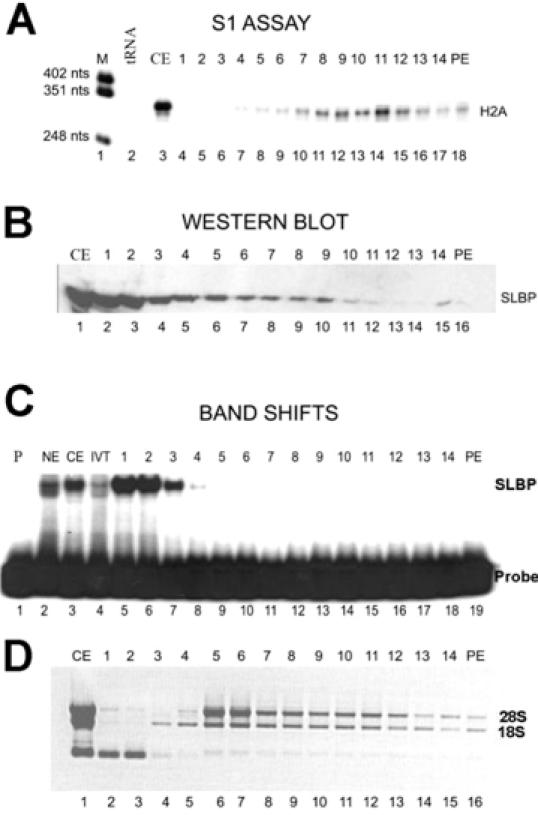
Association of SLBP with polyribosomes. Cytoplasmic lysate prepared from exponentially growing HeLa cells was fractionated by sucrose gradient centrifugation as described in Materials and Methods. The fraction numbers from the sucrose gradients are indicated above each lane; PE is the sucrose gradient pellet. The monoribosome peak (fractions 6 and 7 in each panel) was identified by ethidium bromide staining of total RNA. (A) Total RNA isolated from each fraction was analyzed for histone H2a mRNA by S1 nuclease mapping. Lane 1, marker (pUC18 digested with HpaII); lane 2, 10 μg of yeast tRNA; lane 3, is analysis of 5 μg of RNA from the cytoplasmic extract; lanes 4–18, sucrose gradient fractions. (B) An aliquot of each fraction sample was analyzed by western blotting for SLBP. Lane 1, 10 μg of cytoplasmic extract; lanes 2–16, sucrose gradient fractions. (C) An aliquot of each fraction was analyzed for RNA binding activity with a mobility shift assay using the radiolabeled stem–loop RNA as a probe. Lane 1, probe; lane 2, nuclear extract; lane 3, cytoplasmic extract; lane 4, in vitro-translated SLBP; Lanes 5–19, sucrose gradient fractions. (D) An aliquot of each fraction was analyzed by agarose gel electrophoresis and the RNAs detected by ethidium bromide staining. Lane 1, RNA from cytoplasmic extract; lanes 2–16: sucrose gradient fractions 1-14 and the pellet (PE).
Figure 6.
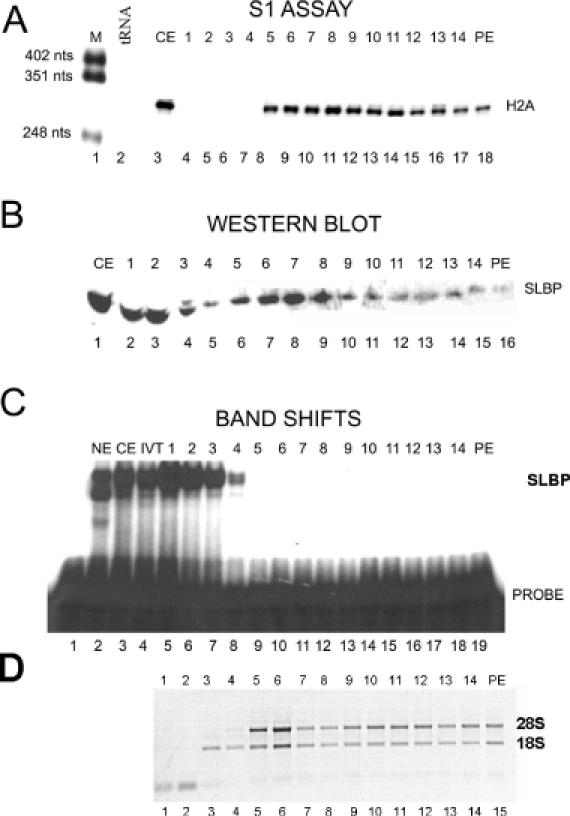
SLBP is associated with histone mRNA on ribosomes from CHX-treated cells. Cytoplasmic lysate prepared from HeLa cells treated with CHX for 45 min were fractionated by sucrose gradient centrifugation. The fraction numbers from the sucrose gradients are indicated above each lane; PE is the sucrose gradient pellet. Fractions 6 and 7 contain the monoribosome peak as determined by ethidium bromide staining to total RNA. (A) Total RNA isolated from each fraction was analyzed for histone H2a mRNA by S1 nuclease mapping. Lane 1, marker (pUC18 digested with HpaII); lane 2, 10 μg of yeast tRNA; lane 3, 5 μg of RNA from the cytoplasmic extract; lanes 4–18, sucrose gradient fractions. (B) Protein from each fraction was analyzed by western blotting for SLBP. Lane 1, 10 μg of cytoplasmic extract; lanes 2–16, sucrose gradient fractions; PE, sucrose gradient pellet. (C) Each fraction was analyzed for RNA binding activity by a mobility shift assay using the radiolabeled stem–loop RNA probe. Lane 1, probe; lane 2, nuclear extract; lane 3, cytoplasmic extract; lane 4, in vitro translated SLBP; lanes 5–19, sucrose gradient fractions. (D) An aliquot of each fraction was analyzed by agarose gel electrophoresis and the RNAs detected by ethidium bromide staining. Lanes 1–15, sucrose gradient fractions 1–14 and the pellet (PE).
Since histone mRNAs are small (coding region only 400 nt, total length ∼500 nt) they are associated with small polyribosomes (Figure 5A). Under these conditions the mRNAs for larger mRNAs such as GAPDH subunits (>1200 nt encoding a 30–40 kDa protein) are concentrated near the bottom of the gradient (22). In the presence of CHX, histone mRNAs are stabilized and are resistant to degradation by HU (12,39). There is continued production of histone mRNA in the presence of CHX, resulting in an increase in histone mRNA levels (12,34). Since CHX blocks the elongation of the polypeptide chain, mRNAs synthesized and recruited for translation after addition of CHX are associated with monoribosomes, resulting in a shift in the overall population of histone mRNA toward the top of the gradient (Figure 6A).
We analyzed each fraction for the presence of SLBP by western blotting. In exponentially growing cells, much of the SLBP was not tightly associated with polyribosomes but remained at the top of the gradient (Figure 5B). There was a peak of SLBP present in fractions 6–11, which include the monoribosomes and small polyribosomes (Figure 5B). There was clearly less SLBP associated with the larger polyribosomes that contained the largest amounts of histone mRNA (Figure 5B, lanes 10–12). In cells treated with HU, all of the cytoplasmic SLBP remained at the top of the gradient (data not shown, Supplementary Figure A), and there was very little histone mRNA in any of the fractions, demonstrating that the observed association with ribosomes is dependent on the presence of histone mRNA.
In CHX-treated cells, a much larger fraction of the total SLBP in the cytosolic fraction was associated with polyribosomes, and there was a clear peak of SLBP in fractions 6–8 (Figure 6B). There was also a much larger proportion of the total histone mRNA associated with fractions 6–8, the positions of monosomes, in CHX-treated cells (Figure 6A). Since CHX blocks elongation of translation but not initiation, we anticipate that histone mRNAs synthesized in the presence of CHX would associate with only one ribosome. These newly synthesized mRNAs are associated with SLBP when they are associated with monoribosomes consistent with a role for SLBP in the initiation of translation (8).
Importantly, the SLBP which co-fractionated with histone mRNA in CHX-treated cells was unable to bind with an exogenously added stem–loop RNA, consistent with the interpretation that it is bound to the histone mRNA in the initial rounds of translation (Figure 6B, fractions 8–10). In contrast, the SLBP at the top of the gradient, which did not co-fractionate with histone mRNA, bound to the stem–loop very efficiently (Figures 5C and 6C, fractions 1–3). We have previously shown that under conditions used for the mobility shift assay, once SLBP is bound to the stem–loop, it is not readily exchanged onto another stem–loop RNA (40). Therefore, we interpret these results to indicate that the SLBP tightly associated with ribosomes is in a stable complex with histone mRNA and not available for binding to the exogenously added probe, consistent with SLBP being bound to histone mRNA as a component of the mRNP.
DISCUSSION
The proteins that associate with pre-mRNAs in the nucleus are critical factors for the regulation of mRNA processing and transport to the cytoplasm. The regulation of mRNA stability and translation is also mediated by proteins in the cytoplasmic mRNPs, many of which may become associated with the mRNA during the initial pre-mRNA processing steps in the nucleus.
Polyadenylated mRNAs exist as an mRNP with the PABP bound to their 3′ ends. PABP is not required for pre-mRNA processing but only binds to the mature mRNA at the time the RNA enters the cytoplasm (41), replacing PAB2, which is involved in polyadenylation (42). PABP stimulates initiation of translation of an mRNA by interaction with the eIF4F cap binding complex (eIF4G, eIF4E and eIF3) through interactions with the N-terminal portion of eIF4G (43,44). These interactions effectively circularize the mRNA allowing the 3′ end to affect events at the 5′ end of the message. Degradation of polyadenylated mRNAs is initiated by deadenylation of the mRNA, breaking the interaction between the 5′ and 3′ ends of the mRNA.
There are proteins bound to the 3′-untranslated region (3′-UTR) of many mRNAs. These include protein complexes which stabilize the globin (45,46) and collagen (47) mRNAs, as well as proteins that bind to AU-rich sequences and either stabilize (48–51) or destabilize (52,53) the mRNA. The rev protein binds to the 3′-UTR of full-length retroviral mRNAs (which synthesize gag and gag/pol) and is required for export of these mRNAs to the cytoplasm and remains associated with the mRNA in the cytoplasm (54,55). These and/or other proteins bound to the 3′-UTR often participate in regulation of translation of the mRNA (48).
There are also proteins that associate with the coding region of the mRNA. Some of these bind during splicing and remain associated with the mRNA possibly marking it for export into the cytoplasm (56,57), and others affect the stability of the mRNA (58,59). It is not known when these latter proteins associate with the mRNA, but many of them are present both in the cytoplasm and in the nucleus, and it is likely that they associate with the mRNA during biosynthesis in the nucleus or export into the cytoplasm.
During histone pre-mRNA processing in the nucleus, SLBP becomes associated with the mature message and the data presented here suggests that it accompanies the mature message to the cytoplasm. S-phase cells, which have large amounts of histone mRNA, contain SLBP in both the nucleus and the cytoplasm (Figure 3). When DNA synthesis is inhibited and histone mRNAs are degraded, SLBP is not degraded, but relocates to the nucleus. This result suggests that the SLBP present in the cytoplasm is bound to histone mRNA, and this interpretation is supported by the precipitation of histone mRNAs with anti-SLBP (Figure 1).
SLBP has been shown to participate in translation initiation (8) in the cytoplasm consistent with its association with histone mRNA on ribosomes. We find that SLBP is tightly associated with the histone mRNPs immediately after initiation of translation, as evidenced by the association of the histone mRNA synthesized in the presence of CHX with SLBP on monoribosomes. In CHX-treated cells, there is a substantial increase in the amount of SLBP associated with ribosomes; the bulk of this SLBP is associated with monoribosomes, coincident with the location of histone mRNAs synthesized in the presence of CHX. SLBP is not associated with histone mRNPs with multiple ribosomes for reasons that we do not understand. Our data support the hypothesis that SLBP is tightly bound during the initial round of translation and is less tightly associated with or is lost from the mRNP after several rounds of translation.
In addition to a role in histone mRNA translation, it is likely that SLBP plays a role in regulating histone mRNA degradation, as PABP does for polyadenylated mRNAs (60). The initial step in histone mRNA degradation is likely to be trimming by an exonuclease starting from the 3′ end of the mRNA (14,17,61); consistent with this hypothesis, SLBP protects histone mRNA from degradation by a purified 3′–5′ exonuclease in vitro (17). Rapid degradation of histone mRNA in response to inhibition of DNA synthesis occurs without any reduction in SLBP levels. We have never observed any free SLBP that was not capable of binding to the stem–loop. Indeed depletion of either whole-cell (23) or nuclear extracts (20) with a biotinylated stem–loop RNA removes all the SLBP as analyzed by western blotting. Thus, if there is a form of SLBP that cannot bind RNA it must only exist transiently. It is possible that the SLBP/RNA complex could be cleaved off the mRNA by an endonucleolytic cleavage. Alternatively, the SLBP may be displaced by modification of the SLBP resulting in recruitment or activation of an exonuclease.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
α-p48 was a gift from Drs Alain Verrault and Bruce Stillman (Cold Spring Harbor Labs). αCUL1 was a gift from Dr Yue Xiong (University of North Carolina). This work was supported by NIH grant GM29832 to W.F.M. M.L.W. was supported by HHMI Biomedical Research Support Award #76200-560801 to Dartmouth College.
REFERENCES
- 1.Marzluff W.F. (1992) Histone 3′ ends: essential and regulatory functions. Gene Expr., 2, 93–97. [PMC free article] [PubMed] [Google Scholar]
- 2.Dominski Z. and Marzluff,W.F. (1999) Formation of the 3′ end of histone mRNA. Gene, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- 3.Gallie D.R. (1998) A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- 4.Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson A. (1996) PolyA metabolism and translation: the closed loop model. In Hershey,J.W., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–480. [Google Scholar]
- 6.Sun J.-H., Pilch,D.R. and Marzluff,W.F. (1992) The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res., 20, 6057–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallie D.R., Lewis,N.J. and Marzluff,W.F. (1996) The histone 3′-terminal stem–loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res., 24, 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez R. and Marzluff,W.F. (2002) The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol., 22, 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris M.E., Böhni,R., Schneiderman,M.H., Ramamurthy,L., Schümperli,D. and Marzluff,W.F. (1991) Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol., 11, 2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sariban E., Wu,R.S., Erickson,L.C. and Bonner,W.M. (1985) Interrelationships of protein and DNA syntheses during replication of mammalian cells. Mol. Cell. Biol., 5, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sittman D.B., Graves,R.A. and Marzluff,W.F. (1983) Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl Acad. Sci. USA, 80, 1849–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves R.A. and Marzluff,W.F. (1984) Rapid, reversible alterations in histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol. Cell. Biol., 4, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey N.B. and Marzluff,W.F. (1987) The stem–loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol., 7, 4557–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross J., Peltz,S.W., Kobs,G. and Brewer,G. (1986) Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3′ terminus. Mol. Cell. Biol., 6, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross J., Kobs,G., Brewer,G. and Peltz,S.W. (1987) Properties of the exonuclease activity that degrades H4 histone mRNA. J. Biol. Chem., 262, 9374–9381. [PubMed] [Google Scholar]
- 16.Caruccio N. and Ross,J. (1994) Purification of a human polyribosome-associated 3′ to 5′ exonuclease. J. Biol. Chem., 269, 31814–31821. [PubMed] [Google Scholar]
- 17.Dominski Z., Yang,X., Kaygun,H. and Marzluff,W.F. (2003) A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell, 12, 295–305. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z.-F., Whitfield,M.L., Ingledue,T.I., Dominski,Z. and Marzluff,W.F. (1996) The protein which binds the 3′ end of histone mRNA: a novel RNA- binding protein required for histone pre-mRNA processing. Genes Dev., 10, 3028–3040. [DOI] [PubMed] [Google Scholar]
- 19.Martin F., Schaller,A., Eglite,S., Schümperli,D. and Müller,B. (1997) The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J., 16, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominski Z., Zheng,L.-X., Sanchez,R. and Marzluff,W.F. (1999) The stem–loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol. Cell. Biol., 19, 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marzluff W.F. and Duronio,R.J. (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol., 14, 692–699. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield M.L., Zheng,L.-X., Baldwin,A., Ohta,T., Hurt,M.M. and Marzluff,W.F. (2000) Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol., 20, 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L.-X., Dominski,Z., Yang,X., Elms,P., Raska,C.S., Borchers,C.H. and Marzluff,W.F. (2003) Phosphorylation of SLBP on two threonines triggers degradation of SLBP, the sole cell-cycle regulated factor required for regulation of histone mRNA processing, at the end of S-phase. Mol. Cell. Biol., 23, 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson R.J., Sun,J.-H., Willis,D.G. and Marzluff,W.F. (1996) Efficient extraction and partial purification of the polyribosomal-associated stem–loop binding protein bound to the 3′ end of histone mRNA. Biochemistry, 35, 2146–2156. [DOI] [PubMed] [Google Scholar]
- 25.Marzluff W.F., Whitfield,M.L., Dominski,Z. and Wang,Z.-F. (1997) Identification of the protein that interacts with the 3′ end of histone mRNA. In Richter,J.D. (ed.), mRNA Formation and Function. Academic Press, NY, pp. 163–193. [Google Scholar]
- 26.Graves R.A., Wellman,S.E., Chiu,I.-M. and Marzluff,W.F. (1985) Differential expression of two clusters of mouse histone genes. J. Mol. Biol., 183, 179–194. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z.-F., Sirotkin,A.M., Buchold,G.M., Skoultchi,A.I. and Marzluff,W.F. (1997) The mouse histone H1 genes: gene organization and differential regulation. J. Mol. Biol., 271, 124–138. [DOI] [PubMed] [Google Scholar]
- 28.Pandey N.B., Sun,J.-H. and Marzluff,W.F. (1991) Different complexes are formed on the 3′ end of histone mRNA in nuclear and polysomal extracts. Nucleic Acids Res., 19, 5653–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schochetman G. and Perry,R.P. (1972) Early appearance of histone messenger RNA in polyribosomes of cultured L cells. J. Mol. Biol., 63, 591–596. [DOI] [PubMed] [Google Scholar]
- 30.Michel J.J. and Xiong,Y. (1998) Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ., 9, 435–449. [PubMed] [Google Scholar]
- 31.Wells D. and Kedes,L. (1985) Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylated mRNAs. Proc. Natl Acad. Sci. USA, 82, 2834–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzluff W.F., Gongidi,P., Woods,K.R., Jin,J.P. and Maltais,L. (2002) The human and mouse replication-dependent histone genes. Genomics, 80, 487–498. [PubMed] [Google Scholar]
- 33.Pilch D.R. and Marzluff,W.F. (1991) Expression of histone-U1 snRNA genes: U1 promoters are compatible with histone 3′ end formation. Gene Expr., 1, 41–53. [PMC free article] [PubMed] [Google Scholar]
- 34.Stimac E., Groppi,V.E.,Jr and Coffino,P. (1984) Inhibition of protein synthesis stabilizes histone mRNA. Mol. Cell. Biol., 4, 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumbach L.L., Marashi,F., Plumb,M., Stein,G. and Stein,J. (1984) Inhibition of DNA replication coordinately reduces cellular levels of core and H1 histone mRNAs: requirement for protein synthesis. Biochemistry, 23, 1618–1625. [DOI] [PubMed] [Google Scholar]
- 36.Lüscher B., Stauber,C., Schindler,R. and Schümperli,D. (1985) Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc. Natl Acad. Sci. USA, 82, 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stauber C., Lüscher,B., Eckner,R., Lotscher,E. and Schümperli,D. (1986) A signal regulating mouse histone H4 mRNA levels in a mammalian cell cycle mutant and sequences controlling RNA 3′ processing are both contained within the same 80-bp fragment. EMBO J., 5, 3297–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauber C. and Schümperli,D. (1988) 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res., 16, 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler W.B. and Mueller,G.C. (1973) Control of histone synthesis in HeLa cells. Biochim. Biophys. Acta, 294, 481–496. [DOI] [PubMed] [Google Scholar]
- 40.Williams A.S. and Marzluff,W.F. (1995) The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem–loop binding protein. Nucleic Acids Res., 23, 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishigaki Y., Li,X.J., Serin,G. and Maquat,L.E. (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell, 106, 607–617. [DOI] [PubMed] [Google Scholar]
- 42.Wahle E. (1991) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- 43.Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kessler S.H. and Sachs,A.B. (1998) RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol., 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chkheidze A.N., Lyakhov,D.L., Makeyev,A.V., Morales,J., Kong,J. and Liebhaber,S.A. (1999) Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol. Cell. Biol., 19, 4572–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiledjian M., Wang,X.M. and Liebhaber,S.A. (1995) Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J., 14, 4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindquist J.N., Kauschke,S.G., Stefanovic,B., Burchardt,E.R. and Brenner,D.A. (2000) Characterization of the interaction between αCP2 and the 3′-untranslated region of collagen α1(I) mRNA. Nucleic Acids Res., 28, 4306–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antic D., Lu,N. and Keene,J.D. (1999) ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev., 13, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain R.G., Andrews,L.G., McGowan,K.M., Pekala,P.H. and Keene,J.D. (1997) Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol., 17, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan X.H. and Steitz,J.A. (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng S.S., Chen,C.Y., Xu,N.H. and Shyu,A.B. (1998) RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loflin P., Chen,C.Y.A. and Shyu,A.B. (1999) Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeMaria C.T. and Brewer,G. (1996) AUF1 binding affinity to A + U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem., 271, 12179–12184. [DOI] [PubMed] [Google Scholar]
- 54.D'Agostino D.M., Felber,B.K., Harrison,J.E. and Pavlakis,G.N. (1992) The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol., 12, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalland K.-H., Szilvay,A.M., Brokstad,K.A., Sætrevik,W. and Haukenes,G. (1994) The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell. Biol., 14, 7436–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Hir H., Moore,M.J. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 57.Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grosset C., Chen,C.Y.A., Xu,N.H., Sonenberg,N., Jacquemin-Sablon,H. and Shyu,A.B. (2000) A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell, 103, 29–40. [DOI] [PubMed] [Google Scholar]
- 59.Shyu A.B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein P. and Ross,J. (1989) Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci., 14, 373–377. [DOI] [PubMed] [Google Scholar]
- 61.Ross J. and Kobs,G. (1986) H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J. Mol. Biol., 188, 579–593. [DOI] [PubMed] [Google Scholar]
- 62.Whitfield M.L., Sherlock,G., Saldanha,A.J., Murray,J.I., Ball,C.A., Alexander,K.E., Matese,J.C., Perou,C.M., Hurt,M.M., Brown,P.O. et al. (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell, 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


